Genomic profiling of T cell activation reveals dependency of memory T cells on CD28 costimulation Article Swipe
 YOU?
·
· 2018
· Open Access
·
· DOI: https://doi.org/10.1101/421099
YOU?
·
· 2018
· Open Access
·
· DOI: https://doi.org/10.1101/421099
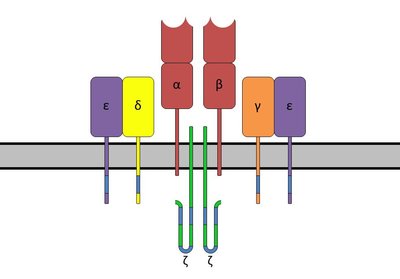
Summary T cell activation is a critical driver of immune response and if uncontrolled, it can result in failure to respond to infection or in excessive inflammation and autoimmunity. CD28 costimulatory pathway is an essential regulator of CD4 T cell responses. To deconvolute how T cell receptor (TCR) and CD28 orchestrate activation of human CD4 T cells we stimulated cells using varying intensities of TCR and CD28 signals followed by gene expression profiling. We demonstrate that T-helper differentiation and cytokine expression are controlled by CD28. Strikingly, cell cycle and cell division are sensitive to CD28 in memory cells, but under TCR control in naive cells, in contrast to the paradigm that memory cells are CD28-independent. Using a combination of chromatin accessibility and enhancer profiling, we observe that IRFs and Blimp-1 (PRDM1) motifs are enriched in naive and memory T cells in response to TCR. In contrast, memory cells initiate AP1 transcriptional regulation only when both TCR and CD28 are engaged, implicating CD28 as an amplifier of transcriptional programmes in memory cells. Lastly, we show that CD28-sensitive genes are enriched in autoimmune disease loci, pointing towards the role of memory cells and the regulation of T cell activation through CD28 in autoimmune disease development. This study provides important insights into the differential role of CD28 in naive and memory T cell responses and offers a new platform for design and interpretation of costimulatory based therapies. One-sentence summary Genomic profiling of CD4 T cell activation reveals a sensitivity switch from TCR in naive to CD28 in memory cells.
Related Topics
- Type
- preprint
- Language
- en
- Landing Page
- https://doi.org/10.1101/421099
- https://www.biorxiv.org/content/biorxiv/early/2018/09/18/421099.full.pdf
- OA Status
- green
- Cited By
- 6
- References
- 75
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W2890242504
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W2890242504Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1101/421099Digital Object Identifier
- Title
-
Genomic profiling of T cell activation reveals dependency of memory T cells on CD28 costimulationWork title
- Type
-
preprintOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2018Year of publication
- Publication date
-
2018-09-18Full publication date if available
- Authors
-
Dafni A. Glinos, Blagoje Soskic, Luke Jostins, David M. Sansom, Gosia TrynkaList of authors in order
- Landing page
-
https://doi.org/10.1101/421099Publisher landing page
- PDF URL
-
https://www.biorxiv.org/content/biorxiv/early/2018/09/18/421099.full.pdfDirect link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
greenOpen access status per OpenAlex
- OA URL
-
https://www.biorxiv.org/content/biorxiv/early/2018/09/18/421099.full.pdfDirect OA link when available
- Concepts
-
CD28, Biology, T-cell receptor, T cell, Cytotoxic T cell, Cell biology, Jurkat cells, Priming (agriculture), Immune system, Immunology, Genetics, In vitro, Botany, GerminationTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
6Total citation count in OpenAlex
- Citations by year (recent)
-
2021: 2, 2020: 1, 2019: 3Per-year citation counts (last 5 years)
- References (count)
-
75Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W2890242504 |
|---|---|
| doi | https://doi.org/10.1101/421099 |
| ids.doi | https://doi.org/10.1101/421099 |
| ids.mag | 2890242504 |
| ids.openalex | https://openalex.org/W2890242504 |
| fwci | 0.72887015 |
| type | preprint |
| title | Genomic profiling of T cell activation reveals dependency of memory T cells on CD28 costimulation |
| biblio.issue | |
| biblio.volume | |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T10031 |
| topics[0].field.id | https://openalex.org/fields/24 |
| topics[0].field.display_name | Immunology and Microbiology |
| topics[0].score | 0.9998999834060669 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/2403 |
| topics[0].subfield.display_name | Immunology |
| topics[0].display_name | T-cell and B-cell Immunology |
| topics[1].id | https://openalex.org/T11020 |
| topics[1].field.id | https://openalex.org/fields/24 |
| topics[1].field.display_name | Immunology and Microbiology |
| topics[1].score | 0.9998000264167786 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/2403 |
| topics[1].subfield.display_name | Immunology |
| topics[1].display_name | Immune Cell Function and Interaction |
| topics[2].id | https://openalex.org/T10580 |
| topics[2].field.id | https://openalex.org/fields/24 |
| topics[2].field.display_name | Immunology and Microbiology |
| topics[2].score | 0.9977999925613403 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/2403 |
| topics[2].subfield.display_name | Immunology |
| topics[2].display_name | Immunotherapy and Immune Responses |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C148125776 |
| concepts[0].level | 4 |
| concepts[0].score | 0.863266110420227 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q1622348 |
| concepts[0].display_name | CD28 |
| concepts[1].id | https://openalex.org/C86803240 |
| concepts[1].level | 0 |
| concepts[1].score | 0.6241803169250488 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[1].display_name | Biology |
| concepts[2].id | https://openalex.org/C19317047 |
| concepts[2].level | 4 |
| concepts[2].score | 0.6016310453414917 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q412037 |
| concepts[2].display_name | T-cell receptor |
| concepts[3].id | https://openalex.org/C2776090121 |
| concepts[3].level | 3 |
| concepts[3].score | 0.5122956037521362 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q193529 |
| concepts[3].display_name | T cell |
| concepts[4].id | https://openalex.org/C154317977 |
| concepts[4].level | 3 |
| concepts[4].score | 0.5044065713882446 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q376266 |
| concepts[4].display_name | Cytotoxic T cell |
| concepts[5].id | https://openalex.org/C95444343 |
| concepts[5].level | 1 |
| concepts[5].score | 0.46950411796569824 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q7141 |
| concepts[5].display_name | Cell biology |
| concepts[6].id | https://openalex.org/C179464577 |
| concepts[6].level | 4 |
| concepts[6].score | 0.4440387487411499 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q1632589 |
| concepts[6].display_name | Jurkat cells |
| concepts[7].id | https://openalex.org/C81444415 |
| concepts[7].level | 3 |
| concepts[7].score | 0.4312947988510132 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q7243535 |
| concepts[7].display_name | Priming (agriculture) |
| concepts[8].id | https://openalex.org/C8891405 |
| concepts[8].level | 2 |
| concepts[8].score | 0.31569600105285645 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q1059 |
| concepts[8].display_name | Immune system |
| concepts[9].id | https://openalex.org/C203014093 |
| concepts[9].level | 1 |
| concepts[9].score | 0.30177927017211914 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q101929 |
| concepts[9].display_name | Immunology |
| concepts[10].id | https://openalex.org/C54355233 |
| concepts[10].level | 1 |
| concepts[10].score | 0.1593581736087799 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[10].display_name | Genetics |
| concepts[11].id | https://openalex.org/C202751555 |
| concepts[11].level | 2 |
| concepts[11].score | 0.08339563012123108 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q221681 |
| concepts[11].display_name | In vitro |
| concepts[12].id | https://openalex.org/C59822182 |
| concepts[12].level | 1 |
| concepts[12].score | 0.0 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q441 |
| concepts[12].display_name | Botany |
| concepts[13].id | https://openalex.org/C100701293 |
| concepts[13].level | 2 |
| concepts[13].score | 0.0 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q193838 |
| concepts[13].display_name | Germination |
| keywords[0].id | https://openalex.org/keywords/cd28 |
| keywords[0].score | 0.863266110420227 |
| keywords[0].display_name | CD28 |
| keywords[1].id | https://openalex.org/keywords/biology |
| keywords[1].score | 0.6241803169250488 |
| keywords[1].display_name | Biology |
| keywords[2].id | https://openalex.org/keywords/t-cell-receptor |
| keywords[2].score | 0.6016310453414917 |
| keywords[2].display_name | T-cell receptor |
| keywords[3].id | https://openalex.org/keywords/t-cell |
| keywords[3].score | 0.5122956037521362 |
| keywords[3].display_name | T cell |
| keywords[4].id | https://openalex.org/keywords/cytotoxic-t-cell |
| keywords[4].score | 0.5044065713882446 |
| keywords[4].display_name | Cytotoxic T cell |
| keywords[5].id | https://openalex.org/keywords/cell-biology |
| keywords[5].score | 0.46950411796569824 |
| keywords[5].display_name | Cell biology |
| keywords[6].id | https://openalex.org/keywords/jurkat-cells |
| keywords[6].score | 0.4440387487411499 |
| keywords[6].display_name | Jurkat cells |
| keywords[7].id | https://openalex.org/keywords/priming |
| keywords[7].score | 0.4312947988510132 |
| keywords[7].display_name | Priming (agriculture) |
| keywords[8].id | https://openalex.org/keywords/immune-system |
| keywords[8].score | 0.31569600105285645 |
| keywords[8].display_name | Immune system |
| keywords[9].id | https://openalex.org/keywords/immunology |
| keywords[9].score | 0.30177927017211914 |
| keywords[9].display_name | Immunology |
| keywords[10].id | https://openalex.org/keywords/genetics |
| keywords[10].score | 0.1593581736087799 |
| keywords[10].display_name | Genetics |
| keywords[11].id | https://openalex.org/keywords/in-vitro |
| keywords[11].score | 0.08339563012123108 |
| keywords[11].display_name | In vitro |
| language | en |
| locations[0].id | doi:10.1101/421099 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4306402567 |
| locations[0].source.issn | |
| locations[0].source.type | repository |
| locations[0].source.is_oa | False |
| locations[0].source.issn_l | |
| locations[0].source.is_core | False |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| locations[0].source.host_organization | https://openalex.org/I2750212522 |
| locations[0].source.host_organization_name | Cold Spring Harbor Laboratory |
| locations[0].source.host_organization_lineage | https://openalex.org/I2750212522 |
| locations[0].license | cc-by |
| locations[0].pdf_url | https://www.biorxiv.org/content/biorxiv/early/2018/09/18/421099.full.pdf |
| locations[0].version | acceptedVersion |
| locations[0].raw_type | posted-content |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | True |
| locations[0].is_published | False |
| locations[0].raw_source_name | |
| locations[0].landing_page_url | https://doi.org/10.1101/421099 |
| indexed_in | crossref |
| authorships[0].author.id | https://openalex.org/A5053171932 |
| authorships[0].author.orcid | https://orcid.org/0000-0001-5556-0222 |
| authorships[0].author.display_name | Dafni A. Glinos |
| authorships[0].countries | GB |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I2802476451 |
| authorships[0].affiliations[0].raw_affiliation_string | Wellcome Sanger Institute, Wellcome Trust Genome Campus, Hinxton CB0 SA, UK. |
| authorships[0].institutions[0].id | https://openalex.org/I2802476451 |
| authorships[0].institutions[0].ror | https://ror.org/05cy4wa09 |
| authorships[0].institutions[0].type | nonprofit |
| authorships[0].institutions[0].lineage | https://openalex.org/I2802476451, https://openalex.org/I87048295 |
| authorships[0].institutions[0].country_code | GB |
| authorships[0].institutions[0].display_name | Wellcome Sanger Institute |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Dafni A. Glinos |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Wellcome Sanger Institute, Wellcome Trust Genome Campus, Hinxton CB0 SA, UK. |
| authorships[1].author.id | https://openalex.org/A5060234756 |
| authorships[1].author.orcid | https://orcid.org/0000-0003-3173-4528 |
| authorships[1].author.display_name | Blagoje Soskic |
| authorships[1].countries | GB |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I2802476451 |
| authorships[1].affiliations[0].raw_affiliation_string | Wellcome Sanger Institute, Wellcome Trust Genome Campus, Hinxton CB0 SA, UK. |
| authorships[1].institutions[0].id | https://openalex.org/I2802476451 |
| authorships[1].institutions[0].ror | https://ror.org/05cy4wa09 |
| authorships[1].institutions[0].type | nonprofit |
| authorships[1].institutions[0].lineage | https://openalex.org/I2802476451, https://openalex.org/I87048295 |
| authorships[1].institutions[0].country_code | GB |
| authorships[1].institutions[0].display_name | Wellcome Sanger Institute |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Blagoje Soskic |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | Wellcome Sanger Institute, Wellcome Trust Genome Campus, Hinxton CB0 SA, UK. |
| authorships[2].author.id | https://openalex.org/A5049890044 |
| authorships[2].author.orcid | |
| authorships[2].author.display_name | Luke Jostins |
| authorships[2].countries | GB |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I119474363 |
| authorships[2].affiliations[0].raw_affiliation_string | Christ Church, St. Aldates, Oxford OX1 1DP, UK. |
| authorships[2].affiliations[1].institution_ids | https://openalex.org/I2802952180, https://openalex.org/I40120149 |
| authorships[2].affiliations[1].raw_affiliation_string | Big Data Institute, University of Oxford, Roosevelt Drive, Oxford OX 7FZ, UK. |
| authorships[2].institutions[0].id | https://openalex.org/I119474363 |
| authorships[2].institutions[0].ror | https://ror.org/0489ggv38 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I119474363 |
| authorships[2].institutions[0].country_code | GB |
| authorships[2].institutions[0].display_name | Canterbury Christ Church University |
| authorships[2].institutions[1].id | https://openalex.org/I2802952180 |
| authorships[2].institutions[1].ror | https://ror.org/02hr72y72 |
| authorships[2].institutions[1].type | nonprofit |
| authorships[2].institutions[1].lineage | https://openalex.org/I2802952180 |
| authorships[2].institutions[1].country_code | GB |
| authorships[2].institutions[1].display_name | Open Data Institute |
| authorships[2].institutions[2].id | https://openalex.org/I40120149 |
| authorships[2].institutions[2].ror | https://ror.org/052gg0110 |
| authorships[2].institutions[2].type | education |
| authorships[2].institutions[2].lineage | https://openalex.org/I40120149 |
| authorships[2].institutions[2].country_code | GB |
| authorships[2].institutions[2].display_name | University of Oxford |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Luke Jostins |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | Big Data Institute, University of Oxford, Roosevelt Drive, Oxford OX 7FZ, UK., Christ Church, St. Aldates, Oxford OX1 1DP, UK. |
| authorships[3].author.id | https://openalex.org/A5042350027 |
| authorships[3].author.orcid | https://orcid.org/0000-0001-6506-3115 |
| authorships[3].author.display_name | David M. Sansom |
| authorships[3].countries | GB |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I2801138448, https://openalex.org/I45129253 |
| authorships[3].affiliations[0].raw_affiliation_string | UCL Institute of Immunity and Transplantation, Royal Free Hospital, London NW3 2PF, UK. |
| authorships[3].institutions[0].id | https://openalex.org/I2801138448 |
| authorships[3].institutions[0].ror | https://ror.org/01ge67z96 |
| authorships[3].institutions[0].type | healthcare |
| authorships[3].institutions[0].lineage | https://openalex.org/I169248724, https://openalex.org/I2801138448 |
| authorships[3].institutions[0].country_code | GB |
| authorships[3].institutions[0].display_name | The Royal Free Hospital |
| authorships[3].institutions[1].id | https://openalex.org/I45129253 |
| authorships[3].institutions[1].ror | https://ror.org/02jx3x895 |
| authorships[3].institutions[1].type | education |
| authorships[3].institutions[1].lineage | https://openalex.org/I124357947, https://openalex.org/I45129253 |
| authorships[3].institutions[1].country_code | GB |
| authorships[3].institutions[1].display_name | University College London |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | David M. Sansom |
| authorships[3].is_corresponding | True |
| authorships[3].raw_affiliation_strings | UCL Institute of Immunity and Transplantation, Royal Free Hospital, London NW3 2PF, UK. |
| authorships[4].author.id | https://openalex.org/A5091121983 |
| authorships[4].author.orcid | https://orcid.org/0000-0002-6955-9529 |
| authorships[4].author.display_name | Gosia Trynka |
| authorships[4].countries | GB |
| authorships[4].affiliations[0].institution_ids | https://openalex.org/I2802476451 |
| authorships[4].affiliations[0].raw_affiliation_string | Wellcome Sanger Institute, Wellcome Trust Genome Campus, Hinxton CB0 SA, UK. |
| authorships[4].institutions[0].id | https://openalex.org/I2802476451 |
| authorships[4].institutions[0].ror | https://ror.org/05cy4wa09 |
| authorships[4].institutions[0].type | nonprofit |
| authorships[4].institutions[0].lineage | https://openalex.org/I2802476451, https://openalex.org/I87048295 |
| authorships[4].institutions[0].country_code | GB |
| authorships[4].institutions[0].display_name | Wellcome Sanger Institute |
| authorships[4].author_position | last |
| authorships[4].raw_author_name | Gosia Trynka |
| authorships[4].is_corresponding | True |
| authorships[4].raw_affiliation_strings | Wellcome Sanger Institute, Wellcome Trust Genome Campus, Hinxton CB0 SA, UK. |
| has_content.pdf | True |
| has_content.grobid_xml | True |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://www.biorxiv.org/content/biorxiv/early/2018/09/18/421099.full.pdf |
| open_access.oa_status | green |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Genomic profiling of T cell activation reveals dependency of memory T cells on CD28 costimulation |
| has_fulltext | True |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T10031 |
| primary_topic.field.id | https://openalex.org/fields/24 |
| primary_topic.field.display_name | Immunology and Microbiology |
| primary_topic.score | 0.9998999834060669 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/2403 |
| primary_topic.subfield.display_name | Immunology |
| primary_topic.display_name | T-cell and B-cell Immunology |
| related_works | https://openalex.org/W1995507496, https://openalex.org/W1906673334, https://openalex.org/W2090555440, https://openalex.org/W2096118127, https://openalex.org/W2043153946, https://openalex.org/W2944432736, https://openalex.org/W2081520685, https://openalex.org/W1971240700, https://openalex.org/W2153762242, https://openalex.org/W2058811157 |
| cited_by_count | 6 |
| counts_by_year[0].year | 2021 |
| counts_by_year[0].cited_by_count | 2 |
| counts_by_year[1].year | 2020 |
| counts_by_year[1].cited_by_count | 1 |
| counts_by_year[2].year | 2019 |
| counts_by_year[2].cited_by_count | 3 |
| locations_count | 1 |
| best_oa_location.id | doi:10.1101/421099 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4306402567 |
| best_oa_location.source.issn | |
| best_oa_location.source.type | repository |
| best_oa_location.source.is_oa | False |
| best_oa_location.source.issn_l | |
| best_oa_location.source.is_core | False |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| best_oa_location.source.host_organization | https://openalex.org/I2750212522 |
| best_oa_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| best_oa_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2018/09/18/421099.full.pdf |
| best_oa_location.version | acceptedVersion |
| best_oa_location.raw_type | posted-content |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | False |
| best_oa_location.raw_source_name | |
| best_oa_location.landing_page_url | https://doi.org/10.1101/421099 |
| primary_location.id | doi:10.1101/421099 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4306402567 |
| primary_location.source.issn | |
| primary_location.source.type | repository |
| primary_location.source.is_oa | False |
| primary_location.source.issn_l | |
| primary_location.source.is_core | False |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| primary_location.source.host_organization | https://openalex.org/I2750212522 |
| primary_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| primary_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| primary_location.license | cc-by |
| primary_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2018/09/18/421099.full.pdf |
| primary_location.version | acceptedVersion |
| primary_location.raw_type | posted-content |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | True |
| primary_location.is_published | False |
| primary_location.raw_source_name | |
| primary_location.landing_page_url | https://doi.org/10.1101/421099 |
| publication_date | 2018-09-18 |
| publication_year | 2018 |
| referenced_works | https://openalex.org/W2465973416, https://openalex.org/W2569270101, https://openalex.org/W2769764904, https://openalex.org/W1914776013, https://openalex.org/W2103224648, https://openalex.org/W1993219448, https://openalex.org/W2117757143, https://openalex.org/W2130473166, https://openalex.org/W1485596855, https://openalex.org/W1995181168, https://openalex.org/W2169456326, https://openalex.org/W2009445020, https://openalex.org/W1565814966, https://openalex.org/W2112409097, https://openalex.org/W1490736032, https://openalex.org/W1510071762, https://openalex.org/W2395845086, https://openalex.org/W2012697072, https://openalex.org/W2056198580, https://openalex.org/W2953141386, https://openalex.org/W2086776723, https://openalex.org/W2343736575, https://openalex.org/W2045311357, https://openalex.org/W2590096315, https://openalex.org/W2473063121, https://openalex.org/W1975528320, https://openalex.org/W2592133876, https://openalex.org/W2080341180, https://openalex.org/W2553838260, https://openalex.org/W2160734881, https://openalex.org/W2592666036, https://openalex.org/W1516446271, https://openalex.org/W2140497145, https://openalex.org/W1863501054, https://openalex.org/W2081255498, https://openalex.org/W2559699708, https://openalex.org/W2169353806, https://openalex.org/W4247053599, https://openalex.org/W2108234281, https://openalex.org/W2001042507, https://openalex.org/W2138207763, https://openalex.org/W2214074259, https://openalex.org/W2171998058, https://openalex.org/W1574766328, https://openalex.org/W1647115901, https://openalex.org/W1527802692, https://openalex.org/W2145026575, https://openalex.org/W2004951070, https://openalex.org/W2951158909, https://openalex.org/W2952947531, https://openalex.org/W2138837186, https://openalex.org/W2041588091, https://openalex.org/W1989590412, https://openalex.org/W2077955816, https://openalex.org/W2102619694, https://openalex.org/W2087482409, https://openalex.org/W2767343890, https://openalex.org/W1925000049, https://openalex.org/W1967067685, https://openalex.org/W2122353285, https://openalex.org/W2004619659, https://openalex.org/W2076067786, https://openalex.org/W1534774246, https://openalex.org/W2133009026, https://openalex.org/W1865582025, https://openalex.org/W1559425537, https://openalex.org/W2030656636, https://openalex.org/W2073776582, https://openalex.org/W2019638031, https://openalex.org/W2000672459, https://openalex.org/W2604780989, https://openalex.org/W2135199713, https://openalex.org/W2171808845, https://openalex.org/W3145739264, https://openalex.org/W1529287480 |
| referenced_works_count | 75 |
| abstract_inverted_index.T | 1, 38, 44, 55, 138, 194, 218, 240 |
| abstract_inverted_index.a | 5, 116, 223, 244 |
| abstract_inverted_index.In | 144 |
| abstract_inverted_index.To | 41 |
| abstract_inverted_index.We | 73 |
| abstract_inverted_index.an | 33, 163 |
| abstract_inverted_index.as | 162 |
| abstract_inverted_index.by | 69, 83 |
| abstract_inverted_index.if | 12 |
| abstract_inverted_index.in | 17, 24, 95, 102, 105, 134, 140, 168, 179, 199, 214, 249, 253 |
| abstract_inverted_index.is | 4, 32 |
| abstract_inverted_index.it | 14 |
| abstract_inverted_index.of | 8, 36, 52, 63, 118, 165, 187, 193, 212, 230, 238 |
| abstract_inverted_index.or | 23 |
| abstract_inverted_index.to | 19, 21, 93, 107, 142, 251 |
| abstract_inverted_index.we | 57, 124, 172 |
| abstract_inverted_index.AP1 | 149 |
| abstract_inverted_index.CD4 | 37, 54, 239 |
| abstract_inverted_index.TCR | 64, 100, 155, 248 |
| abstract_inverted_index.and | 11, 27, 48, 65, 78, 88, 121, 128, 136, 156, 190, 216, 221, 228 |
| abstract_inverted_index.are | 81, 91, 113, 132, 158, 177 |
| abstract_inverted_index.but | 98 |
| abstract_inverted_index.can | 15 |
| abstract_inverted_index.for | 226 |
| abstract_inverted_index.how | 43 |
| abstract_inverted_index.new | 224 |
| abstract_inverted_index.the | 108, 185, 191, 209 |
| abstract_inverted_index.CD28 | 29, 49, 66, 94, 157, 161, 198, 213, 252 |
| abstract_inverted_index.IRFs | 127 |
| abstract_inverted_index.TCR. | 143 |
| abstract_inverted_index.This | 203 |
| abstract_inverted_index.both | 154 |
| abstract_inverted_index.cell | 2, 39, 45, 86, 89, 195, 219, 241 |
| abstract_inverted_index.from | 247 |
| abstract_inverted_index.gene | 70 |
| abstract_inverted_index.into | 208 |
| abstract_inverted_index.only | 152 |
| abstract_inverted_index.role | 186, 211 |
| abstract_inverted_index.show | 173 |
| abstract_inverted_index.that | 75, 110, 126, 174 |
| abstract_inverted_index.when | 153 |
| abstract_inverted_index.(TCR) | 47 |
| abstract_inverted_index.CD28. | 84 |
| abstract_inverted_index.Using | 115 |
| abstract_inverted_index.based | 232 |
| abstract_inverted_index.cells | 56, 59, 112, 139, 147, 189 |
| abstract_inverted_index.cycle | 87 |
| abstract_inverted_index.genes | 176 |
| abstract_inverted_index.human | 53 |
| abstract_inverted_index.loci, | 182 |
| abstract_inverted_index.naive | 103, 135, 215, 250 |
| abstract_inverted_index.study | 204 |
| abstract_inverted_index.under | 99 |
| abstract_inverted_index.using | 60 |
| abstract_inverted_index.cells, | 97, 104 |
| abstract_inverted_index.cells. | 170, 255 |
| abstract_inverted_index.design | 227 |
| abstract_inverted_index.driver | 7 |
| abstract_inverted_index.immune | 9 |
| abstract_inverted_index.memory | 96, 111, 137, 146, 169, 188, 217, 254 |
| abstract_inverted_index.motifs | 131 |
| abstract_inverted_index.offers | 222 |
| abstract_inverted_index.result | 16 |
| abstract_inverted_index.switch | 246 |
| abstract_inverted_index.(PRDM1) | 130 |
| abstract_inverted_index.Blimp-1 | 129 |
| abstract_inverted_index.Genomic | 236 |
| abstract_inverted_index.Lastly, | 171 |
| abstract_inverted_index.Summary | 0 |
| abstract_inverted_index.control | 101 |
| abstract_inverted_index.disease | 181, 201 |
| abstract_inverted_index.failure | 18 |
| abstract_inverted_index.observe | 125 |
| abstract_inverted_index.pathway | 31 |
| abstract_inverted_index.respond | 20 |
| abstract_inverted_index.reveals | 243 |
| abstract_inverted_index.signals | 67 |
| abstract_inverted_index.summary | 235 |
| abstract_inverted_index.through | 197 |
| abstract_inverted_index.towards | 184 |
| abstract_inverted_index.varying | 61 |
| abstract_inverted_index.T-helper | 76 |
| abstract_inverted_index.contrast | 106 |
| abstract_inverted_index.critical | 6 |
| abstract_inverted_index.cytokine | 79 |
| abstract_inverted_index.division | 90 |
| abstract_inverted_index.engaged, | 159 |
| abstract_inverted_index.enhancer | 122 |
| abstract_inverted_index.enriched | 133, 178 |
| abstract_inverted_index.followed | 68 |
| abstract_inverted_index.initiate | 148 |
| abstract_inverted_index.insights | 207 |
| abstract_inverted_index.paradigm | 109 |
| abstract_inverted_index.platform | 225 |
| abstract_inverted_index.pointing | 183 |
| abstract_inverted_index.provides | 205 |
| abstract_inverted_index.receptor | 46 |
| abstract_inverted_index.response | 10, 141 |
| abstract_inverted_index.amplifier | 164 |
| abstract_inverted_index.chromatin | 119 |
| abstract_inverted_index.contrast, | 145 |
| abstract_inverted_index.essential | 34 |
| abstract_inverted_index.excessive | 25 |
| abstract_inverted_index.important | 206 |
| abstract_inverted_index.infection | 22 |
| abstract_inverted_index.profiling | 237 |
| abstract_inverted_index.regulator | 35 |
| abstract_inverted_index.responses | 220 |
| abstract_inverted_index.sensitive | 92 |
| abstract_inverted_index.activation | 3, 51, 196, 242 |
| abstract_inverted_index.autoimmune | 180, 200 |
| abstract_inverted_index.controlled | 82 |
| abstract_inverted_index.expression | 71, 80 |
| abstract_inverted_index.profiling, | 123 |
| abstract_inverted_index.profiling. | 72 |
| abstract_inverted_index.programmes | 167 |
| abstract_inverted_index.regulation | 151, 192 |
| abstract_inverted_index.responses. | 40 |
| abstract_inverted_index.stimulated | 58 |
| abstract_inverted_index.therapies. | 233 |
| abstract_inverted_index.Strikingly, | 85 |
| abstract_inverted_index.combination | 117 |
| abstract_inverted_index.deconvolute | 42 |
| abstract_inverted_index.demonstrate | 74 |
| abstract_inverted_index.implicating | 160 |
| abstract_inverted_index.intensities | 62 |
| abstract_inverted_index.orchestrate | 50 |
| abstract_inverted_index.sensitivity | 245 |
| abstract_inverted_index.One-sentence | 234 |
| abstract_inverted_index.development. | 202 |
| abstract_inverted_index.differential | 210 |
| abstract_inverted_index.inflammation | 26 |
| abstract_inverted_index.accessibility | 120 |
| abstract_inverted_index.autoimmunity. | 28 |
| abstract_inverted_index.costimulatory | 30, 231 |
| abstract_inverted_index.uncontrolled, | 13 |
| abstract_inverted_index.CD28-sensitive | 175 |
| abstract_inverted_index.interpretation | 229 |
| abstract_inverted_index.differentiation | 77 |
| abstract_inverted_index.transcriptional | 150, 166 |
| abstract_inverted_index.CD28-independent. | 114 |
| cited_by_percentile_year.max | 97 |
| cited_by_percentile_year.min | 89 |
| corresponding_author_ids | https://openalex.org/A5091121983, https://openalex.org/A5042350027 |
| countries_distinct_count | 1 |
| institutions_distinct_count | 5 |
| corresponding_institution_ids | https://openalex.org/I2801138448, https://openalex.org/I2802476451, https://openalex.org/I45129253 |
| sustainable_development_goals[0].id | https://metadata.un.org/sdg/3 |
| sustainable_development_goals[0].score | 0.699999988079071 |
| sustainable_development_goals[0].display_name | Good health and well-being |
| citation_normalized_percentile.value | 0.6548806 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |