Genome-wide Association Study (GWAS) and Its Application for Improving the Genomic Estimated Breeding Values (GEBV) of the Berkshire Pork Quality Traits Article Swipe
 YOU?
·
· 2015
· Open Access
·
· DOI: https://doi.org/10.5713/ajas.15.0287
YOU?
·
· 2015
· Open Access
·
· DOI: https://doi.org/10.5713/ajas.15.0287
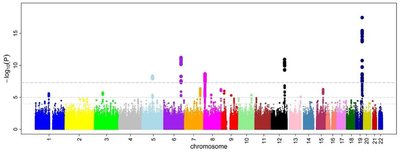
The missing heritability has been a major problem in the analysis of best linear unbiased prediction (BLUP). We introduced the traditional genome-wide association study (GWAS) into the BLUP to improve the heritability estimation. We analyzed eight pork quality traits of the Berkshire breeds using GWAS and BLUP. GWAS detects the putative quantitative trait loci regions given traits. The single nucleotide polymorphisms (SNPs) were obtained using GWAS results with p value <0.01. BLUP analyzed with significant SNPs was much more accurate than that with total genotyped SNPs in terms of narrow-sense heritability. It implies that genomic estimated breeding values (GEBVs) of pork quality traits can be calculated by BLUP via GWAS. The GWAS model was the linear regression using PLINK and BLUP model was the G-BLUP and SNP-GBLUP. The SNP-GBLUP uses SNP-SNP relationship matrix. The BLUP analysis using preprocessing of GWAS can be one of the possible alternatives of solving the missing heritability problem and it can provide alternative BLUP method which can find more accurate GEBVs.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.5713/ajas.15.0287
- https://www.animbiosci.org/upload/pdf/ajas-28-11-1551.pdf
- OA Status
- gold
- Cited By
- 4
- References
- 22
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W2204650909
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W2204650909Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.5713/ajas.15.0287Digital Object Identifier
- Title
-
Genome-wide Association Study (GWAS) and Its Application for Improving the Genomic Estimated Breeding Values (GEBV) of the Berkshire Pork Quality TraitsWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2015Year of publication
- Publication date
-
2015-09-03Full publication date if available
- Authors
-
Young Sup Lee, Hyeonsoo Jeong, Mengistie Taye, Hyeon Jeong Kim, Sojeong Ka, Youn‐Chul Ryu, Seoae ChoList of authors in order
- Landing page
-
https://doi.org/10.5713/ajas.15.0287Publisher landing page
- PDF URL
-
https://www.animbiosci.org/upload/pdf/ajas-28-11-1551.pdfDirect link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
goldOpen access status per OpenAlex
- OA URL
-
https://www.animbiosci.org/upload/pdf/ajas-28-11-1551.pdfDirect OA link when available
- Concepts
-
Best linear unbiased prediction, Genome-wide association study, Heritability, Single-nucleotide polymorphism, Missing heritability problem, Statistics, Genetic association, Biology, SNP, Genetics, Mathematics, Selection (genetic algorithm), Computer science, Genotype, Gene, Artificial intelligenceTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
4Total citation count in OpenAlex
- Citations by year (recent)
-
2023: 1, 2021: 1, 2019: 1, 2018: 1Per-year citation counts (last 5 years)
- References (count)
-
22Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W2204650909 |
|---|---|
| doi | https://doi.org/10.5713/ajas.15.0287 |
| ids.doi | https://doi.org/10.5713/ajas.15.0287 |
| ids.mag | 2204650909 |
| ids.pmid | https://pubmed.ncbi.nlm.nih.gov/26580278 |
| ids.openalex | https://openalex.org/W2204650909 |
| fwci | 0.2198695 |
| type | article |
| title | Genome-wide Association Study (GWAS) and Its Application for Improving the Genomic Estimated Breeding Values (GEBV) of the Berkshire Pork Quality Traits |
| biblio.issue | 11 |
| biblio.volume | 28 |
| biblio.last_page | 1557 |
| biblio.first_page | 1551 |
| topics[0].id | https://openalex.org/T10594 |
| topics[0].field.id | https://openalex.org/fields/13 |
| topics[0].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[0].score | 0.9998999834060669 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1311 |
| topics[0].subfield.display_name | Genetics |
| topics[0].display_name | Genetic and phenotypic traits in livestock |
| topics[1].id | https://openalex.org/T11468 |
| topics[1].field.id | https://openalex.org/fields/13 |
| topics[1].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[1].score | 0.9990000128746033 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1311 |
| topics[1].subfield.display_name | Genetics |
| topics[1].display_name | Genetic Mapping and Diversity in Plants and Animals |
| topics[2].id | https://openalex.org/T11229 |
| topics[2].field.id | https://openalex.org/fields/11 |
| topics[2].field.display_name | Agricultural and Biological Sciences |
| topics[2].score | 0.9912999868392944 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1110 |
| topics[2].subfield.display_name | Plant Science |
| topics[2].display_name | Genetics and Plant Breeding |
| is_xpac | False |
| apc_list.value | 370 |
| apc_list.currency | USD |
| apc_list.value_usd | 370 |
| apc_paid.value | 370 |
| apc_paid.currency | USD |
| apc_paid.value_usd | 370 |
| concepts[0].id | https://openalex.org/C103545067 |
| concepts[0].level | 3 |
| concepts[0].score | 0.8848710656166077 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q796265 |
| concepts[0].display_name | Best linear unbiased prediction |
| concepts[1].id | https://openalex.org/C106208931 |
| concepts[1].level | 5 |
| concepts[1].score | 0.7475833892822266 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q1098876 |
| concepts[1].display_name | Genome-wide association study |
| concepts[2].id | https://openalex.org/C161890455 |
| concepts[2].level | 2 |
| concepts[2].score | 0.7345113754272461 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q1503548 |
| concepts[2].display_name | Heritability |
| concepts[3].id | https://openalex.org/C153209595 |
| concepts[3].level | 4 |
| concepts[3].score | 0.5236893892288208 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q501128 |
| concepts[3].display_name | Single-nucleotide polymorphism |
| concepts[4].id | https://openalex.org/C144621757 |
| concepts[4].level | 5 |
| concepts[4].score | 0.5198795795440674 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q3083720 |
| concepts[4].display_name | Missing heritability problem |
| concepts[5].id | https://openalex.org/C105795698 |
| concepts[5].level | 1 |
| concepts[5].score | 0.49337974190711975 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q12483 |
| concepts[5].display_name | Statistics |
| concepts[6].id | https://openalex.org/C186413461 |
| concepts[6].level | 5 |
| concepts[6].score | 0.4613959491252899 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q744727 |
| concepts[6].display_name | Genetic association |
| concepts[7].id | https://openalex.org/C86803240 |
| concepts[7].level | 0 |
| concepts[7].score | 0.44784843921661377 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[7].display_name | Biology |
| concepts[8].id | https://openalex.org/C139275648 |
| concepts[8].level | 5 |
| concepts[8].score | 0.4364285171031952 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q17134011 |
| concepts[8].display_name | SNP |
| concepts[9].id | https://openalex.org/C54355233 |
| concepts[9].level | 1 |
| concepts[9].score | 0.30896204710006714 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[9].display_name | Genetics |
| concepts[10].id | https://openalex.org/C33923547 |
| concepts[10].level | 0 |
| concepts[10].score | 0.253362774848938 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q395 |
| concepts[10].display_name | Mathematics |
| concepts[11].id | https://openalex.org/C81917197 |
| concepts[11].level | 2 |
| concepts[11].score | 0.1891867220401764 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q628760 |
| concepts[11].display_name | Selection (genetic algorithm) |
| concepts[12].id | https://openalex.org/C41008148 |
| concepts[12].level | 0 |
| concepts[12].score | 0.14960163831710815 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q21198 |
| concepts[12].display_name | Computer science |
