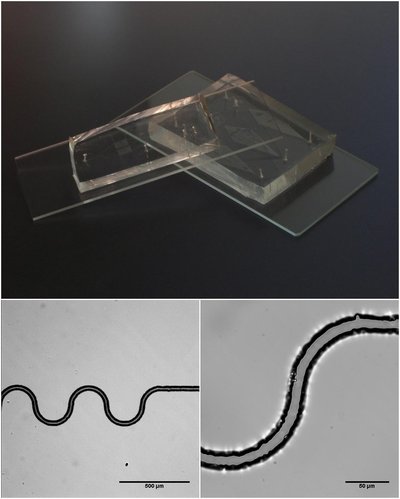
High-throughput generic single-entity sequencing using droplet microfluidics Article Swipe
 YOU?
·
· 2023
· Open Access
·
· DOI: https://doi.org/10.1101/2023.08.15.549386
YOU?
·
· 2023
· Open Access
·
· DOI: https://doi.org/10.1101/2023.08.15.549386
Summary Single-cell sequencing has revolutionized our understanding of cellular heterogeneity by providing a micro-level perspective over the past decades. Although heterogeneity is essential for various biological communities, the currently demonstrated platform predominantly focuses on eukaryotic cells without cell walls and their transcriptomics 1,2 , leaving significant gaps in the study of omics from other single biological entities such as bacteria and viruses. Due to the difficulty of isolating and acquiring their DNA 3 , contemporary methodologies for the characterization of generic biological entities remain conspicuously constrained, with low throughput 4 , compromised lysis efficiency 5 , and highly fragmented genomes 6 . Herein, we present the Generic Single Entity Sequencing platform (GSE-Seq), which boasts ample versatility, high throughput, and high coverage, and is enabled by an innovative workflow, addressing the critical challenges in single entities sequencing: (1) one-step manufacturing of massive barcodes, (2) degradable hydrogel-based in situ sample processing and whole genome amplification, (3) integrated in-drop library preparation, (4) compatible long-read sequencing. By GSE-Seq, we have achieved a significant milestone by enabling high-throughput, long-read single-entity profiling of dsDNA and ssDNA from single virus sequencing (SV-seq) and single bacteria sequencing (SB-seq) of the human gut and marine sediment for the first time. Notably, our analysis uncovered previously overlooked viral and bacterial dark matter and phage-host interactions. In summary, the presented conceptually new workflow offers a toolbox based on droplet microfluidics to tackle the persistent challenges in high-throughput profiling to generic applications, which hold immense promise for diverse biological entities, especially hard-to-lyse cells.
Related Topics
- Type
- preprint
- Language
- en
- Landing Page
- https://doi.org/10.1101/2023.08.15.549386
- https://www.biorxiv.org/content/biorxiv/early/2023/08/15/2023.08.15.549386.full.pdf
- OA Status
- green
- Cited By
- 2
- References
- 35
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4385875706
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4385875706Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1101/2023.08.15.549386Digital Object Identifier
- Title
-
High-throughput generic single-entity sequencing using droplet microfluidicsWork title
- Type
-
preprintOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2023Year of publication
- Publication date
-
2023-08-15Full publication date if available
- Authors
-
Guoping Wang, Liuyang Zhao, Yu Shi, Fuyang Qu, Yanqiang Ding, Weixin Liu, Changan Liu, Gang Luo, Meiyi Li, Xiaowu Bai, Luoquan Li, Yi‐Ping Ho, Jun YuList of authors in order
- Landing page
-
https://doi.org/10.1101/2023.08.15.549386Publisher landing page
- PDF URL
-
https://www.biorxiv.org/content/biorxiv/early/2023/08/15/2023.08.15.549386.full.pdfDirect link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
greenOpen access status per OpenAlex
- OA URL
-
https://www.biorxiv.org/content/biorxiv/early/2023/08/15/2023.08.15.549386.full.pdfDirect OA link when available
- Concepts
-
Workflow, DNA sequencing, Computational biology, Microfluidics, Profiling (computer programming), Throughput, Biology, Single-cell analysis, Genome, Computer science, Nanotechnology, DNA, Genetics, Gene, Cell, Database, Telecommunications, Wireless, Materials science, Operating systemTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
2Total citation count in OpenAlex
- Citations by year (recent)
-
2025: 2Per-year citation counts (last 5 years)
- References (count)
-
35Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4385875706 |
|---|---|
| doi | https://doi.org/10.1101/2023.08.15.549386 |
| ids.doi | https://doi.org/10.1101/2023.08.15.549386 |
| ids.openalex | https://openalex.org/W4385875706 |
| fwci | 0.60501648 |
| type | preprint |
| title | High-throughput generic single-entity sequencing using droplet microfluidics |
| biblio.issue | |
| biblio.volume | |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T11048 |
| topics[0].field.id | https://openalex.org/fields/23 |
| topics[0].field.display_name | Environmental Science |
| topics[0].score | 0.9947999715805054 |
| topics[0].domain.id | https://openalex.org/domains/3 |
| topics[0].domain.display_name | Physical Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/2303 |
| topics[0].subfield.display_name | Ecology |
| topics[0].display_name | Bacteriophages and microbial interactions |
| topics[1].id | https://openalex.org/T11289 |
| topics[1].field.id | https://openalex.org/fields/13 |
| topics[1].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[1].score | 0.9864000082015991 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1312 |
| topics[1].subfield.display_name | Molecular Biology |
| topics[1].display_name | Single-cell and spatial transcriptomics |
| topics[2].id | https://openalex.org/T10015 |
| topics[2].field.id | https://openalex.org/fields/13 |
| topics[2].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[2].score | 0.9726999998092651 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1312 |
| topics[2].subfield.display_name | Molecular Biology |
| topics[2].display_name | Genomics and Phylogenetic Studies |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C177212765 |
| concepts[0].level | 2 |
| concepts[0].score | 0.7279012203216553 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q627335 |
| concepts[0].display_name | Workflow |
| concepts[1].id | https://openalex.org/C51679486 |
| concepts[1].level | 3 |
| concepts[1].score | 0.5917695164680481 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q380546 |
| concepts[1].display_name | DNA sequencing |
| concepts[2].id | https://openalex.org/C70721500 |
| concepts[2].level | 1 |
| concepts[2].score | 0.5809140205383301 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q177005 |
| concepts[2].display_name | Computational biology |
| concepts[3].id | https://openalex.org/C8673954 |
| concepts[3].level | 2 |
| concepts[3].score | 0.5204913020133972 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q138845 |
| concepts[3].display_name | Microfluidics |
| concepts[4].id | https://openalex.org/C187191949 |
| concepts[4].level | 2 |
| concepts[4].score | 0.4627220034599304 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q1138496 |
| concepts[4].display_name | Profiling (computer programming) |
| concepts[5].id | https://openalex.org/C157764524 |
| concepts[5].level | 3 |
| concepts[5].score | 0.46035003662109375 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q1383412 |
| concepts[5].display_name | Throughput |
| concepts[6].id | https://openalex.org/C86803240 |
| concepts[6].level | 0 |
| concepts[6].score | 0.4150891602039337 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[6].display_name | Biology |
| concepts[7].id | https://openalex.org/C2776950831 |
| concepts[7].level | 3 |
| concepts[7].score | 0.4144879877567291 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q15635432 |
| concepts[7].display_name | Single-cell analysis |
| concepts[8].id | https://openalex.org/C141231307 |
| concepts[8].level | 3 |
| concepts[8].score | 0.41379544138908386 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q7020 |
| concepts[8].display_name | Genome |
| concepts[9].id | https://openalex.org/C41008148 |
| concepts[9].level | 0 |
| concepts[9].score | 0.4013136029243469 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q21198 |
| concepts[9].display_name | Computer science |
| concepts[10].id | https://openalex.org/C171250308 |
| concepts[10].level | 1 |
| concepts[10].score | 0.28350546956062317 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q11468 |
| concepts[10].display_name | Nanotechnology |
| concepts[11].id | https://openalex.org/C552990157 |
| concepts[11].level | 2 |
| concepts[11].score | 0.18663722276687622 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q7430 |
| concepts[11].display_name | DNA |
| concepts[12].id | https://openalex.org/C54355233 |
| concepts[12].level | 1 |
| concepts[12].score | 0.16502097249031067 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[12].display_name | Genetics |
| concepts[13].id | https://openalex.org/C104317684 |
| concepts[13].level | 2 |
| concepts[13].score | 0.11572381854057312 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[13].display_name | Gene |
| concepts[14].id | https://openalex.org/C1491633281 |
| concepts[14].level | 2 |
| concepts[14].score | 0.10939076542854309 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q7868 |
| concepts[14].display_name | Cell |
| concepts[15].id | https://openalex.org/C77088390 |
| concepts[15].level | 1 |
| concepts[15].score | 0.0871688723564148 |
| concepts[15].wikidata | https://www.wikidata.org/wiki/Q8513 |
| concepts[15].display_name | Database |
| concepts[16].id | https://openalex.org/C76155785 |
| concepts[16].level | 1 |
| concepts[16].score | 0.0 |
| concepts[16].wikidata | https://www.wikidata.org/wiki/Q418 |
| concepts[16].display_name | Telecommunications |
| concepts[17].id | https://openalex.org/C555944384 |
| concepts[17].level | 2 |
| concepts[17].score | 0.0 |
| concepts[17].wikidata | https://www.wikidata.org/wiki/Q249 |
| concepts[17].display_name | Wireless |
| concepts[18].id | https://openalex.org/C192562407 |
| concepts[18].level | 0 |
| concepts[18].score | 0.0 |
| concepts[18].wikidata | https://www.wikidata.org/wiki/Q228736 |
| concepts[18].display_name | Materials science |
| concepts[19].id | https://openalex.org/C111919701 |
| concepts[19].level | 1 |
| concepts[19].score | 0.0 |
| concepts[19].wikidata | https://www.wikidata.org/wiki/Q9135 |
| concepts[19].display_name | Operating system |
| keywords[0].id | https://openalex.org/keywords/workflow |
| keywords[0].score | 0.7279012203216553 |
| keywords[0].display_name | Workflow |
| keywords[1].id | https://openalex.org/keywords/dna-sequencing |
| keywords[1].score | 0.5917695164680481 |
| keywords[1].display_name | DNA sequencing |
| keywords[2].id | https://openalex.org/keywords/computational-biology |
| keywords[2].score | 0.5809140205383301 |
| keywords[2].display_name | Computational biology |
| keywords[3].id | https://openalex.org/keywords/microfluidics |
| keywords[3].score | 0.5204913020133972 |
| keywords[3].display_name | Microfluidics |
| keywords[4].id | https://openalex.org/keywords/profiling |
| keywords[4].score | 0.4627220034599304 |
| keywords[4].display_name | Profiling (computer programming) |
| keywords[5].id | https://openalex.org/keywords/throughput |
| keywords[5].score | 0.46035003662109375 |
| keywords[5].display_name | Throughput |
| keywords[6].id | https://openalex.org/keywords/biology |
| keywords[6].score | 0.4150891602039337 |
| keywords[6].display_name | Biology |
| keywords[7].id | https://openalex.org/keywords/single-cell-analysis |
| keywords[7].score | 0.4144879877567291 |
| keywords[7].display_name | Single-cell analysis |
| keywords[8].id | https://openalex.org/keywords/genome |
| keywords[8].score | 0.41379544138908386 |
| keywords[8].display_name | Genome |
| keywords[9].id | https://openalex.org/keywords/computer-science |
| keywords[9].score | 0.4013136029243469 |
| keywords[9].display_name | Computer science |
| keywords[10].id | https://openalex.org/keywords/nanotechnology |
| keywords[10].score | 0.28350546956062317 |
| keywords[10].display_name | Nanotechnology |
| keywords[11].id | https://openalex.org/keywords/dna |
| keywords[11].score | 0.18663722276687622 |
| keywords[11].display_name | DNA |
| keywords[12].id | https://openalex.org/keywords/genetics |
| keywords[12].score | 0.16502097249031067 |
| keywords[12].display_name | Genetics |
| keywords[13].id | https://openalex.org/keywords/gene |
| keywords[13].score | 0.11572381854057312 |
| keywords[13].display_name | Gene |
| keywords[14].id | https://openalex.org/keywords/cell |
| keywords[14].score | 0.10939076542854309 |
| keywords[14].display_name | Cell |
| keywords[15].id | https://openalex.org/keywords/database |
| keywords[15].score | 0.0871688723564148 |
| keywords[15].display_name | Database |
| language | en |
| locations[0].id | doi:10.1101/2023.08.15.549386 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4306402567 |
| locations[0].source.issn | |
| locations[0].source.type | repository |
| locations[0].source.is_oa | False |
| locations[0].source.issn_l | |
| locations[0].source.is_core | False |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| locations[0].source.host_organization | https://openalex.org/I2750212522 |
| locations[0].source.host_organization_name | Cold Spring Harbor Laboratory |
| locations[0].source.host_organization_lineage | https://openalex.org/I2750212522 |
| locations[0].license | |
| locations[0].pdf_url | https://www.biorxiv.org/content/biorxiv/early/2023/08/15/2023.08.15.549386.full.pdf |
| locations[0].version | acceptedVersion |
| locations[0].raw_type | posted-content |
| locations[0].license_id | |
| locations[0].is_accepted | True |
| locations[0].is_published | False |
| locations[0].raw_source_name | |
| locations[0].landing_page_url | https://doi.org/10.1101/2023.08.15.549386 |
| indexed_in | crossref |
| authorships[0].author.id | https://openalex.org/A5043327776 |
| authorships[0].author.orcid | https://orcid.org/0000-0002-2635-6932 |
| authorships[0].author.display_name | Guoping Wang |
| authorships[0].countries | HK |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I177725633 |
| authorships[0].affiliations[0].raw_affiliation_string | Institute of Digestive Diseases and Department of Medicine and Therapeutics, State Key Laboratory of Digestive Disease, Li Ka Shing Institute of Health Sciences, The Chinese University of Hong Kong, Hong Kong SAR, China |
| authorships[0].institutions[0].id | https://openalex.org/I177725633 |
| authorships[0].institutions[0].ror | https://ror.org/00t33hh48 |
| authorships[0].institutions[0].type | education |
| authorships[0].institutions[0].lineage | https://openalex.org/I177725633 |
| authorships[0].institutions[0].country_code | HK |
| authorships[0].institutions[0].display_name | Chinese University of Hong Kong |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Guoping Wang |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Institute of Digestive Diseases and Department of Medicine and Therapeutics, State Key Laboratory of Digestive Disease, Li Ka Shing Institute of Health Sciences, The Chinese University of Hong Kong, Hong Kong SAR, China |
| authorships[1].author.id | https://openalex.org/A5000059613 |
| authorships[1].author.orcid | https://orcid.org/0000-0001-7559-5131 |
| authorships[1].author.display_name | Liuyang Zhao |
| authorships[1].countries | HK |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I177725633 |
| authorships[1].affiliations[0].raw_affiliation_string | Institute of Digestive Diseases and Department of Medicine and Therapeutics, State Key Laboratory of Digestive Disease, Li Ka Shing Institute of Health Sciences, The Chinese University of Hong Kong, Hong Kong SAR, China |
| authorships[1].institutions[0].id | https://openalex.org/I177725633 |
| authorships[1].institutions[0].ror | https://ror.org/00t33hh48 |
| authorships[1].institutions[0].type | education |
| authorships[1].institutions[0].lineage | https://openalex.org/I177725633 |
| authorships[1].institutions[0].country_code | HK |
| authorships[1].institutions[0].display_name | Chinese University of Hong Kong |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Liuyang Zhao |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | Institute of Digestive Diseases and Department of Medicine and Therapeutics, State Key Laboratory of Digestive Disease, Li Ka Shing Institute of Health Sciences, The Chinese University of Hong Kong, Hong Kong SAR, China |
| authorships[2].author.id | https://openalex.org/A5101783298 |
| authorships[2].author.orcid | https://orcid.org/0009-0004-5886-2487 |
| authorships[2].author.display_name | Yu Shi |
| authorships[2].countries | HK |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I177725633 |
| authorships[2].affiliations[0].raw_affiliation_string | Institute of Digestive Diseases and Department of Medicine and Therapeutics, State Key Laboratory of Digestive Disease, Li Ka Shing Institute of Health Sciences, The Chinese University of Hong Kong, Hong Kong SAR, China |
| authorships[2].institutions[0].id | https://openalex.org/I177725633 |
| authorships[2].institutions[0].ror | https://ror.org/00t33hh48 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I177725633 |
| authorships[2].institutions[0].country_code | HK |
| authorships[2].institutions[0].display_name | Chinese University of Hong Kong |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Yu Shi |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | Institute of Digestive Diseases and Department of Medicine and Therapeutics, State Key Laboratory of Digestive Disease, Li Ka Shing Institute of Health Sciences, The Chinese University of Hong Kong, Hong Kong SAR, China |
| authorships[3].author.id | https://openalex.org/A5008040394 |
| authorships[3].author.orcid | https://orcid.org/0000-0002-4641-0289 |
| authorships[3].author.display_name | Fuyang Qu |
| authorships[3].countries | HK |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I177725633 |
| authorships[3].affiliations[0].raw_affiliation_string | Department of Biomedical Engineering, The Chinese University of Hong Kong, Hong Kong SAR, China |
| authorships[3].institutions[0].id | https://openalex.org/I177725633 |
| authorships[3].institutions[0].ror | https://ror.org/00t33hh48 |
| authorships[3].institutions[0].type | education |
| authorships[3].institutions[0].lineage | https://openalex.org/I177725633 |
| authorships[3].institutions[0].country_code | HK |
| authorships[3].institutions[0].display_name | Chinese University of Hong Kong |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Fuyang Qu |
| authorships[3].is_corresponding | False |
| authorships[3].raw_affiliation_strings | Department of Biomedical Engineering, The Chinese University of Hong Kong, Hong Kong SAR, China |
| authorships[4].author.id | https://openalex.org/A5073940603 |
| authorships[4].author.orcid | https://orcid.org/0000-0002-7740-6696 |
| authorships[4].author.display_name | Yanqiang Ding |
| authorships[4].countries | HK |
| authorships[4].affiliations[0].institution_ids | https://openalex.org/I177725633 |
| authorships[4].affiliations[0].raw_affiliation_string | Institute of Digestive Diseases and Department of Medicine and Therapeutics, State Key Laboratory of Digestive Disease, Li Ka Shing Institute of Health Sciences, The Chinese University of Hong Kong, Hong Kong SAR, China |
| authorships[4].institutions[0].id | https://openalex.org/I177725633 |
| authorships[4].institutions[0].ror | https://ror.org/00t33hh48 |
| authorships[4].institutions[0].type | education |
| authorships[4].institutions[0].lineage | https://openalex.org/I177725633 |
| authorships[4].institutions[0].country_code | HK |
| authorships[4].institutions[0].display_name | Chinese University of Hong Kong |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Yanqiang Ding |
| authorships[4].is_corresponding | False |
| authorships[4].raw_affiliation_strings | Institute of Digestive Diseases and Department of Medicine and Therapeutics, State Key Laboratory of Digestive Disease, Li Ka Shing Institute of Health Sciences, The Chinese University of Hong Kong, Hong Kong SAR, China |
| authorships[5].author.id | https://openalex.org/A5100683895 |
| authorships[5].author.orcid | https://orcid.org/0000-0002-1650-0371 |
| authorships[5].author.display_name | Weixin Liu |
| authorships[5].countries | HK |
| authorships[5].affiliations[0].institution_ids | https://openalex.org/I177725633 |
| authorships[5].affiliations[0].raw_affiliation_string | Institute of Digestive Diseases and Department of Medicine and Therapeutics, State Key Laboratory of Digestive Disease, Li Ka Shing Institute of Health Sciences, The Chinese University of Hong Kong, Hong Kong SAR, China |
| authorships[5].institutions[0].id | https://openalex.org/I177725633 |
| authorships[5].institutions[0].ror | https://ror.org/00t33hh48 |
| authorships[5].institutions[0].type | education |
| authorships[5].institutions[0].lineage | https://openalex.org/I177725633 |
| authorships[5].institutions[0].country_code | HK |
| authorships[5].institutions[0].display_name | Chinese University of Hong Kong |
| authorships[5].author_position | middle |
| authorships[5].raw_author_name | Weixin Liu |
| authorships[5].is_corresponding | False |
| authorships[5].raw_affiliation_strings | Institute of Digestive Diseases and Department of Medicine and Therapeutics, State Key Laboratory of Digestive Disease, Li Ka Shing Institute of Health Sciences, The Chinese University of Hong Kong, Hong Kong SAR, China |
| authorships[6].author.id | https://openalex.org/A5103006561 |
| authorships[6].author.orcid | https://orcid.org/0009-0001-3437-468X |
| authorships[6].author.display_name | Changan Liu |
| authorships[6].countries | HK |
| authorships[6].affiliations[0].institution_ids | https://openalex.org/I177725633 |
| authorships[6].affiliations[0].raw_affiliation_string | Institute of Digestive Diseases and Department of Medicine and Therapeutics, State Key Laboratory of Digestive Disease, Li Ka Shing Institute of Health Sciences, The Chinese University of Hong Kong, Hong Kong SAR, China |
| authorships[6].institutions[0].id | https://openalex.org/I177725633 |
| authorships[6].institutions[0].ror | https://ror.org/00t33hh48 |
| authorships[6].institutions[0].type | education |
| authorships[6].institutions[0].lineage | https://openalex.org/I177725633 |
| authorships[6].institutions[0].country_code | HK |
| authorships[6].institutions[0].display_name | Chinese University of Hong Kong |
| authorships[6].author_position | middle |
| authorships[6].raw_author_name | Changan Liu |
| authorships[6].is_corresponding | False |
| authorships[6].raw_affiliation_strings | Institute of Digestive Diseases and Department of Medicine and Therapeutics, State Key Laboratory of Digestive Disease, Li Ka Shing Institute of Health Sciences, The Chinese University of Hong Kong, Hong Kong SAR, China |
| authorships[7].author.id | https://openalex.org/A5102812675 |
| authorships[7].author.orcid | https://orcid.org/0009-0000-4677-4923 |
| authorships[7].author.display_name | Gang Luo |
| authorships[7].countries | HK |
| authorships[7].affiliations[0].institution_ids | https://openalex.org/I177725633 |
| authorships[7].affiliations[0].raw_affiliation_string | Department of Electronic Engineering, The Chinese University of Hong Kong, Hong Kong SAR, China |
| authorships[7].institutions[0].id | https://openalex.org/I177725633 |
| authorships[7].institutions[0].ror | https://ror.org/00t33hh48 |
| authorships[7].institutions[0].type | education |
| authorships[7].institutions[0].lineage | https://openalex.org/I177725633 |
| authorships[7].institutions[0].country_code | HK |
| authorships[7].institutions[0].display_name | Chinese University of Hong Kong |
| authorships[7].author_position | middle |
| authorships[7].raw_author_name | Gang Luo |
| authorships[7].is_corresponding | False |
| authorships[7].raw_affiliation_strings | Department of Electronic Engineering, The Chinese University of Hong Kong, Hong Kong SAR, China |
| authorships[8].author.id | https://openalex.org/A5032046795 |
| authorships[8].author.orcid | https://orcid.org/0000-0002-9309-6149 |
| authorships[8].author.display_name | Meiyi Li |
| authorships[8].countries | HK |
| authorships[8].affiliations[0].institution_ids | https://openalex.org/I177725633 |
| authorships[8].affiliations[0].raw_affiliation_string | Institute of Digestive Diseases and Department of Medicine and Therapeutics, State Key Laboratory of Digestive Disease, Li Ka Shing Institute of Health Sciences, The Chinese University of Hong Kong, Hong Kong SAR, China |
| authorships[8].institutions[0].id | https://openalex.org/I177725633 |
| authorships[8].institutions[0].ror | https://ror.org/00t33hh48 |
| authorships[8].institutions[0].type | education |
| authorships[8].institutions[0].lineage | https://openalex.org/I177725633 |
| authorships[8].institutions[0].country_code | HK |
| authorships[8].institutions[0].display_name | Chinese University of Hong Kong |
| authorships[8].author_position | middle |
| authorships[8].raw_author_name | Meiyi Li |
| authorships[8].is_corresponding | False |
| authorships[8].raw_affiliation_strings | Institute of Digestive Diseases and Department of Medicine and Therapeutics, State Key Laboratory of Digestive Disease, Li Ka Shing Institute of Health Sciences, The Chinese University of Hong Kong, Hong Kong SAR, China |
| authorships[9].author.id | https://openalex.org/A5103241807 |
| authorships[9].author.orcid | https://orcid.org/0000-0001-8377-0518 |
| authorships[9].author.display_name | Xiaowu Bai |
| authorships[9].countries | HK |
| authorships[9].affiliations[0].institution_ids | https://openalex.org/I177725633 |
| authorships[9].affiliations[0].raw_affiliation_string | Institute of Digestive Diseases and Department of Medicine and Therapeutics, State Key Laboratory of Digestive Disease, Li Ka Shing Institute of Health Sciences, The Chinese University of Hong Kong, Hong Kong SAR, China |
| authorships[9].institutions[0].id | https://openalex.org/I177725633 |
| authorships[9].institutions[0].ror | https://ror.org/00t33hh48 |
| authorships[9].institutions[0].type | education |
| authorships[9].institutions[0].lineage | https://openalex.org/I177725633 |
| authorships[9].institutions[0].country_code | HK |
| authorships[9].institutions[0].display_name | Chinese University of Hong Kong |
| authorships[9].author_position | middle |
| authorships[9].raw_author_name | Xiaowu Bai |
| authorships[9].is_corresponding | False |
| authorships[9].raw_affiliation_strings | Institute of Digestive Diseases and Department of Medicine and Therapeutics, State Key Laboratory of Digestive Disease, Li Ka Shing Institute of Health Sciences, The Chinese University of Hong Kong, Hong Kong SAR, China |
| authorships[10].author.id | https://openalex.org/A5065818596 |
| authorships[10].author.orcid | https://orcid.org/0000-0002-7571-816X |
| authorships[10].author.display_name | Luoquan Li |
| authorships[10].countries | HK |
| authorships[10].affiliations[0].institution_ids | https://openalex.org/I177725633 |
| authorships[10].affiliations[0].raw_affiliation_string | Department of Biomedical Engineering, The Chinese University of Hong Kong, Hong Kong SAR, China |
| authorships[10].institutions[0].id | https://openalex.org/I177725633 |
| authorships[10].institutions[0].ror | https://ror.org/00t33hh48 |
| authorships[10].institutions[0].type | education |
| authorships[10].institutions[0].lineage | https://openalex.org/I177725633 |
| authorships[10].institutions[0].country_code | HK |
| authorships[10].institutions[0].display_name | Chinese University of Hong Kong |
| authorships[10].author_position | middle |
| authorships[10].raw_author_name | Luoquan Li |
| authorships[10].is_corresponding | False |
| authorships[10].raw_affiliation_strings | Department of Biomedical Engineering, The Chinese University of Hong Kong, Hong Kong SAR, China |
| authorships[11].author.id | https://openalex.org/A5026619357 |
| authorships[11].author.orcid | https://orcid.org/0000-0002-2052-9724 |
| authorships[11].author.display_name | Yi‐Ping Ho |
| authorships[11].countries | HK |
| authorships[11].affiliations[0].institution_ids | https://openalex.org/I177725633 |
| authorships[11].affiliations[0].raw_affiliation_string | Department of Biomedical Engineering, The Chinese University of Hong Kong, Hong Kong SAR, China |
| authorships[11].affiliations[1].institution_ids | https://openalex.org/I177725633 |
| authorships[11].affiliations[1].raw_affiliation_string | The Ministry of Education Key Laboratory of Regeneration Medicine, The Chinese University of Hong Kong, Hong Kong SAR, China |
| authorships[11].affiliations[2].institution_ids | https://openalex.org/I177725633 |
| authorships[11].affiliations[2].raw_affiliation_string | Hong Kong Branch of CAS Center for Excellence in Animal Evolution and Genetics, The Chinese University of Hong Kong, Hong Kong SAR, China |
| authorships[11].institutions[0].id | https://openalex.org/I177725633 |
| authorships[11].institutions[0].ror | https://ror.org/00t33hh48 |
| authorships[11].institutions[0].type | education |
| authorships[11].institutions[0].lineage | https://openalex.org/I177725633 |
| authorships[11].institutions[0].country_code | HK |
| authorships[11].institutions[0].display_name | Chinese University of Hong Kong |
| authorships[11].author_position | middle |
| authorships[11].raw_author_name | Yi-Ping Ho |
| authorships[11].is_corresponding | True |
| authorships[11].raw_affiliation_strings | Department of Biomedical Engineering, The Chinese University of Hong Kong, Hong Kong SAR, China, Hong Kong Branch of CAS Center for Excellence in Animal Evolution and Genetics, The Chinese University of Hong Kong, Hong Kong SAR, China, The Ministry of Education Key Laboratory of Regeneration Medicine, The Chinese University of Hong Kong, Hong Kong SAR, China |
| authorships[12].author.id | https://openalex.org/A5100614702 |
| authorships[12].author.orcid | https://orcid.org/0000-0001-5008-2153 |
| authorships[12].author.display_name | Jun Yu |
| authorships[12].countries | HK |
| authorships[12].affiliations[0].institution_ids | https://openalex.org/I177725633 |
| authorships[12].affiliations[0].raw_affiliation_string | Institute of Digestive Diseases and Department of Medicine and Therapeutics, State Key Laboratory of Digestive Disease, Li Ka Shing Institute of Health Sciences, The Chinese University of Hong Kong, Hong Kong SAR, China |
| authorships[12].institutions[0].id | https://openalex.org/I177725633 |
| authorships[12].institutions[0].ror | https://ror.org/00t33hh48 |
| authorships[12].institutions[0].type | education |
| authorships[12].institutions[0].lineage | https://openalex.org/I177725633 |
| authorships[12].institutions[0].country_code | HK |
| authorships[12].institutions[0].display_name | Chinese University of Hong Kong |
| authorships[12].author_position | last |
| authorships[12].raw_author_name | Jun Yu |
| authorships[12].is_corresponding | True |
| authorships[12].raw_affiliation_strings | Institute of Digestive Diseases and Department of Medicine and Therapeutics, State Key Laboratory of Digestive Disease, Li Ka Shing Institute of Health Sciences, The Chinese University of Hong Kong, Hong Kong SAR, China |
| has_content.pdf | True |
| has_content.grobid_xml | True |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://www.biorxiv.org/content/biorxiv/early/2023/08/15/2023.08.15.549386.full.pdf |
| open_access.oa_status | green |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | High-throughput generic single-entity sequencing using droplet microfluidics |
| has_fulltext | True |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T11048 |
| primary_topic.field.id | https://openalex.org/fields/23 |
| primary_topic.field.display_name | Environmental Science |
| primary_topic.score | 0.9947999715805054 |
| primary_topic.domain.id | https://openalex.org/domains/3 |
| primary_topic.domain.display_name | Physical Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/2303 |
| primary_topic.subfield.display_name | Ecology |
| primary_topic.display_name | Bacteriophages and microbial interactions |
| related_works | https://openalex.org/W2348361596, https://openalex.org/W2752303597, https://openalex.org/W2130706252, https://openalex.org/W2255506873, https://openalex.org/W3209838000, https://openalex.org/W2126573584, https://openalex.org/W1558071211, https://openalex.org/W4377098284, https://openalex.org/W2099851046, https://openalex.org/W3035610982 |
| cited_by_count | 2 |
| counts_by_year[0].year | 2025 |
| counts_by_year[0].cited_by_count | 2 |
| locations_count | 1 |
| best_oa_location.id | doi:10.1101/2023.08.15.549386 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4306402567 |
| best_oa_location.source.issn | |
| best_oa_location.source.type | repository |
| best_oa_location.source.is_oa | False |
| best_oa_location.source.issn_l | |
| best_oa_location.source.is_core | False |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| best_oa_location.source.host_organization | https://openalex.org/I2750212522 |
| best_oa_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| best_oa_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| best_oa_location.license | |
| best_oa_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2023/08/15/2023.08.15.549386.full.pdf |
| best_oa_location.version | acceptedVersion |
| best_oa_location.raw_type | posted-content |
| best_oa_location.license_id | |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | False |
| best_oa_location.raw_source_name | |
| best_oa_location.landing_page_url | https://doi.org/10.1101/2023.08.15.549386 |
| primary_location.id | doi:10.1101/2023.08.15.549386 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4306402567 |
| primary_location.source.issn | |
| primary_location.source.type | repository |
| primary_location.source.is_oa | False |
| primary_location.source.issn_l | |
| primary_location.source.is_core | False |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| primary_location.source.host_organization | https://openalex.org/I2750212522 |
| primary_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| primary_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| primary_location.license | |
| primary_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2023/08/15/2023.08.15.549386.full.pdf |
| primary_location.version | acceptedVersion |
| primary_location.raw_type | posted-content |
| primary_location.license_id | |
| primary_location.is_accepted | True |
| primary_location.is_published | False |
| primary_location.raw_source_name | |
| primary_location.landing_page_url | https://doi.org/10.1101/2023.08.15.549386 |
| publication_date | 2023-08-15 |
| publication_year | 2023 |
| referenced_works | https://openalex.org/W3013151148, https://openalex.org/W4286510420, https://openalex.org/W2765135073, https://openalex.org/W2638784546, https://openalex.org/W4281716380, https://openalex.org/W2620147451, https://openalex.org/W2102212449, https://openalex.org/W2135937351, https://openalex.org/W2029583307, https://openalex.org/W1975631835, https://openalex.org/W2559325775, https://openalex.org/W2517568995, https://openalex.org/W2809646241, https://openalex.org/W4297990241, https://openalex.org/W3204872827, https://openalex.org/W2742647527, https://openalex.org/W2995649573, https://openalex.org/W3092429684, https://openalex.org/W2792384873, https://openalex.org/W2169899471, https://openalex.org/W4285097835, https://openalex.org/W2940459311, https://openalex.org/W2979358136, https://openalex.org/W4313656753, https://openalex.org/W4288096194, https://openalex.org/W1966912927, https://openalex.org/W3129897473, https://openalex.org/W4319293985, https://openalex.org/W2125826054, https://openalex.org/W3042305844, https://openalex.org/W1939004772, https://openalex.org/W1974288263, https://openalex.org/W2245444176, https://openalex.org/W4300861479, https://openalex.org/W3104596207 |
| referenced_works_count | 35 |
| abstract_inverted_index., | 43, 73, 90, 95 |
| abstract_inverted_index.. | 101 |
| abstract_inverted_index.3 | 72 |
| abstract_inverted_index.4 | 89 |
| abstract_inverted_index.5 | 94 |
| abstract_inverted_index.6 | 100 |
| abstract_inverted_index.a | 12, 167, 223 |
| abstract_inverted_index.By | 162 |
| abstract_inverted_index.In | 215 |
| abstract_inverted_index.an | 125 |
| abstract_inverted_index.as | 58 |
| abstract_inverted_index.by | 10, 124, 170 |
| abstract_inverted_index.in | 47, 132, 145, 234 |
| abstract_inverted_index.is | 21, 122 |
| abstract_inverted_index.of | 7, 50, 66, 79, 139, 176, 190 |
| abstract_inverted_index.on | 33, 226 |
| abstract_inverted_index.to | 63, 229, 237 |
| abstract_inverted_index.we | 103, 164 |
| abstract_inverted_index.(1) | 136 |
| abstract_inverted_index.(2) | 142 |
| abstract_inverted_index.(3) | 153 |
| abstract_inverted_index.(4) | 158 |
| abstract_inverted_index.1,2 | 42 |
| abstract_inverted_index.DNA | 71 |
| abstract_inverted_index.Due | 62 |
| abstract_inverted_index.and | 39, 60, 68, 96, 118, 121, 149, 178, 185, 194, 208, 212 |
| abstract_inverted_index.for | 23, 76, 197, 244 |
| abstract_inverted_index.gut | 193 |
| abstract_inverted_index.has | 3 |
| abstract_inverted_index.low | 87 |
| abstract_inverted_index.new | 220 |
| abstract_inverted_index.our | 5, 202 |
| abstract_inverted_index.the | 16, 27, 48, 64, 77, 105, 129, 191, 198, 217, 231 |
| abstract_inverted_index.cell | 37 |
| abstract_inverted_index.dark | 210 |
| abstract_inverted_index.from | 52, 180 |
| abstract_inverted_index.gaps | 46 |
| abstract_inverted_index.have | 165 |
| abstract_inverted_index.high | 116, 119 |
| abstract_inverted_index.hold | 241 |
| abstract_inverted_index.over | 15 |
| abstract_inverted_index.past | 17 |
| abstract_inverted_index.situ | 146 |
| abstract_inverted_index.such | 57 |
| abstract_inverted_index.with | 86 |
| abstract_inverted_index.ample | 114 |
| abstract_inverted_index.based | 225 |
| abstract_inverted_index.cells | 35 |
| abstract_inverted_index.dsDNA | 177 |
| abstract_inverted_index.first | 199 |
| abstract_inverted_index.human | 192 |
| abstract_inverted_index.lysis | 92 |
| abstract_inverted_index.omics | 51 |
| abstract_inverted_index.other | 53 |
| abstract_inverted_index.ssDNA | 179 |
| abstract_inverted_index.study | 49 |
| abstract_inverted_index.their | 40, 70 |
| abstract_inverted_index.time. | 200 |
| abstract_inverted_index.viral | 207 |
| abstract_inverted_index.virus | 182 |
| abstract_inverted_index.walls | 38 |
| abstract_inverted_index.which | 112, 240 |
| abstract_inverted_index.whole | 150 |
| abstract_inverted_index.Entity | 108 |
| abstract_inverted_index.Single | 107 |
| abstract_inverted_index.boasts | 113 |
| abstract_inverted_index.cells. | 250 |
| abstract_inverted_index.genome | 151 |
| abstract_inverted_index.highly | 97 |
| abstract_inverted_index.marine | 195 |
| abstract_inverted_index.matter | 211 |
| abstract_inverted_index.offers | 222 |
| abstract_inverted_index.remain | 83 |
| abstract_inverted_index.sample | 147 |
| abstract_inverted_index.single | 54, 133, 181, 186 |
| abstract_inverted_index.tackle | 230 |
| abstract_inverted_index.Generic | 106 |
| abstract_inverted_index.Herein, | 102 |
| abstract_inverted_index.Summary | 0 |
| abstract_inverted_index.diverse | 245 |
| abstract_inverted_index.droplet | 227 |
| abstract_inverted_index.enabled | 123 |
| abstract_inverted_index.focuses | 32 |
| abstract_inverted_index.generic | 80, 238 |
| abstract_inverted_index.genomes | 99 |
| abstract_inverted_index.immense | 242 |
| abstract_inverted_index.in-drop | 155 |
| abstract_inverted_index.leaving | 44 |
| abstract_inverted_index.library | 156 |
| abstract_inverted_index.massive | 140 |
| abstract_inverted_index.present | 104 |
| abstract_inverted_index.promise | 243 |
| abstract_inverted_index.toolbox | 224 |
| abstract_inverted_index.various | 24 |
| abstract_inverted_index.without | 36 |
| abstract_inverted_index.(SB-seq) | 189 |
| abstract_inverted_index.(SV-seq) | 184 |
| abstract_inverted_index.Although | 19 |
| abstract_inverted_index.GSE-Seq, | 163 |
| abstract_inverted_index.Notably, | 201 |
| abstract_inverted_index.achieved | 166 |
| abstract_inverted_index.analysis | 203 |
| abstract_inverted_index.bacteria | 59, 187 |
| abstract_inverted_index.cellular | 8 |
| abstract_inverted_index.critical | 130 |
| abstract_inverted_index.decades. | 18 |
| abstract_inverted_index.enabling | 171 |
| abstract_inverted_index.entities | 56, 82, 134 |
| abstract_inverted_index.one-step | 137 |
| abstract_inverted_index.platform | 30, 110 |
| abstract_inverted_index.sediment | 196 |
| abstract_inverted_index.summary, | 216 |
| abstract_inverted_index.viruses. | 61 |
| abstract_inverted_index.workflow | 221 |
| abstract_inverted_index.acquiring | 69 |
| abstract_inverted_index.bacterial | 209 |
| abstract_inverted_index.barcodes, | 141 |
| abstract_inverted_index.coverage, | 120 |
| abstract_inverted_index.currently | 28 |
| abstract_inverted_index.entities, | 247 |
| abstract_inverted_index.essential | 22 |
| abstract_inverted_index.isolating | 67 |
| abstract_inverted_index.long-read | 160, 173 |
| abstract_inverted_index.milestone | 169 |
| abstract_inverted_index.presented | 218 |
| abstract_inverted_index.profiling | 175, 236 |
| abstract_inverted_index.providing | 11 |
| abstract_inverted_index.uncovered | 204 |
| abstract_inverted_index.workflow, | 127 |
| abstract_inverted_index.(GSE-Seq), | 111 |
| abstract_inverted_index.Sequencing | 109 |
| abstract_inverted_index.addressing | 128 |
| abstract_inverted_index.biological | 25, 55, 81, 246 |
| abstract_inverted_index.challenges | 131, 233 |
| abstract_inverted_index.compatible | 159 |
| abstract_inverted_index.degradable | 143 |
| abstract_inverted_index.difficulty | 65 |
| abstract_inverted_index.efficiency | 93 |
| abstract_inverted_index.especially | 248 |
| abstract_inverted_index.eukaryotic | 34 |
| abstract_inverted_index.fragmented | 98 |
| abstract_inverted_index.innovative | 126 |
| abstract_inverted_index.integrated | 154 |
| abstract_inverted_index.overlooked | 206 |
| abstract_inverted_index.persistent | 232 |
| abstract_inverted_index.phage-host | 213 |
| abstract_inverted_index.previously | 205 |
| abstract_inverted_index.processing | 148 |
| abstract_inverted_index.sequencing | 2, 183, 188 |
| abstract_inverted_index.throughput | 88 |
| abstract_inverted_index.Single-cell | 1 |
| abstract_inverted_index.compromised | 91 |
| abstract_inverted_index.micro-level | 13 |
| abstract_inverted_index.perspective | 14 |
| abstract_inverted_index.sequencing. | 161 |
| abstract_inverted_index.sequencing: | 135 |
| abstract_inverted_index.significant | 45, 168 |
| abstract_inverted_index.throughput, | 117 |
| abstract_inverted_index.communities, | 26 |
| abstract_inverted_index.conceptually | 219 |
| abstract_inverted_index.constrained, | 85 |
| abstract_inverted_index.contemporary | 74 |
| abstract_inverted_index.demonstrated | 29 |
| abstract_inverted_index.hard-to-lyse | 249 |
| abstract_inverted_index.preparation, | 157 |
| abstract_inverted_index.versatility, | 115 |
| abstract_inverted_index.applications, | 239 |
| abstract_inverted_index.conspicuously | 84 |
| abstract_inverted_index.heterogeneity | 9, 20 |
| abstract_inverted_index.interactions. | 214 |
| abstract_inverted_index.manufacturing | 138 |
| abstract_inverted_index.methodologies | 75 |
| abstract_inverted_index.microfluidics | 228 |
| abstract_inverted_index.predominantly | 31 |
| abstract_inverted_index.single-entity | 174 |
| abstract_inverted_index.understanding | 6 |
| abstract_inverted_index.amplification, | 152 |
| abstract_inverted_index.hydrogel-based | 144 |
| abstract_inverted_index.revolutionized | 4 |
| abstract_inverted_index.high-throughput | 235 |
| abstract_inverted_index.transcriptomics | 41 |
| abstract_inverted_index.characterization | 78 |
| abstract_inverted_index.high-throughput, | 172 |
| cited_by_percentile_year.max | 97 |
| cited_by_percentile_year.min | 95 |
| corresponding_author_ids | https://openalex.org/A5100614702, https://openalex.org/A5026619357 |
| countries_distinct_count | 1 |
| institutions_distinct_count | 13 |
| corresponding_institution_ids | https://openalex.org/I177725633 |
| sustainable_development_goals[0].id | https://metadata.un.org/sdg/14 |
| sustainable_development_goals[0].score | 0.8199999928474426 |
| sustainable_development_goals[0].display_name | Life below water |
| citation_normalized_percentile.value | 0.70957042 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |