GWAS Procedures for Gene Mapping in Diverse Populations With Complex Structures Article Swipe
 YOU?
·
· 2025
· Open Access
·
· DOI: https://doi.org/10.21769/bioprotoc.5284
YOU?
·
· 2025
· Open Access
·
· DOI: https://doi.org/10.21769/bioprotoc.5284
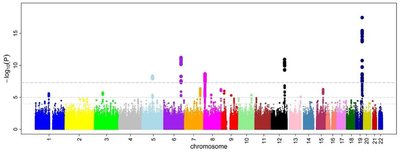
With reduced genotyping costs, genome-wide association studies (GWAS) face more challenges in diverse populations with complex structures to map genes of interest. The complex structure demands sophisticated statistical models, and increased marker density and population size require efficient computing tools. Many statistical models and computing tools have been developed with varied properties in statistical power, computing efficiency, and user-friendly accessibility. Some statistical models were developed with dedicated computing tools, such as efficient mixed model analysis (EMMA), multiple loci mixed model (MLMM), fixed and random model circulating probability unification (FarmCPU), and Bayesian-information and linkage-disequilibrium iteratively nested keyway (BLINK). However, there are computing tools (e.g., GAPIT) that implement multiple statistical models, retain a constant user interface, and maintain enhancement on input data and result interpretation. In this study, we developed a protocol utilizing a minimal set of software tools (BEAGLE, BLINK, and GAPIT) to perform a variety of analyses including file format conversion, missing genotype imputation, GWAS, and interpretation of input data and outcome results. We demonstrated the protocol by reanalyzing data from the Rice 3000 Genomes Project and highlighting advancements in GWAS model development.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.21769/bioprotoc.5284
- OA Status
- diamond
- Cited By
- 2
- References
- 1
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4409115241
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4409115241Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.21769/bioprotoc.5284Digital Object Identifier
- Title
-
GWAS Procedures for Gene Mapping in Diverse Populations With Complex StructuresWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2025Year of publication
- Publication date
-
2025-01-01Full publication date if available
- Authors
-
Zhen Zuo, Mingliang Li, Defu Liu, Qi Li, Bin Huang, G. Ye, Jiabo Wang, You Tang, Zhiwu ZhangList of authors in order
- Landing page
-
https://doi.org/10.21769/bioprotoc.5284Publisher landing page
- Open access
-
YesWhether a free full text is available
- OA status
-
diamondOpen access status per OpenAlex
- OA URL
-
https://doi.org/10.21769/bioprotoc.5284Direct OA link when available
- Concepts
-
Genome-wide association study, Computational biology, Data science, Biology, Evolutionary biology, Geography, Computer science, Gene, Genetics, Single-nucleotide polymorphism, GenotypeTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
2Total citation count in OpenAlex
- Citations by year (recent)
-
2025: 2Per-year citation counts (last 5 years)
- References (count)
-
1Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4409115241 |
|---|---|
| doi | https://doi.org/10.21769/bioprotoc.5284 |
| ids.doi | https://doi.org/10.21769/bioprotoc.5284 |
| ids.pmid | https://pubmed.ncbi.nlm.nih.gov/40291431 |
| ids.openalex | https://openalex.org/W4409115241 |
| fwci | 5.36588926 |
| type | article |
| title | GWAS Procedures for Gene Mapping in Diverse Populations With Complex Structures |
| biblio.issue | 1369 |
| biblio.volume | 15 |
| biblio.last_page | e5284 |
| biblio.first_page | e5284 |
| topics[0].id | https://openalex.org/T10885 |
| topics[0].field.id | https://openalex.org/fields/13 |
| topics[0].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[0].score | 0.9265999794006348 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1312 |
| topics[0].subfield.display_name | Molecular Biology |
| topics[0].display_name | Gene expression and cancer classification |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C106208931 |
| concepts[0].level | 5 |
| concepts[0].score | 0.7134928107261658 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q1098876 |
| concepts[0].display_name | Genome-wide association study |
| concepts[1].id | https://openalex.org/C70721500 |
| concepts[1].level | 1 |
| concepts[1].score | 0.5752235651016235 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q177005 |
| concepts[1].display_name | Computational biology |
| concepts[2].id | https://openalex.org/C2522767166 |
| concepts[2].level | 1 |
| concepts[2].score | 0.44473645091056824 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q2374463 |
| concepts[2].display_name | Data science |
| concepts[3].id | https://openalex.org/C86803240 |
| concepts[3].level | 0 |
| concepts[3].score | 0.3710251450538635 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[3].display_name | Biology |
| concepts[4].id | https://openalex.org/C78458016 |
| concepts[4].level | 1 |
| concepts[4].score | 0.3570537567138672 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q840400 |
| concepts[4].display_name | Evolutionary biology |
| concepts[5].id | https://openalex.org/C205649164 |
| concepts[5].level | 0 |
| concepts[5].score | 0.3336861729621887 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q1071 |
| concepts[5].display_name | Geography |
| concepts[6].id | https://openalex.org/C41008148 |
| concepts[6].level | 0 |
| concepts[6].score | 0.31631484627723694 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q21198 |
| concepts[6].display_name | Computer science |
| concepts[7].id | https://openalex.org/C104317684 |
| concepts[7].level | 2 |
| concepts[7].score | 0.2840179204940796 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[7].display_name | Gene |
| concepts[8].id | https://openalex.org/C54355233 |
| concepts[8].level | 1 |
| concepts[8].score | 0.2567777633666992 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[8].display_name | Genetics |
| concepts[9].id | https://openalex.org/C153209595 |
| concepts[9].level | 4 |
| concepts[9].score | 0.11480683088302612 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q501128 |
| concepts[9].display_name | Single-nucleotide polymorphism |
| concepts[10].id | https://openalex.org/C135763542 |
| concepts[10].level | 3 |
| concepts[10].score | 0.0 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q106016 |
| concepts[10].display_name | Genotype |
| keywords[0].id | https://openalex.org/keywords/genome-wide-association-study |
| keywords[0].score | 0.7134928107261658 |
| keywords[0].display_name | Genome-wide association study |
| keywords[1].id | https://openalex.org/keywords/computational-biology |
| keywords[1].score | 0.5752235651016235 |
| keywords[1].display_name | Computational biology |
| keywords[2].id | https://openalex.org/keywords/data-science |
| keywords[2].score | 0.44473645091056824 |
| keywords[2].display_name | Data science |
| keywords[3].id | https://openalex.org/keywords/biology |
| keywords[3].score | 0.3710251450538635 |
| keywords[3].display_name | Biology |
| keywords[4].id | https://openalex.org/keywords/evolutionary-biology |
| keywords[4].score | 0.3570537567138672 |
| keywords[4].display_name | Evolutionary biology |
| keywords[5].id | https://openalex.org/keywords/geography |
| keywords[5].score | 0.3336861729621887 |
| keywords[5].display_name | Geography |
| keywords[6].id | https://openalex.org/keywords/computer-science |
| keywords[6].score | 0.31631484627723694 |
| keywords[6].display_name | Computer science |
| keywords[7].id | https://openalex.org/keywords/gene |
| keywords[7].score | 0.2840179204940796 |
| keywords[7].display_name | Gene |
| keywords[8].id | https://openalex.org/keywords/genetics |
| keywords[8].score | 0.2567777633666992 |
| keywords[8].display_name | Genetics |
| keywords[9].id | https://openalex.org/keywords/single-nucleotide-polymorphism |
| keywords[9].score | 0.11480683088302612 |
| keywords[9].display_name | Single-nucleotide polymorphism |
| language | en |
| locations[0].id | doi:10.21769/bioprotoc.5284 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4210179406 |
| locations[0].source.issn | 2331-8325 |
| locations[0].source.type | journal |
| locations[0].source.is_oa | True |
| locations[0].source.issn_l | 2331-8325 |
| locations[0].source.is_core | True |
| locations[0].source.is_in_doaj | True |
| locations[0].source.display_name | BIO-PROTOCOL |
| locations[0].source.host_organization | https://openalex.org/P4310320252 |
| locations[0].source.host_organization_name | American Academy of Arts and Sciences |
| locations[0].source.host_organization_lineage | https://openalex.org/P4310320252 |
| locations[0].source.host_organization_lineage_names | American Academy of Arts and Sciences |
| locations[0].license | |
| locations[0].pdf_url | |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | BIO-PROTOCOL |
| locations[0].landing_page_url | https://doi.org/10.21769/bioprotoc.5284 |
| locations[1].id | pmid:40291431 |
| locations[1].is_oa | False |
| locations[1].source.id | https://openalex.org/S4306525036 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | PubMed |
| locations[1].source.host_organization | https://openalex.org/I1299303238 |
| locations[1].source.host_organization_name | National Institutes of Health |
| locations[1].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[1].license | |
| locations[1].pdf_url | |
| locations[1].version | publishedVersion |
| locations[1].raw_type | |
| locations[1].license_id | |
| locations[1].is_accepted | True |
| locations[1].is_published | True |
| locations[1].raw_source_name | Bio-protocol |
| locations[1].landing_page_url | https://pubmed.ncbi.nlm.nih.gov/40291431 |
| locations[2].id | pmh:oai:doaj.org/article:3653312c7d494e6990d9818e135495ab |
| locations[2].is_oa | False |
| locations[2].source.id | https://openalex.org/S4306401280 |
| locations[2].source.issn | |
| locations[2].source.type | repository |
| locations[2].source.is_oa | False |
| locations[2].source.issn_l | |
| locations[2].source.is_core | False |
| locations[2].source.is_in_doaj | False |
| locations[2].source.display_name | DOAJ (DOAJ: Directory of Open Access Journals) |
| locations[2].source.host_organization | |
| locations[2].source.host_organization_name | |
| locations[2].license | |
| locations[2].pdf_url | |
| locations[2].version | submittedVersion |
| locations[2].raw_type | article |
| locations[2].license_id | |
| locations[2].is_accepted | False |
| locations[2].is_published | False |
| locations[2].raw_source_name | Bio-Protocol, Vol 15, Iss 8 (2025) |
| locations[2].landing_page_url | https://doaj.org/article/3653312c7d494e6990d9818e135495ab |
| locations[3].id | pmh:oai:europepmc.org:10831924 |
| locations[3].is_oa | True |
| locations[3].source.id | https://openalex.org/S4306400806 |
| locations[3].source.issn | |
| locations[3].source.type | repository |
| locations[3].source.is_oa | False |
| locations[3].source.issn_l | |
| locations[3].source.is_core | False |
| locations[3].source.is_in_doaj | False |
| locations[3].source.display_name | Europe PMC (PubMed Central) |
| locations[3].source.host_organization | https://openalex.org/I1303153112 |
| locations[3].source.host_organization_name | European Bioinformatics Institute |
| locations[3].source.host_organization_lineage | https://openalex.org/I1303153112 |
| locations[3].license | other-oa |
| locations[3].pdf_url | |
| locations[3].version | submittedVersion |
| locations[3].raw_type | Text |
| locations[3].license_id | https://openalex.org/licenses/other-oa |
| locations[3].is_accepted | False |
| locations[3].is_published | False |
| locations[3].raw_source_name | |
| locations[3].landing_page_url | https://www.ncbi.nlm.nih.gov/pmc/articles/12021685 |
| indexed_in | crossref, doaj, pubmed |
| authorships[0].author.id | https://openalex.org/A5006662591 |
| authorships[0].author.orcid | https://orcid.org/0000-0003-1555-1756 |
| authorships[0].author.display_name | Zhen Zuo |
| authorships[0].countries | CN |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I4210141933 |
| authorships[0].affiliations[0].raw_affiliation_string | Electrical and Information Engineering College, Jilin Agricultural Science and Technology University, Jilin, Jilin, China. |
| authorships[0].institutions[0].id | https://openalex.org/I4210141933 |
| authorships[0].institutions[0].ror | https://ror.org/04w5zb891 |
| authorships[0].institutions[0].type | education |
| authorships[0].institutions[0].lineage | https://openalex.org/I4210141933 |
| authorships[0].institutions[0].country_code | CN |
| authorships[0].institutions[0].display_name | Jilin Agricultural Science and Technology University |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Zhen Zuo |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Electrical and Information Engineering College, Jilin Agricultural Science and Technology University, Jilin, Jilin, China. |
| authorships[1].author.id | https://openalex.org/A5100762551 |
| authorships[1].author.orcid | https://orcid.org/0000-0001-6201-2232 |
| authorships[1].author.display_name | Mingliang Li |
| authorships[1].countries | CN |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I4210141933 |
| authorships[1].affiliations[0].raw_affiliation_string | Electrical and Information Engineering College, Jilin Agricultural Science and Technology University, Jilin, Jilin, China. |
| authorships[1].institutions[0].id | https://openalex.org/I4210141933 |
| authorships[1].institutions[0].ror | https://ror.org/04w5zb891 |
| authorships[1].institutions[0].type | education |
| authorships[1].institutions[0].lineage | https://openalex.org/I4210141933 |
| authorships[1].institutions[0].country_code | CN |
| authorships[1].institutions[0].display_name | Jilin Agricultural Science and Technology University |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Mingliang Li |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | Electrical and Information Engineering College, Jilin Agricultural Science and Technology University, Jilin, Jilin, China. |
| authorships[2].author.id | https://openalex.org/A5113858706 |
| authorships[2].author.orcid | https://orcid.org/0000-0002-8251-2733 |
| authorships[2].author.display_name | Defu Liu |
| authorships[2].countries | CN |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I4210152006 |
| authorships[2].affiliations[0].raw_affiliation_string | Information Technology Academy, Jilin Agricultural University, Changchun, Jilin, China. |
| authorships[2].institutions[0].id | https://openalex.org/I4210152006 |
| authorships[2].institutions[0].ror | https://ror.org/05dmhhd41 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I4210152006 |
| authorships[2].institutions[0].country_code | CN |
| authorships[2].institutions[0].display_name | Jilin Agricultural University |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Defu Liu |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | Information Technology Academy, Jilin Agricultural University, Changchun, Jilin, China. |
| authorships[3].author.id | https://openalex.org/A5100350269 |
| authorships[3].author.orcid | https://orcid.org/0000-0003-2112-8452 |
| authorships[3].author.display_name | Qi Li |
| authorships[3].countries | CN |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I4210141933 |
| authorships[3].affiliations[0].raw_affiliation_string | Electrical and Information Engineering College, Jilin Agricultural Science and Technology University, Jilin, Jilin, China. |
| authorships[3].institutions[0].id | https://openalex.org/I4210141933 |
| authorships[3].institutions[0].ror | https://ror.org/04w5zb891 |
| authorships[3].institutions[0].type | education |
| authorships[3].institutions[0].lineage | https://openalex.org/I4210141933 |
| authorships[3].institutions[0].country_code | CN |
| authorships[3].institutions[0].display_name | Jilin Agricultural Science and Technology University |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Qi Li |
| authorships[3].is_corresponding | False |
| authorships[3].raw_affiliation_strings | Electrical and Information Engineering College, Jilin Agricultural Science and Technology University, Jilin, Jilin, China. |
| authorships[4].author.id | https://openalex.org/A5101481519 |
| authorships[4].author.orcid | https://orcid.org/0000-0002-6668-4885 |
| authorships[4].author.display_name | Bin Huang |
| authorships[4].countries | CN |
| authorships[4].affiliations[0].institution_ids | https://openalex.org/I4210141933 |
| authorships[4].affiliations[0].raw_affiliation_string | Electrical and Information Engineering College, Jilin Agricultural Science and Technology University, Jilin, Jilin, China. |
| authorships[4].institutions[0].id | https://openalex.org/I4210141933 |
| authorships[4].institutions[0].ror | https://ror.org/04w5zb891 |
| authorships[4].institutions[0].type | education |
| authorships[4].institutions[0].lineage | https://openalex.org/I4210141933 |
| authorships[4].institutions[0].country_code | CN |
| authorships[4].institutions[0].display_name | Jilin Agricultural Science and Technology University |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Bin Huang |
| authorships[4].is_corresponding | False |
| authorships[4].raw_affiliation_strings | Electrical and Information Engineering College, Jilin Agricultural Science and Technology University, Jilin, Jilin, China. |
| authorships[5].author.id | https://openalex.org/A5012009186 |
| authorships[5].author.orcid | |
| authorships[5].author.display_name | G. Ye |
| authorships[5].countries | CN |
| authorships[5].affiliations[0].institution_ids | https://openalex.org/I4210141933 |
| authorships[5].affiliations[0].raw_affiliation_string | Electrical and Information Engineering College, Jilin Agricultural Science and Technology University, Jilin, Jilin, China. |
| authorships[5].institutions[0].id | https://openalex.org/I4210141933 |
| authorships[5].institutions[0].ror | https://ror.org/04w5zb891 |
| authorships[5].institutions[0].type | education |
| authorships[5].institutions[0].lineage | https://openalex.org/I4210141933 |
| authorships[5].institutions[0].country_code | CN |
| authorships[5].institutions[0].display_name | Jilin Agricultural Science and Technology University |
| authorships[5].author_position | middle |
| authorships[5].raw_author_name | Guanshi Ye |
| authorships[5].is_corresponding | False |
| authorships[5].raw_affiliation_strings | Electrical and Information Engineering College, Jilin Agricultural Science and Technology University, Jilin, Jilin, China. |
| authorships[6].author.id | https://openalex.org/A5100665802 |
| authorships[6].author.orcid | https://orcid.org/0000-0002-1386-0435 |
| authorships[6].author.display_name | Jiabo Wang |
| authorships[6].countries | CN |
| authorships[6].affiliations[0].institution_ids | https://openalex.org/I6645349 |
| authorships[6].affiliations[0].raw_affiliation_string | Key Laboratory of Qinghai-Tibetan Plateau Animal Genetic Resource Reservation and Utilization, Sichuan Province and Ministry of Education, Southwest Minzu University, Chengdu, Sichuan, China. |
| authorships[6].institutions[0].id | https://openalex.org/I6645349 |
| authorships[6].institutions[0].ror | https://ror.org/04gaexw88 |
| authorships[6].institutions[0].type | education |
| authorships[6].institutions[0].lineage | https://openalex.org/I6645349 |
| authorships[6].institutions[0].country_code | CN |
| authorships[6].institutions[0].display_name | Southwest Minzu University |
| authorships[6].author_position | middle |
| authorships[6].raw_author_name | Jiabo Wang |
| authorships[6].is_corresponding | False |
| authorships[6].raw_affiliation_strings | Key Laboratory of Qinghai-Tibetan Plateau Animal Genetic Resource Reservation and Utilization, Sichuan Province and Ministry of Education, Southwest Minzu University, Chengdu, Sichuan, China. |
| authorships[7].author.id | https://openalex.org/A5101440100 |
| authorships[7].author.orcid | https://orcid.org/0000-0002-2873-4212 |
| authorships[7].author.display_name | You Tang |
| authorships[7].countries | CN |
| authorships[7].affiliations[0].institution_ids | https://openalex.org/I4210141933 |
| authorships[7].affiliations[0].raw_affiliation_string | Electrical and Information Engineering College, Jilin Agricultural Science and Technology University, Jilin, Jilin, China. |
| authorships[7].affiliations[1].institution_ids | https://openalex.org/I4210152006 |
| authorships[7].affiliations[1].raw_affiliation_string | Information Technology Academy, Jilin Agricultural University, Changchun, Jilin, China. |
| authorships[7].institutions[0].id | https://openalex.org/I4210141933 |
| authorships[7].institutions[0].ror | https://ror.org/04w5zb891 |
| authorships[7].institutions[0].type | education |
| authorships[7].institutions[0].lineage | https://openalex.org/I4210141933 |
| authorships[7].institutions[0].country_code | CN |
| authorships[7].institutions[0].display_name | Jilin Agricultural Science and Technology University |
| authorships[7].institutions[1].id | https://openalex.org/I4210152006 |
| authorships[7].institutions[1].ror | https://ror.org/05dmhhd41 |
| authorships[7].institutions[1].type | education |
| authorships[7].institutions[1].lineage | https://openalex.org/I4210152006 |
| authorships[7].institutions[1].country_code | CN |
| authorships[7].institutions[1].display_name | Jilin Agricultural University |
| authorships[7].author_position | middle |
| authorships[7].raw_author_name | You Tang |
| authorships[7].is_corresponding | False |
| authorships[7].raw_affiliation_strings | Electrical and Information Engineering College, Jilin Agricultural Science and Technology University, Jilin, Jilin, China., Information Technology Academy, Jilin Agricultural University, Changchun, Jilin, China. |
| authorships[8].author.id | https://openalex.org/A5065237213 |
| authorships[8].author.orcid | https://orcid.org/0000-0002-5784-9684 |
| authorships[8].author.display_name | Zhiwu Zhang |
| authorships[8].countries | US |
| authorships[8].affiliations[0].institution_ids | https://openalex.org/I72951846 |
| authorships[8].affiliations[0].raw_affiliation_string | Department of Crop and Soil Sciences, Washington State University, Pullman, WA, USA. |
| authorships[8].institutions[0].id | https://openalex.org/I72951846 |
| authorships[8].institutions[0].ror | https://ror.org/05dk0ce17 |
| authorships[8].institutions[0].type | education |
| authorships[8].institutions[0].lineage | https://openalex.org/I72951846 |
| authorships[8].institutions[0].country_code | US |
| authorships[8].institutions[0].display_name | Washington State University |
| authorships[8].author_position | last |
| authorships[8].raw_author_name | Zhiwu Zhang |
| authorships[8].is_corresponding | False |
| authorships[8].raw_affiliation_strings | Department of Crop and Soil Sciences, Washington State University, Pullman, WA, USA. |
| has_content.pdf | False |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://doi.org/10.21769/bioprotoc.5284 |
| open_access.oa_status | diamond |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | GWAS Procedures for Gene Mapping in Diverse Populations With Complex Structures |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T10885 |
| primary_topic.field.id | https://openalex.org/fields/13 |
| primary_topic.field.display_name | Biochemistry, Genetics and Molecular Biology |
| primary_topic.score | 0.9265999794006348 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1312 |
| primary_topic.subfield.display_name | Molecular Biology |
| primary_topic.display_name | Gene expression and cancer classification |
| related_works | https://openalex.org/W2748952813, https://openalex.org/W1539935214, https://openalex.org/W4390666975, https://openalex.org/W1864400744, https://openalex.org/W2736520836, https://openalex.org/W3093265873, https://openalex.org/W3209211130, https://openalex.org/W1966248182, https://openalex.org/W2971313479, https://openalex.org/W2116256947 |
| cited_by_count | 2 |
| counts_by_year[0].year | 2025 |
| counts_by_year[0].cited_by_count | 2 |
| locations_count | 4 |
| best_oa_location.id | doi:10.21769/bioprotoc.5284 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4210179406 |
| best_oa_location.source.issn | 2331-8325 |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | True |
| best_oa_location.source.issn_l | 2331-8325 |
| best_oa_location.source.is_core | True |
| best_oa_location.source.is_in_doaj | True |
| best_oa_location.source.display_name | BIO-PROTOCOL |
| best_oa_location.source.host_organization | https://openalex.org/P4310320252 |
| best_oa_location.source.host_organization_name | American Academy of Arts and Sciences |
| best_oa_location.source.host_organization_lineage | https://openalex.org/P4310320252 |
| best_oa_location.source.host_organization_lineage_names | American Academy of Arts and Sciences |
| best_oa_location.license | |
| best_oa_location.pdf_url | |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | journal-article |
| best_oa_location.license_id | |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | BIO-PROTOCOL |
| best_oa_location.landing_page_url | https://doi.org/10.21769/bioprotoc.5284 |
| primary_location.id | doi:10.21769/bioprotoc.5284 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4210179406 |
| primary_location.source.issn | 2331-8325 |
| primary_location.source.type | journal |
| primary_location.source.is_oa | True |
| primary_location.source.issn_l | 2331-8325 |
| primary_location.source.is_core | True |
| primary_location.source.is_in_doaj | True |
| primary_location.source.display_name | BIO-PROTOCOL |
| primary_location.source.host_organization | https://openalex.org/P4310320252 |
| primary_location.source.host_organization_name | American Academy of Arts and Sciences |
| primary_location.source.host_organization_lineage | https://openalex.org/P4310320252 |
| primary_location.source.host_organization_lineage_names | American Academy of Arts and Sciences |
| primary_location.license | |
| primary_location.pdf_url | |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | BIO-PROTOCOL |
| primary_location.landing_page_url | https://doi.org/10.21769/bioprotoc.5284 |
| publication_date | 2025-01-01 |
| publication_year | 2025 |
| referenced_works | https://openalex.org/W1547202054 |
| referenced_works_count | 1 |
| abstract_inverted_index.a | 110, 128, 131, 143 |
| abstract_inverted_index.In | 123 |
| abstract_inverted_index.We | 163 |
| abstract_inverted_index.as | 70 |
| abstract_inverted_index.by | 167 |
| abstract_inverted_index.in | 11, 52, 179 |
| abstract_inverted_index.of | 20, 134, 145, 157 |
| abstract_inverted_index.on | 117 |
| abstract_inverted_index.to | 17, 141 |
| abstract_inverted_index.we | 126 |
| abstract_inverted_index.The | 22 |
| abstract_inverted_index.and | 29, 33, 43, 57, 82, 89, 91, 114, 120, 139, 155, 160, 176 |
| abstract_inverted_index.are | 99 |
| abstract_inverted_index.map | 18 |
| abstract_inverted_index.set | 133 |
| abstract_inverted_index.the | 165, 171 |
| abstract_inverted_index.3000 | 173 |
| abstract_inverted_index.GWAS | 180 |
| abstract_inverted_index.Many | 40 |
| abstract_inverted_index.Rice | 172 |
| abstract_inverted_index.Some | 60 |
| abstract_inverted_index.With | 0 |
| abstract_inverted_index.been | 47 |
| abstract_inverted_index.data | 119, 159, 169 |
| abstract_inverted_index.face | 8 |
| abstract_inverted_index.file | 148 |
| abstract_inverted_index.from | 170 |
| abstract_inverted_index.have | 46 |
| abstract_inverted_index.loci | 77 |
| abstract_inverted_index.more | 9 |
| abstract_inverted_index.size | 35 |
| abstract_inverted_index.such | 69 |
| abstract_inverted_index.that | 104 |
| abstract_inverted_index.this | 124 |
| abstract_inverted_index.user | 112 |
| abstract_inverted_index.were | 63 |
| abstract_inverted_index.with | 14, 49, 65 |
| abstract_inverted_index.GWAS, | 154 |
| abstract_inverted_index.fixed | 81 |
| abstract_inverted_index.genes | 19 |
| abstract_inverted_index.input | 118, 158 |
| abstract_inverted_index.mixed | 72, 78 |
| abstract_inverted_index.model | 73, 79, 84, 181 |
| abstract_inverted_index.there | 98 |
| abstract_inverted_index.tools | 45, 101, 136 |
| abstract_inverted_index.(GWAS) | 7 |
| abstract_inverted_index.(e.g., | 102 |
| abstract_inverted_index.BLINK, | 138 |
| abstract_inverted_index.GAPIT) | 103, 140 |
| abstract_inverted_index.costs, | 3 |
| abstract_inverted_index.format | 149 |
| abstract_inverted_index.keyway | 95 |
| abstract_inverted_index.marker | 31 |
| abstract_inverted_index.models | 42, 62 |
| abstract_inverted_index.nested | 94 |
| abstract_inverted_index.power, | 54 |
| abstract_inverted_index.random | 83 |
| abstract_inverted_index.result | 121 |
| abstract_inverted_index.retain | 109 |
| abstract_inverted_index.study, | 125 |
| abstract_inverted_index.tools, | 68 |
| abstract_inverted_index.tools. | 39 |
| abstract_inverted_index.varied | 50 |
| abstract_inverted_index.(EMMA), | 75 |
| abstract_inverted_index.(MLMM), | 80 |
| abstract_inverted_index.Genomes | 174 |
| abstract_inverted_index.Project | 175 |
| abstract_inverted_index.complex | 15, 23 |
| abstract_inverted_index.demands | 25 |
| abstract_inverted_index.density | 32 |
| abstract_inverted_index.diverse | 12 |
| abstract_inverted_index.minimal | 132 |
| abstract_inverted_index.missing | 151 |
| abstract_inverted_index.models, | 28, 108 |
| abstract_inverted_index.outcome | 161 |
| abstract_inverted_index.perform | 142 |
| abstract_inverted_index.reduced | 1 |
| abstract_inverted_index.require | 36 |
| abstract_inverted_index.studies | 6 |
| abstract_inverted_index.variety | 144 |
| abstract_inverted_index.(BEAGLE, | 137 |
| abstract_inverted_index.(BLINK). | 96 |
| abstract_inverted_index.However, | 97 |
| abstract_inverted_index.analyses | 146 |
| abstract_inverted_index.analysis | 74 |
| abstract_inverted_index.constant | 111 |
| abstract_inverted_index.genotype | 152 |
| abstract_inverted_index.maintain | 115 |
| abstract_inverted_index.multiple | 76, 106 |
| abstract_inverted_index.protocol | 129, 166 |
| abstract_inverted_index.results. | 162 |
| abstract_inverted_index.software | 135 |
| abstract_inverted_index.computing | 38, 44, 55, 67, 100 |
| abstract_inverted_index.dedicated | 66 |
| abstract_inverted_index.developed | 48, 64, 127 |
| abstract_inverted_index.efficient | 37, 71 |
| abstract_inverted_index.implement | 105 |
| abstract_inverted_index.including | 147 |
| abstract_inverted_index.increased | 30 |
| abstract_inverted_index.interest. | 21 |
| abstract_inverted_index.structure | 24 |
| abstract_inverted_index.utilizing | 130 |
| abstract_inverted_index.(FarmCPU), | 88 |
| abstract_inverted_index.challenges | 10 |
| abstract_inverted_index.genotyping | 2 |
| abstract_inverted_index.interface, | 113 |
| abstract_inverted_index.population | 34 |
| abstract_inverted_index.properties | 51 |
| abstract_inverted_index.structures | 16 |
| abstract_inverted_index.association | 5 |
| abstract_inverted_index.circulating | 85 |
| abstract_inverted_index.conversion, | 150 |
| abstract_inverted_index.efficiency, | 56 |
| abstract_inverted_index.enhancement | 116 |
| abstract_inverted_index.genome-wide | 4 |
| abstract_inverted_index.imputation, | 153 |
| abstract_inverted_index.iteratively | 93 |
| abstract_inverted_index.populations | 13 |
| abstract_inverted_index.probability | 86 |
| abstract_inverted_index.reanalyzing | 168 |
| abstract_inverted_index.statistical | 27, 41, 53, 61, 107 |
| abstract_inverted_index.unification | 87 |
| abstract_inverted_index.advancements | 178 |
| abstract_inverted_index.demonstrated | 164 |
| abstract_inverted_index.development. | 182 |
| abstract_inverted_index.highlighting | 177 |
| abstract_inverted_index.sophisticated | 26 |
| abstract_inverted_index.user-friendly | 58 |
| abstract_inverted_index.accessibility. | 59 |
| abstract_inverted_index.interpretation | 156 |
| abstract_inverted_index.interpretation. | 122 |
| abstract_inverted_index.Bayesian-information | 90 |
| abstract_inverted_index.linkage-disequilibrium | 92 |
| cited_by_percentile_year.max | 97 |
| cited_by_percentile_year.min | 95 |
| countries_distinct_count | 2 |
| institutions_distinct_count | 9 |
| sustainable_development_goals[0].id | https://metadata.un.org/sdg/2 |
| sustainable_development_goals[0].score | 0.4099999964237213 |
| sustainable_development_goals[0].display_name | Zero hunger |
| citation_normalized_percentile.value | 0.88809111 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | True |