| concepts[13].id | https://openalex.org/C135763542 |
| concepts[13].level | 3 |
| concepts[13].score | 0.13940861821174622 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q106016 |
| concepts[13].display_name | Genotype |
| concepts[14].id | https://openalex.org/C104317684 |
| concepts[14].level | 2 |
| concepts[14].score | 0.06386956572532654 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[14].display_name | Gene |
| concepts[15].id | https://openalex.org/C154945302 |
| concepts[15].level | 1 |
| concepts[15].score | 0.0 |
| concepts[15].wikidata | https://www.wikidata.org/wiki/Q11660 |
| concepts[15].display_name | Artificial intelligence |
| keywords[0].id | https://openalex.org/keywords/best-linear-unbiased-prediction |
| keywords[0].score | 0.8848710656166077 |
| keywords[0].display_name | Best linear unbiased prediction |
| keywords[1].id | https://openalex.org/keywords/genome-wide-association-study |
| keywords[1].score | 0.7475833892822266 |
| keywords[1].display_name | Genome-wide association study |
| keywords[2].id | https://openalex.org/keywords/heritability |
| keywords[2].score | 0.7345113754272461 |
| keywords[2].display_name | Heritability |
| keywords[3].id | https://openalex.org/keywords/single-nucleotide-polymorphism |
| keywords[3].score | 0.5236893892288208 |
| keywords[3].display_name | Single-nucleotide polymorphism |
| keywords[4].id | https://openalex.org/keywords/missing-heritability-problem |
| keywords[4].score | 0.5198795795440674 |
| keywords[4].display_name | Missing heritability problem |
| keywords[5].id | https://openalex.org/keywords/statistics |
| keywords[5].score | 0.49337974190711975 |
| keywords[5].display_name | Statistics |
| keywords[6].id | https://openalex.org/keywords/genetic-association |
| keywords[6].score | 0.4613959491252899 |
| keywords[6].display_name | Genetic association |
| keywords[7].id | https://openalex.org/keywords/biology |
| keywords[7].score | 0.44784843921661377 |
| keywords[7].display_name | Biology |
| keywords[8].id | https://openalex.org/keywords/snp |
| keywords[8].score | 0.4364285171031952 |
| keywords[8].display_name | SNP |
| keywords[9].id | https://openalex.org/keywords/genetics |
| keywords[9].score | 0.30896204710006714 |
| keywords[9].display_name | Genetics |
| keywords[10].id | https://openalex.org/keywords/mathematics |
| keywords[10].score | 0.253362774848938 |
| keywords[10].display_name | Mathematics |
| keywords[11].id | https://openalex.org/keywords/selection |
| keywords[11].score | 0.1891867220401764 |
| keywords[11].display_name | Selection (genetic algorithm) |
| keywords[12].id | https://openalex.org/keywords/computer-science |
| keywords[12].score | 0.14960163831710815 |
| keywords[12].display_name | Computer science |
| keywords[13].id | https://openalex.org/keywords/genotype |
| keywords[13].score | 0.13940861821174622 |
| keywords[13].display_name | Genotype |
| keywords[14].id | https://openalex.org/keywords/gene |
| keywords[14].score | 0.06386956572532654 |
| keywords[14].display_name | Gene |
| language | en |
| locations[0].id | doi:10.5713/ajas.15.0287 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S118716465 |
| locations[0].source.issn | 1011-2367, 1976-5517 |
| locations[0].source.type | journal |
| locations[0].source.is_oa | True |
| locations[0].source.issn_l | 1011-2367 |
| locations[0].source.is_core | True |
| locations[0].source.is_in_doaj | True |
| locations[0].source.display_name | Asian-Australasian Journal of Animal Sciences |
| locations[0].source.host_organization | https://openalex.org/P4310319462 |
| locations[0].source.host_organization_name | Asian-Australasian Association of Animal Production Societies |
| locations[0].source.host_organization_lineage | https://openalex.org/P4310319462 |
| locations[0].license | cc-by |
| locations[0].pdf_url | https://www.animbiosci.org/upload/pdf/ajas-28-11-1551.pdf |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | Asian-Australasian Journal of Animal Sciences |
| locations[0].landing_page_url | https://doi.org/10.5713/ajas.15.0287 |
| locations[1].id | pmid:26580278 |
| locations[1].is_oa | False |
| locations[1].source.id | https://openalex.org/S4306525036 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | PubMed |
| locations[1].source.host_organization | https://openalex.org/I1299303238 |
| locations[1].source.host_organization_name | National Institutes of Health |
| locations[1].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[1].license | |
| locations[1].pdf_url | |
| locations[1].version | publishedVersion |
| locations[1].raw_type | |
| locations[1].license_id | |
| locations[1].is_accepted | True |
| locations[1].is_published | True |
| locations[1].raw_source_name | Asian-Australasian journal of animal sciences |
| locations[1].landing_page_url | https://pubmed.ncbi.nlm.nih.gov/26580278 |
| locations[2].id | pmh:oai:europepmc.org:3626019 |
| locations[2].is_oa | False |
| locations[2].source.id | https://openalex.org/S4306400806 |
| locations[2].source.issn | |
| locations[2].source.type | repository |
| locations[2].source.is_oa | False |
| locations[2].source.issn_l | |
| locations[2].source.is_core | False |
| locations[2].source.is_in_doaj | False |
| locations[2].source.display_name | Europe PMC (PubMed Central) |
| locations[2].source.host_organization | https://openalex.org/I1303153112 |
| locations[2].source.host_organization_name | European Bioinformatics Institute |
| locations[2].source.host_organization_lineage | https://openalex.org/I1303153112 |
| locations[2].license | |
| locations[2].pdf_url | |
| locations[2].version | submittedVersion |
| locations[2].raw_type | Text |
| locations[2].license_id | |
| locations[2].is_accepted | False |
| locations[2].is_published | False |
| locations[2].raw_source_name | |
| locations[2].landing_page_url | https://www.ncbi.nlm.nih.gov/pmc/articles/4647094 |
| locations[3].id | pmh:oai:doaj.org/article:e39095cbbdd44b7c81bceebfad69941f |
| locations[3].is_oa | False |
| locations[3].source.id | https://openalex.org/S4306401280 |
| locations[3].source.issn | |
| locations[3].source.type | repository |
| locations[3].source.is_oa | False |
| locations[3].source.issn_l | |
| locations[3].source.is_core | False |
| locations[3].source.is_in_doaj | False |
| locations[3].source.display_name | DOAJ (DOAJ: Directory of Open Access Journals) |
| locations[3].source.host_organization | |
| locations[3].source.host_organization_name | |
| locations[3].source.host_organization_lineage | |
| locations[3].license | |
| locations[3].pdf_url | |
| locations[3].version | submittedVersion |
| locations[3].raw_type | article |
| locations[3].license_id | |
| locations[3].is_accepted | False |
| locations[3].is_published | False |
| locations[3].raw_source_name | Asian-Australasian Journal of Animal Sciences, Vol 28, Iss 11, Pp 1551-1557 (2015) |
| locations[3].landing_page_url | https://doaj.org/article/e39095cbbdd44b7c81bceebfad69941f |
| indexed_in | crossref, doaj, pubmed |
| authorships[0].author.id | https://openalex.org/A5021258379 |
| authorships[0].author.orcid | https://orcid.org/0000-0002-5841-6506 |
| authorships[0].author.display_name | Young Sup Lee |
| authorships[0].countries | KR |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I139264467 |
| authorships[0].affiliations[0].raw_affiliation_string | Department of Agricultural Biotechnology, Animal Biotechnology, and Research Institute for Agriculture and Life Sciences, Seoul National University, Seoul 151-921, #N#Korea |
| authorships[0].institutions[0].id | https://openalex.org/I139264467 |
| authorships[0].institutions[0].ror | https://ror.org/04h9pn542 |
| authorships[0].institutions[0].type | education |
| authorships[0].institutions[0].lineage | https://openalex.org/I139264467 |
| authorships[0].institutions[0].country_code | KR |
| authorships[0].institutions[0].display_name | Seoul National University |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Young-Sup Lee |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Department of Agricultural Biotechnology, Animal Biotechnology, and Research Institute for Agriculture and Life Sciences, Seoul National University, Seoul 151-921, #N#Korea |
| authorships[1].author.id | https://openalex.org/A5021740365 |
| authorships[1].author.orcid | https://orcid.org/0000-0002-5565-2685 |
| authorships[1].author.display_name | Hyeonsoo Jeong |
| authorships[1].countries | KR |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I139264467 |
| authorships[1].affiliations[0].raw_affiliation_string | Department of Agricultural Biotechnology, Animal Biotechnology, and Research Institute for Agriculture and Life Sciences, Seoul National University, Seoul 151-921, #N#Korea |
| authorships[1].institutions[0].id | https://openalex.org/I139264467 |
| authorships[1].institutions[0].ror | https://ror.org/04h9pn542 |
| authorships[1].institutions[0].type | education |
| authorships[1].institutions[0].lineage | https://openalex.org/I139264467 |
| authorships[1].institutions[0].country_code | KR |
| authorships[1].institutions[0].display_name | Seoul National University |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Hyeonsoo Jeong |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | Department of Agricultural Biotechnology, Animal Biotechnology, and Research Institute for Agriculture and Life Sciences, Seoul National University, Seoul 151-921, #N#Korea |
| authorships[2].author.id | https://openalex.org/A5041634766 |
| authorships[2].author.orcid | https://orcid.org/0000-0001-6795-9943 |
| authorships[2].author.display_name | Mengistie Taye |
| authorships[2].countries | KR |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I139264467 |
| authorships[2].affiliations[0].raw_affiliation_string | Department of Agricultural Biotechnology, Animal Biotechnology, and Research Institute for Agriculture and Life Sciences, Seoul National University, Seoul 151-921, |
| authorships[2].institutions[0].id | https://openalex.org/I139264467 |
| authorships[2].institutions[0].ror | https://ror.org/04h9pn542 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I139264467 |
| authorships[2].institutions[0].country_code | KR |
| authorships[2].institutions[0].display_name | Seoul National University |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Mengistie Taye |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | Department of Agricultural Biotechnology, Animal Biotechnology, and Research Institute for Agriculture and Life Sciences, Seoul National University, Seoul 151-921, |
| authorships[3].author.id | https://openalex.org/A5107834243 |
| authorships[3].author.orcid | |
| authorships[3].author.display_name | Hyeon Jeong Kim |
| authorships[3].countries | GB |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I4210151791 |
| authorships[3].affiliations[0].raw_affiliation_string | C&K Genomics Inc., Seoul 151-919, |
| authorships[3].institutions[0].id | https://openalex.org/I4210151791 |
| authorships[3].institutions[0].ror | https://ror.org/053a6xa29 |
| authorships[3].institutions[0].type | company |
| authorships[3].institutions[0].lineage | https://openalex.org/I4210151791 |
| authorships[3].institutions[0].country_code | GB |
| authorships[3].institutions[0].display_name | Genomics (United Kingdom) |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Hyeon Jeong Kim |
| authorships[3].is_corresponding | False |
| authorships[3].raw_affiliation_strings | C&K Genomics Inc., Seoul 151-919, |
| authorships[4].author.id | https://openalex.org/A5109362277 |
| authorships[4].author.orcid | |
| authorships[4].author.display_name | Sojeong Ka |
| authorships[4].countries | KR |
| authorships[4].affiliations[0].institution_ids | https://openalex.org/I139264467 |
| authorships[4].affiliations[0].raw_affiliation_string | Department of Agricultural Biotechnology, Animal Biotechnology, and Research Institute for Agriculture and Life Sciences, Seoul National University, Seoul 151-921, |
| authorships[4].institutions[0].id | https://openalex.org/I139264467 |
| authorships[4].institutions[0].ror | https://ror.org/04h9pn542 |
| authorships[4].institutions[0].type | education |
| authorships[4].institutions[0].lineage | https://openalex.org/I139264467 |
| authorships[4].institutions[0].country_code | KR |
| authorships[4].institutions[0].display_name | Seoul National University |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Sojeong Ka |
| authorships[4].is_corresponding | False |
| authorships[4].raw_affiliation_strings | Department of Agricultural Biotechnology, Animal Biotechnology, and Research Institute for Agriculture and Life Sciences, Seoul National University, Seoul 151-921, |
| authorships[5].author.id | https://openalex.org/A5013094555 |
| authorships[5].author.orcid | https://orcid.org/0000-0001-8940-624X |
| authorships[5].author.display_name | Youn‐Chul Ryu |
| authorships[5].countries | KR |
| authorships[5].affiliations[0].institution_ids | https://openalex.org/I83202590 |
| authorships[5].affiliations[0].raw_affiliation_string | Division of Biotechnology, Sustainable Agriculture Research Institute, Jeju National University, Jeju 690-756, Korea. |
| authorships[5].institutions[0].id | https://openalex.org/I83202590 |
| authorships[5].institutions[0].ror | https://ror.org/05hnb4n85 |
| authorships[5].institutions[0].type | education |
| authorships[5].institutions[0].lineage | https://openalex.org/I83202590 |
| authorships[5].institutions[0].country_code | KR |
| authorships[5].institutions[0].display_name | Jeju National University |
| authorships[5].author_position | middle |
| authorships[5].raw_author_name | Youn-Chul Ryu |
| authorships[5].is_corresponding | False |
| authorships[5].raw_affiliation_strings | Division of Biotechnology, Sustainable Agriculture Research Institute, Jeju National University, Jeju 690-756, Korea. |
| authorships[6].author.id | https://openalex.org/A5021758490 |
| authorships[6].author.orcid | |
| authorships[6].author.display_name | Seoae Cho |
| authorships[6].countries | GB |
| authorships[6].affiliations[0].institution_ids | https://openalex.org/I4210151791 |
| authorships[6].affiliations[0].raw_affiliation_string | C&K Genomics Inc., Seoul 151-919, |
| authorships[6].institutions[0].id | https://openalex.org/I4210151791 |
| authorships[6].institutions[0].ror | https://ror.org/053a6xa29 |
| authorships[6].institutions[0].type | company |
| authorships[6].institutions[0].lineage | https://openalex.org/I4210151791 |
| authorships[6].institutions[0].country_code | GB |
| authorships[6].institutions[0].display_name | Genomics (United Kingdom) |
| authorships[6].author_position | last |
| authorships[6].raw_author_name | Seoae Cho |
| authorships[6].is_corresponding | False |
| authorships[6].raw_affiliation_strings | C&K Genomics Inc., Seoul 151-919, |
| has_content.pdf | True |
| has_content.grobid_xml | True |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://www.animbiosci.org/upload/pdf/ajas-28-11-1551.pdf |
| open_access.oa_status | gold |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Genome-wide Association Study (GWAS) and Its Application for Improving the Genomic Estimated Breeding Values (GEBV) of the Berkshire Pork Quality Traits |
| has_fulltext | True |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T10594 |
| primary_topic.field.id | https://openalex.org/fields/13 |
| primary_topic.field.display_name | Biochemistry, Genetics and Molecular Biology |
| primary_topic.score | 0.9998999834060669 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1311 |
| primary_topic.subfield.display_name | Genetics |
| primary_topic.display_name | Genetic and phenotypic traits in livestock |
| related_works | https://openalex.org/W1884571866, https://openalex.org/W159064268, https://openalex.org/W2587112589, https://openalex.org/W1987620105, https://openalex.org/W2060875624, https://openalex.org/W1864715623, https://openalex.org/W3161797857, https://openalex.org/W1582246138, https://openalex.org/W2616015692, https://openalex.org/W2016170077 |
| cited_by_count | 4 |
| counts_by_year[0].year | 2023 |
| counts_by_year[0].cited_by_count | 1 |
| counts_by_year[1].year | 2021 |
| counts_by_year[1].cited_by_count | 1 |
| counts_by_year[2].year | 2019 |
| counts_by_year[2].cited_by_count | 1 |
| counts_by_year[3].year | 2018 |
| counts_by_year[3].cited_by_count | 1 |
| locations_count | 4 |
| best_oa_location.id | doi:10.5713/ajas.15.0287 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S118716465 |
| best_oa_location.source.issn | 1011-2367, 1976-5517 |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | True |
| best_oa_location.source.issn_l | 1011-2367 |
| best_oa_location.source.is_core | True |
| best_oa_location.source.is_in_doaj | True |
| best_oa_location.source.display_name | Asian-Australasian Journal of Animal Sciences |
| best_oa_location.source.host_organization | https://openalex.org/P4310319462 |
| best_oa_location.source.host_organization_name | Asian-Australasian Association of Animal Production Societies |
| best_oa_location.source.host_organization_lineage | https://openalex.org/P4310319462 |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | https://www.animbiosci.org/upload/pdf/ajas-28-11-1551.pdf |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | journal-article |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | Asian-Australasian Journal of Animal Sciences |
| best_oa_location.landing_page_url | https://doi.org/10.5713/ajas.15.0287 |
| primary_location.id | doi:10.5713/ajas.15.0287 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S118716465 |
| primary_location.source.issn | 1011-2367, 1976-5517 |
| primary_location.source.type | journal |
| primary_location.source.is_oa | True |
| primary_location.source.issn_l | 1011-2367 |
| primary_location.source.is_core | True |
| primary_location.source.is_in_doaj | True |
| primary_location.source.display_name | Asian-Australasian Journal of Animal Sciences |
| primary_location.source.host_organization | https://openalex.org/P4310319462 |
| primary_location.source.host_organization_name | Asian-Australasian Association of Animal Production Societies |
| primary_location.source.host_organization_lineage | https://openalex.org/P4310319462 |
| primary_location.license | cc-by |
| primary_location.pdf_url | https://www.animbiosci.org/upload/pdf/ajas-28-11-1551.pdf |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | Asian-Australasian Journal of Animal Sciences |
| primary_location.landing_page_url | https://doi.org/10.5713/ajas.15.0287 |
| publication_date | 2015-09-03 |
| publication_year | 2015 |
| referenced_works | https://openalex.org/W2238355159, https://openalex.org/W2167638767, https://openalex.org/W2064013109, https://openalex.org/W2013866489, https://openalex.org/W2187181884, https://openalex.org/W2161831898, https://openalex.org/W1607760198, https://openalex.org/W4293258550, https://openalex.org/W2030042181, https://openalex.org/W2124879187, https://openalex.org/W2097222342, https://openalex.org/W2164050502, https://openalex.org/W1980991473, https://openalex.org/W1928998639, https://openalex.org/W2161633633, https://openalex.org/W2078394884, https://openalex.org/W2136260322, https://openalex.org/W2155496693, https://openalex.org/W2148965938, https://openalex.org/W100016954, https://openalex.org/W2410698933, https://openalex.org/W2600291984 |
| referenced_works_count | 22 |
| abstract_inverted_index.a | 5 |
| abstract_inverted_index.p | 68 |
| abstract_inverted_index.It | 91 |
| abstract_inverted_index.We | 17, 33 |
| abstract_inverted_index.be | 104, 141 |
| abstract_inverted_index.by | 106 |
| abstract_inverted_index.in | 8, 86 |
| abstract_inverted_index.it | 154 |
| abstract_inverted_index.of | 11, 39, 88, 99, 138, 143, 147 |
| abstract_inverted_index.to | 28 |
| abstract_inverted_index.The | 0, 57, 110, 127, 133 |
| abstract_inverted_index.and | 45, 119, 125, 153 |
| abstract_inverted_index.can | 103, 140, 155, 161 |
| abstract_inverted_index.has | 3 |
| abstract_inverted_index.one | 142 |
| abstract_inverted_index.the | 9, 19, 26, 30, 40, 49, 114, 123, 144, 149 |
| abstract_inverted_index.via | 108 |
| abstract_inverted_index.was | 76, 113, 122 |
| abstract_inverted_index.BLUP | 27, 71, 107, 120, 134, 158 |
| abstract_inverted_index.GWAS | 44, 47, 65, 111, 139 |
| abstract_inverted_index.SNPs | 75, 85 |
| abstract_inverted_index.been | 4 |
| abstract_inverted_index.best | 12 |
| abstract_inverted_index.find | 162 |
| abstract_inverted_index.into | 25 |
| abstract_inverted_index.loci | 53 |
| abstract_inverted_index.more | 78, 163 |
| abstract_inverted_index.much | 77 |
| abstract_inverted_index.pork | 36, 100 |
| abstract_inverted_index.than | 80 |
| abstract_inverted_index.that | 81, 93 |
| abstract_inverted_index.uses | 129 |
| abstract_inverted_index.were | 62 |
| abstract_inverted_index.with | 67, 73, 82 |
| abstract_inverted_index.BLUP. | 46 |
| abstract_inverted_index.GWAS. | 109 |
| abstract_inverted_index.PLINK | 118 |
| abstract_inverted_index.eight | 35 |
| abstract_inverted_index.given | 55 |
| abstract_inverted_index.major | 6 |
| abstract_inverted_index.model | 112, 121 |
| abstract_inverted_index.study | 23 |
| abstract_inverted_index.terms | 87 |
| abstract_inverted_index.total | 83 |
| abstract_inverted_index.trait | 52 |
| abstract_inverted_index.using | 43, 64, 117, 136 |
| abstract_inverted_index.value | 69 |
| abstract_inverted_index.which | 160 |
| abstract_inverted_index.(GWAS) | 24 |
| abstract_inverted_index.(SNPs) | 61 |
| abstract_inverted_index.<0.01. | 70 |
| abstract_inverted_index.G-BLUP | 124 |
| abstract_inverted_index.GEBVs. | 165 |
| abstract_inverted_index.breeds | 42 |
| abstract_inverted_index.linear | 13, 115 |
| abstract_inverted_index.method | 159 |
| abstract_inverted_index.single | 58 |
| abstract_inverted_index.traits | 38, 102 |
| abstract_inverted_index.values | 97 |
| abstract_inverted_index.(BLUP). | 16 |
| abstract_inverted_index.(GEBVs) | 98 |
| abstract_inverted_index.SNP-SNP | 130 |
| abstract_inverted_index.detects | 48 |
| abstract_inverted_index.genomic | 94 |
| abstract_inverted_index.implies | 92 |
| abstract_inverted_index.improve | 29 |
| abstract_inverted_index.matrix. | 132 |
| abstract_inverted_index.missing | 1, 150 |
| abstract_inverted_index.problem | 7, 152 |
| abstract_inverted_index.provide | 156 |
| abstract_inverted_index.quality | 37, 101 |
| abstract_inverted_index.regions | 54 |
| abstract_inverted_index.results | 66 |
| abstract_inverted_index.solving | 148 |
| abstract_inverted_index.traits. | 56 |
| abstract_inverted_index.accurate | 79, 164 |
| abstract_inverted_index.analysis | 10, 135 |
| abstract_inverted_index.analyzed | 34, 72 |
| abstract_inverted_index.breeding | 96 |
| abstract_inverted_index.obtained | 63 |
| abstract_inverted_index.possible | 145 |
| abstract_inverted_index.putative | 50 |
| abstract_inverted_index.unbiased | 14 |
| abstract_inverted_index.Berkshire | 41 |
| abstract_inverted_index.SNP-GBLUP | 128 |
| abstract_inverted_index.estimated | 95 |
| abstract_inverted_index.genotyped | 84 |
| abstract_inverted_index.SNP-GBLUP. | 126 |
| abstract_inverted_index.calculated | 105 |
| abstract_inverted_index.introduced | 18 |
| abstract_inverted_index.nucleotide | 59 |
| abstract_inverted_index.prediction | 15 |
| abstract_inverted_index.regression | 116 |
| abstract_inverted_index.alternative | 157 |
| abstract_inverted_index.association | 22 |
| abstract_inverted_index.estimation. | 32 |
| abstract_inverted_index.genome-wide | 21 |
| abstract_inverted_index.significant | 74 |
| abstract_inverted_index.traditional | 20 |
| abstract_inverted_index.alternatives | 146 |
| abstract_inverted_index.heritability | 2, 31, 151 |
| abstract_inverted_index.narrow-sense | 89 |
| abstract_inverted_index.quantitative | 51 |
| abstract_inverted_index.relationship | 131 |
| abstract_inverted_index.heritability. | 90 |
| abstract_inverted_index.polymorphisms | 60 |
| abstract_inverted_index.preprocessing | 137 |
| cited_by_percentile_year.max | 94 |
| cited_by_percentile_year.min | 89 |
| countries_distinct_count | 2 |
| institutions_distinct_count | 7 |
| citation_normalized_percentile.value | 0.68845544 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |