Highly sensitive molecular assay based on Identical Multi-Repeat Sequence (IMRS) algorithm for the detection of Trichomonas vaginalis infection Article Swipe
 YOU?
·
· 2025
· Open Access
·
· DOI: https://doi.org/10.1371/journal.pone.0317958
YOU?
·
· 2025
· Open Access
·
· DOI: https://doi.org/10.1371/journal.pone.0317958
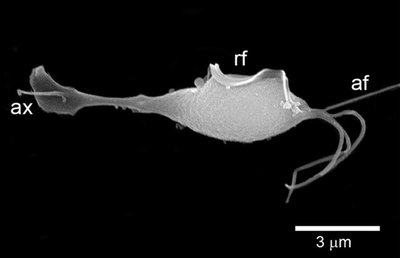
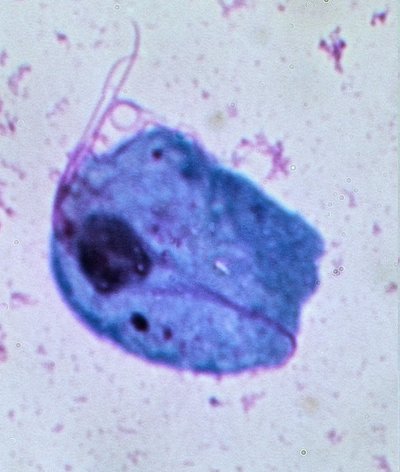
Introduction Annually, approximately 174 million people globally are affected by Trichomonas vaginalis ( T . vaginalis ) infection. Half of these infections occur in resource-limited regions. Untreated T . vaginalis infections are associated with complications such as pelvic inflammatory disease and adverse pregnancy outcomes mostly seen in women. In resource-limited regions, the World Health Organization (WHO) advocates for syndromic case management. However, this can lead to unnecessary treatment. Accurate diagnosis of T . vaginalis is required for effective and prompt treatment. Molecular tests such as Polymerase Chain Reaction (PCR) have the advantage of having a short turn-around time and allow the use of non-invasive specimens such as urine and vaginal swabs. However, these diagnostic techniques have numerous disadvantages such as high infrastructure costs, false negative and positive results, and interstrain variation among others. This study aimed to evaluate the use of identical multi-repeat sequences (IMRS) as amplification primers for developing ultrasensitive diagnostic for T . vaginalis . Methods We used genome-mining approaches based on identical multi-repeat sequences (IMRS) algorithm to identify sequences distributed on the T . vaginalis genome to design a primer pair that targets a total of 69 repeat sequences. Genomic T . vaginalis DNA was diluted from 5.8×10 2 to 5.8×10 −4 genome copies/ μ l and used as a template in the IMRS-based amplification assay. For performance comparison, 18S rRNA PCR assay was employed. Results The T . vaginalis -IMRS primers offered a higher test sensitivity of 0.03 fg/ μ L compared to the 18S rRNA PCR (0.714 pg/ μ L). The limit of detection for the Isothermal assay was 0.58 genome copies/mL. Using real-time PCR, the analytical sensitivity of the T . vaginalis -IMRS primers was <0.01 pg/μL, equivalent to less than one genome copy/μL. Conclusion De novo genome mining of T . vaginalis IMRS as amplification primers serves as a platform for developing ultrasensitive diagnostics for Trichomoniasis and a wide range of infectious pathogens.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.1371/journal.pone.0317958
- OA Status
- gold
- References
- 34
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4407251480
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4407251480Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1371/journal.pone.0317958Digital Object Identifier
- Title
-
Highly sensitive molecular assay based on Identical Multi-Repeat Sequence (IMRS) algorithm for the detection of Trichomonas vaginalis infectionWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2025Year of publication
- Publication date
-
2025-02-07Full publication date if available
- Authors
-
Clement Likhovole, Shwetha Kamath, Bernard N. Kanoi, Racheal Kimani, Bernard Oduor, Hussein M. Abkallo, Michael Maina, Harrison Waweru, Moses Kamita, Nicole Pamme, Joshua Dupaty, Catherine M. Klapperich, Srinivasa Raju Lolabattu, Jesse GitakaList of authors in order
- Landing page
-
https://doi.org/10.1371/journal.pone.0317958Publisher landing page
- Open access
-
YesWhether a free full text is available
- OA status
-
goldOpen access status per OpenAlex
- OA URL
-
https://doi.org/10.1371/journal.pone.0317958Direct OA link when available
- Concepts
-
Trichomonas vaginalis, Trichomonas Vaginitis, Algorithm, Sequence (biology), Biology, Virology, Microbiology, Genetics, Computer scienceTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
0Total citation count in OpenAlex
- References (count)
-
34Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4407251480 |
|---|---|
| doi | https://doi.org/10.1371/journal.pone.0317958 |
| ids.doi | https://doi.org/10.1371/journal.pone.0317958 |
| ids.pmid | https://pubmed.ncbi.nlm.nih.gov/39919090 |
| ids.openalex | https://openalex.org/W4407251480 |
| fwci | 0.0 |
| mesh[0].qualifier_ui | Q000235 |
| mesh[0].descriptor_ui | D014246 |
| mesh[0].is_major_topic | True |
| mesh[0].qualifier_name | genetics |
| mesh[0].descriptor_name | Trichomonas vaginalis |
| mesh[1].qualifier_ui | Q000302 |
| mesh[1].descriptor_ui | D014246 |
| mesh[1].is_major_topic | True |
| mesh[1].qualifier_name | isolation & purification |
| mesh[1].descriptor_name | Trichomonas vaginalis |
| mesh[2].qualifier_ui | |
| mesh[2].descriptor_ui | D006801 |
| mesh[2].is_major_topic | False |
| mesh[2].qualifier_name | |
| mesh[2].descriptor_name | Humans |
| mesh[3].qualifier_ui | |
| mesh[3].descriptor_ui | D000465 |
| mesh[3].is_major_topic | True |
| mesh[3].qualifier_name | |
| mesh[3].descriptor_name | Algorithms |
| mesh[4].qualifier_ui | |
| mesh[4].descriptor_ui | D005260 |
| mesh[4].is_major_topic | False |
| mesh[4].qualifier_name | |
| mesh[4].descriptor_name | Female |
| mesh[5].qualifier_ui | |
| mesh[5].descriptor_ui | D012680 |
| mesh[5].is_major_topic | False |
| mesh[5].qualifier_name | |
| mesh[5].descriptor_name | Sensitivity and Specificity |
| mesh[6].qualifier_ui | Q000175 |
| mesh[6].descriptor_ui | D014247 |
| mesh[6].is_major_topic | False |
| mesh[6].qualifier_name | diagnosis |
| mesh[6].descriptor_name | Trichomonas Vaginitis |
| mesh[7].qualifier_ui | Q000175 |
| mesh[7].descriptor_ui | D014245 |
| mesh[7].is_major_topic | False |
| mesh[7].qualifier_name | diagnosis |
| mesh[7].descriptor_name | Trichomonas Infections |
| mesh[8].qualifier_ui | Q000469 |
| mesh[8].descriptor_ui | D014245 |
| mesh[8].is_major_topic | False |
| mesh[8].qualifier_name | parasitology |
| mesh[8].descriptor_name | Trichomonas Infections |
| mesh[9].qualifier_ui | Q000235 |
| mesh[9].descriptor_ui | D012091 |
| mesh[9].is_major_topic | False |
| mesh[9].qualifier_name | genetics |
| mesh[9].descriptor_name | Repetitive Sequences, Nucleic Acid |
| mesh[10].qualifier_ui | Q000379 |
| mesh[10].descriptor_ui | D016133 |
| mesh[10].is_major_topic | False |
| mesh[10].qualifier_name | methods |
| mesh[10].descriptor_name | Polymerase Chain Reaction |
| mesh[11].qualifier_ui | Q000235 |
| mesh[11].descriptor_ui | D017931 |
| mesh[11].is_major_topic | False |
| mesh[11].qualifier_name | genetics |
| mesh[11].descriptor_name | DNA Primers |
| mesh[12].qualifier_ui | Q000235 |
| mesh[12].descriptor_ui | D016054 |
| mesh[12].is_major_topic | False |
| mesh[12].qualifier_name | genetics |
| mesh[12].descriptor_name | DNA, Protozoan |
| mesh[13].qualifier_ui | Q000235 |
| mesh[13].descriptor_ui | D014246 |
| mesh[13].is_major_topic | True |
| mesh[13].qualifier_name | genetics |
| mesh[13].descriptor_name | Trichomonas vaginalis |
| mesh[14].qualifier_ui | Q000302 |
| mesh[14].descriptor_ui | D014246 |
| mesh[14].is_major_topic | True |
| mesh[14].qualifier_name | isolation & purification |
| mesh[14].descriptor_name | Trichomonas vaginalis |
| mesh[15].qualifier_ui | |
| mesh[15].descriptor_ui | D006801 |
| mesh[15].is_major_topic | False |
| mesh[15].qualifier_name | |
| mesh[15].descriptor_name | Humans |
| mesh[16].qualifier_ui | |
| mesh[16].descriptor_ui | D000465 |
| mesh[16].is_major_topic | True |
| mesh[16].qualifier_name | |
| mesh[16].descriptor_name | Algorithms |
| mesh[17].qualifier_ui | |
| mesh[17].descriptor_ui | D005260 |
| mesh[17].is_major_topic | False |
| mesh[17].qualifier_name | |
| mesh[17].descriptor_name | Female |
| mesh[18].qualifier_ui | |
| mesh[18].descriptor_ui | D012680 |
| mesh[18].is_major_topic | False |
| mesh[18].qualifier_name | |
| mesh[18].descriptor_name | Sensitivity and Specificity |
| mesh[19].qualifier_ui | Q000175 |
| mesh[19].descriptor_ui | D014247 |
| mesh[19].is_major_topic | False |
| mesh[19].qualifier_name | diagnosis |
| mesh[19].descriptor_name | Trichomonas Vaginitis |
| mesh[20].qualifier_ui | Q000175 |
| mesh[20].descriptor_ui | D014245 |
| mesh[20].is_major_topic | False |
| mesh[20].qualifier_name | diagnosis |
| mesh[20].descriptor_name | Trichomonas Infections |
| mesh[21].qualifier_ui | Q000469 |
| mesh[21].descriptor_ui | D014245 |
| mesh[21].is_major_topic | False |
| mesh[21].qualifier_name | parasitology |
| mesh[21].descriptor_name | Trichomonas Infections |
| mesh[22].qualifier_ui | Q000235 |
| mesh[22].descriptor_ui | D012091 |
| mesh[22].is_major_topic | False |
| mesh[22].qualifier_name | genetics |
| mesh[22].descriptor_name | Repetitive Sequences, Nucleic Acid |
| mesh[23].qualifier_ui | Q000379 |
| mesh[23].descriptor_ui | D016133 |
| mesh[23].is_major_topic | False |
| mesh[23].qualifier_name | methods |
| mesh[23].descriptor_name | Polymerase Chain Reaction |
| mesh[24].qualifier_ui | Q000235 |
| mesh[24].descriptor_ui | D017931 |
| mesh[24].is_major_topic | False |
| mesh[24].qualifier_name | genetics |
| mesh[24].descriptor_name | DNA Primers |
| mesh[25].qualifier_ui | Q000235 |
| mesh[25].descriptor_ui | D016054 |
| mesh[25].is_major_topic | False |
| mesh[25].qualifier_name | genetics |
| mesh[25].descriptor_name | DNA, Protozoan |
| mesh[26].qualifier_ui | Q000235 |
| mesh[26].descriptor_ui | D014246 |
| mesh[26].is_major_topic | True |
| mesh[26].qualifier_name | genetics |
| mesh[26].descriptor_name | Trichomonas vaginalis |
| mesh[27].qualifier_ui | Q000302 |
| mesh[27].descriptor_ui | D014246 |
| mesh[27].is_major_topic | True |
| mesh[27].qualifier_name | isolation & purification |
| mesh[27].descriptor_name | Trichomonas vaginalis |
| mesh[28].qualifier_ui | |
| mesh[28].descriptor_ui | D006801 |
| mesh[28].is_major_topic | False |
| mesh[28].qualifier_name | |
| mesh[28].descriptor_name | Humans |
| mesh[29].qualifier_ui | |
| mesh[29].descriptor_ui | D005260 |
| mesh[29].is_major_topic | False |
| mesh[29].qualifier_name | |
| mesh[29].descriptor_name | Female |
| mesh[30].qualifier_ui | |
| mesh[30].descriptor_ui | D000465 |
| mesh[30].is_major_topic | True |
| mesh[30].qualifier_name | |
| mesh[30].descriptor_name | Algorithms |
| mesh[31].qualifier_ui | Q000175 |
| mesh[31].descriptor_ui | D014247 |
| mesh[31].is_major_topic | True |
| mesh[31].qualifier_name | diagnosis |
| mesh[31].descriptor_name | Trichomonas Vaginitis |
| mesh[32].qualifier_ui | Q000469 |
| mesh[32].descriptor_ui | D014247 |
| mesh[32].is_major_topic | True |
| mesh[32].qualifier_name | parasitology |
| mesh[32].descriptor_name | Trichomonas Vaginitis |
| mesh[33].qualifier_ui | Q000379 |
| mesh[33].descriptor_ui | D025202 |
| mesh[33].is_major_topic | True |
| mesh[33].qualifier_name | methods |
| mesh[33].descriptor_name | Molecular Diagnostic Techniques |
| mesh[34].qualifier_ui | Q000175 |
| mesh[34].descriptor_ui | D014245 |
| mesh[34].is_major_topic | True |
| mesh[34].qualifier_name | diagnosis |
| mesh[34].descriptor_name | Trichomonas Infections |
| mesh[35].qualifier_ui | Q000469 |
| mesh[35].descriptor_ui | D014245 |
| mesh[35].is_major_topic | True |
| mesh[35].qualifier_name | parasitology |
| mesh[35].descriptor_name | Trichomonas Infections |
| mesh[36].qualifier_ui | |
| mesh[36].descriptor_ui | D012680 |
| mesh[36].is_major_topic | False |
| mesh[36].qualifier_name | |
| mesh[36].descriptor_name | Sensitivity and Specificity |
| mesh[37].qualifier_ui | Q000235 |
| mesh[37].descriptor_ui | D016054 |
| mesh[37].is_major_topic | False |
| mesh[37].qualifier_name | genetics |
| mesh[37].descriptor_name | DNA, Protozoan |
| mesh[38].qualifier_ui | Q000379 |
| mesh[38].descriptor_ui | D016133 |
| mesh[38].is_major_topic | False |
| mesh[38].qualifier_name | methods |
| mesh[38].descriptor_name | Polymerase Chain Reaction |
| mesh[39].qualifier_ui | |
| mesh[39].descriptor_ui | D012091 |
| mesh[39].is_major_topic | True |
| mesh[39].qualifier_name | |
| mesh[39].descriptor_name | Repetitive Sequences, Nucleic Acid |
| type | article |
| title | Highly sensitive molecular assay based on Identical Multi-Repeat Sequence (IMRS) algorithm for the detection of Trichomonas vaginalis infection |
| awards[0].id | https://openalex.org/G2071068958 |
| awards[0].funder_id | https://openalex.org/F4320320006 |
| awards[0].display_name | |
| awards[0].funder_award_id | FLR\R1\201314 |
| awards[0].funder_display_name | Royal Society |
| biblio.issue | 2 |
| biblio.volume | 20 |
| biblio.last_page | e0317958 |
| biblio.first_page | e0317958 |
| topics[0].id | https://openalex.org/T10625 |
| topics[0].field.id | https://openalex.org/fields/27 |
| topics[0].field.display_name | Medicine |
| topics[0].score | 0.9986000061035156 |
| topics[0].domain.id | https://openalex.org/domains/4 |
| topics[0].domain.display_name | Health Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/2713 |
| topics[0].subfield.display_name | Epidemiology |
| topics[0].display_name | Herpesvirus Infections and Treatments |
| topics[1].id | https://openalex.org/T10428 |
| topics[1].field.id | https://openalex.org/fields/24 |
| topics[1].field.display_name | Immunology and Microbiology |
| topics[1].score | 0.998199999332428 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/2404 |
| topics[1].subfield.display_name | Microbiology |
| topics[1].display_name | Reproductive tract infections research |
| topics[2].id | https://openalex.org/T11048 |
| topics[2].field.id | https://openalex.org/fields/23 |
| topics[2].field.display_name | Environmental Science |
| topics[2].score | 0.9876999855041504 |
| topics[2].domain.id | https://openalex.org/domains/3 |
| topics[2].domain.display_name | Physical Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/2303 |
| topics[2].subfield.display_name | Ecology |
| topics[2].display_name | Bacteriophages and microbial interactions |
| funders[0].id | https://openalex.org/F4320320006 |
| funders[0].ror | https://ror.org/03wnrjx87 |
| funders[0].display_name | Royal Society |
| is_xpac | False |
| apc_list.value | 1805 |
| apc_list.currency | USD |
| apc_list.value_usd | 1805 |
| apc_paid.value | 1805 |
| apc_paid.currency | USD |
| apc_paid.value_usd | 1805 |
| concepts[0].id | https://openalex.org/C2781114625 |
| concepts[0].level | 2 |
| concepts[0].score | 0.868090033531189 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q132595 |
| concepts[0].display_name | Trichomonas vaginalis |
| concepts[1].id | https://openalex.org/C2910222780 |
| concepts[1].level | 3 |
| concepts[1].score | 0.5565541982650757 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q745865 |
| concepts[1].display_name | Trichomonas Vaginitis |
| concepts[2].id | https://openalex.org/C11413529 |
| concepts[2].level | 1 |
| concepts[2].score | 0.47522786259651184 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q8366 |
| concepts[2].display_name | Algorithm |
| concepts[3].id | https://openalex.org/C2778112365 |
| concepts[3].level | 2 |
| concepts[3].score | 0.45397213101387024 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q3511065 |
| concepts[3].display_name | Sequence (biology) |
| concepts[4].id | https://openalex.org/C86803240 |
| concepts[4].level | 0 |
| concepts[4].score | 0.4206393361091614 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[4].display_name | Biology |
| concepts[5].id | https://openalex.org/C159047783 |
| concepts[5].level | 1 |
| concepts[5].score | 0.3897331953048706 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q7215 |
| concepts[5].display_name | Virology |
| concepts[6].id | https://openalex.org/C89423630 |
| concepts[6].level | 1 |
| concepts[6].score | 0.1909434199333191 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q7193 |
| concepts[6].display_name | Microbiology |
| concepts[7].id | https://openalex.org/C54355233 |
| concepts[7].level | 1 |
| concepts[7].score | 0.18567103147506714 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[7].display_name | Genetics |
| concepts[8].id | https://openalex.org/C41008148 |
| concepts[8].level | 0 |
| concepts[8].score | 0.0896364152431488 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q21198 |
| concepts[8].display_name | Computer science |
| keywords[0].id | https://openalex.org/keywords/trichomonas-vaginalis |
| keywords[0].score | 0.868090033531189 |
| keywords[0].display_name | Trichomonas vaginalis |
| keywords[1].id | https://openalex.org/keywords/trichomonas-vaginitis |
| keywords[1].score | 0.5565541982650757 |
| keywords[1].display_name | Trichomonas Vaginitis |
| keywords[2].id | https://openalex.org/keywords/algorithm |
| keywords[2].score | 0.47522786259651184 |
| keywords[2].display_name | Algorithm |
| keywords[3].id | https://openalex.org/keywords/sequence |
| keywords[3].score | 0.45397213101387024 |
| keywords[3].display_name | Sequence (biology) |
| keywords[4].id | https://openalex.org/keywords/biology |
| keywords[4].score | 0.4206393361091614 |
| keywords[4].display_name | Biology |
| keywords[5].id | https://openalex.org/keywords/virology |
| keywords[5].score | 0.3897331953048706 |
| keywords[5].display_name | Virology |
| keywords[6].id | https://openalex.org/keywords/microbiology |
| keywords[6].score | 0.1909434199333191 |
| keywords[6].display_name | Microbiology |
| keywords[7].id | https://openalex.org/keywords/genetics |
| keywords[7].score | 0.18567103147506714 |
| keywords[7].display_name | Genetics |
| keywords[8].id | https://openalex.org/keywords/computer-science |
| keywords[8].score | 0.0896364152431488 |
| keywords[8].display_name | Computer science |
| language | en |
| locations[0].id | doi:10.1371/journal.pone.0317958 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S202381698 |
| locations[0].source.issn | 1932-6203 |
| locations[0].source.type | journal |
| locations[0].source.is_oa | True |
| locations[0].source.issn_l | 1932-6203 |
| locations[0].source.is_core | True |
| locations[0].source.is_in_doaj | True |
| locations[0].source.display_name | PLoS ONE |
| locations[0].source.host_organization | https://openalex.org/P4310315706 |
| locations[0].source.host_organization_name | Public Library of Science |
| locations[0].source.host_organization_lineage | https://openalex.org/P4310315706 |
| locations[0].source.host_organization_lineage_names | Public Library of Science |
| locations[0].license | cc-by |
| locations[0].pdf_url | |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | PLOS ONE |
| locations[0].landing_page_url | https://doi.org/10.1371/journal.pone.0317958 |
| locations[1].id | pmid:39919090 |
| locations[1].is_oa | False |
| locations[1].source.id | https://openalex.org/S4306525036 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | PubMed |
| locations[1].source.host_organization | https://openalex.org/I1299303238 |
| locations[1].source.host_organization_name | National Institutes of Health |
| locations[1].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[1].license | |
| locations[1].pdf_url | |
| locations[1].version | publishedVersion |
| locations[1].raw_type | |
| locations[1].license_id | |
| locations[1].is_accepted | True |
| locations[1].is_published | True |
| locations[1].raw_source_name | PloS one |
| locations[1].landing_page_url | https://pubmed.ncbi.nlm.nih.gov/39919090 |
| locations[2].id | pmh:oai:doaj.org/article:81c3b431853e47f891c33983fe1d525f |
| locations[2].is_oa | False |
| locations[2].source.id | https://openalex.org/S4306401280 |
| locations[2].source.issn | |
| locations[2].source.type | repository |
| locations[2].source.is_oa | False |
| locations[2].source.issn_l | |
| locations[2].source.is_core | False |
| locations[2].source.is_in_doaj | False |
| locations[2].source.display_name | DOAJ (DOAJ: Directory of Open Access Journals) |
| locations[2].source.host_organization | |
| locations[2].source.host_organization_name | |
| locations[2].license | |
| locations[2].pdf_url | |
| locations[2].version | submittedVersion |
| locations[2].raw_type | article |
| locations[2].license_id | |
| locations[2].is_accepted | False |
| locations[2].is_published | False |
| locations[2].raw_source_name | PLoS ONE, Vol 20, Iss 2, p e0317958 (2025) |
| locations[2].landing_page_url | https://doaj.org/article/81c3b431853e47f891c33983fe1d525f |
| locations[3].id | pmh:oai:pubmedcentral.nih.gov:11805422 |
| locations[3].is_oa | True |
| locations[3].source.id | https://openalex.org/S2764455111 |
| locations[3].source.issn | |
| locations[3].source.type | repository |
| locations[3].source.is_oa | False |
| locations[3].source.issn_l | |
| locations[3].source.is_core | False |
| locations[3].source.is_in_doaj | False |
| locations[3].source.display_name | PubMed Central |
| locations[3].source.host_organization | https://openalex.org/I1299303238 |
| locations[3].source.host_organization_name | National Institutes of Health |
| locations[3].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[3].license | other-oa |
| locations[3].pdf_url | |
| locations[3].version | submittedVersion |
| locations[3].raw_type | Text |
| locations[3].license_id | https://openalex.org/licenses/other-oa |
| locations[3].is_accepted | False |
| locations[3].is_published | False |
| locations[3].raw_source_name | PLoS One |
| locations[3].landing_page_url | https://www.ncbi.nlm.nih.gov/pmc/articles/11805422 |
| indexed_in | crossref, doaj, pubmed |
| authorships[0].author.id | https://openalex.org/A5023621728 |
| authorships[0].author.orcid | https://orcid.org/0000-0002-1909-3791 |
| authorships[0].author.display_name | Clement Likhovole |
| authorships[0].countries | KE |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I250207469 |
| authorships[0].affiliations[0].raw_affiliation_string | Centre for Research in Infectious Diseases, College of Graduate Studies and Research, Mount Kenya University, Thika, Kenya |
| authorships[0].institutions[0].id | https://openalex.org/I250207469 |
| authorships[0].institutions[0].ror | https://ror.org/04kq7tf63 |
| authorships[0].institutions[0].type | education |
| authorships[0].institutions[0].lineage | https://openalex.org/I250207469 |
| authorships[0].institutions[0].country_code | KE |
| authorships[0].institutions[0].display_name | Mount Kenya University |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Clement Shiluli |
| authorships[0].is_corresponding | True |
| authorships[0].raw_affiliation_strings | Centre for Research in Infectious Diseases, College of Graduate Studies and Research, Mount Kenya University, Thika, Kenya |
| authorships[1].author.id | https://openalex.org/A5104147491 |
| authorships[1].author.orcid | |
| authorships[1].author.display_name | Shwetha Kamath |
| authorships[1].affiliations[0].raw_affiliation_string | Division of Research and Development, Jigsaw Bio Solutions Private Limited, Bangalore, India |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Shwetha Kamath |
| authorships[1].is_corresponding | True |
| authorships[1].raw_affiliation_strings | Division of Research and Development, Jigsaw Bio Solutions Private Limited, Bangalore, India |
| authorships[2].author.id | https://openalex.org/A5023267458 |
| authorships[2].author.orcid | https://orcid.org/0000-0003-2772-7579 |
| authorships[2].author.display_name | Bernard N. Kanoi |
| authorships[2].countries | KE |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I250207469 |
| authorships[2].affiliations[0].raw_affiliation_string | Centre for Research in Infectious Diseases, College of Graduate Studies and Research, Mount Kenya University, Thika, Kenya |
| authorships[2].institutions[0].id | https://openalex.org/I250207469 |
| authorships[2].institutions[0].ror | https://ror.org/04kq7tf63 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I250207469 |
| authorships[2].institutions[0].country_code | KE |
| authorships[2].institutions[0].display_name | Mount Kenya University |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Bernard N. Kanoi |
| authorships[2].is_corresponding | True |
| authorships[2].raw_affiliation_strings | Centre for Research in Infectious Diseases, College of Graduate Studies and Research, Mount Kenya University, Thika, Kenya |
| authorships[3].author.id | https://openalex.org/A5000772668 |
| authorships[3].author.orcid | https://orcid.org/0000-0002-1614-4637 |
| authorships[3].author.display_name | Racheal Kimani |
| authorships[3].countries | KE |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I250207469 |
| authorships[3].affiliations[0].raw_affiliation_string | Centre for Research in Infectious Diseases, College of Graduate Studies and Research, Mount Kenya University, Thika, Kenya |
| authorships[3].institutions[0].id | https://openalex.org/I250207469 |
| authorships[3].institutions[0].ror | https://ror.org/04kq7tf63 |
| authorships[3].institutions[0].type | education |
| authorships[3].institutions[0].lineage | https://openalex.org/I250207469 |
| authorships[3].institutions[0].country_code | KE |
| authorships[3].institutions[0].display_name | Mount Kenya University |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Racheal Kimani |
| authorships[3].is_corresponding | True |
| authorships[3].raw_affiliation_strings | Centre for Research in Infectious Diseases, College of Graduate Studies and Research, Mount Kenya University, Thika, Kenya |
| authorships[4].author.id | https://openalex.org/A5063699921 |
| authorships[4].author.orcid | https://orcid.org/0009-0004-1205-8792 |
| authorships[4].author.display_name | Bernard Oduor |
| authorships[4].countries | KE |
| authorships[4].affiliations[0].institution_ids | https://openalex.org/I58422712 |
| authorships[4].affiliations[0].raw_affiliation_string | Animal and Human Health Program, International Livestock Research Institute, Nairobi, Kenya |
| authorships[4].institutions[0].id | https://openalex.org/I58422712 |
| authorships[4].institutions[0].ror | https://ror.org/01jxjwb74 |
| authorships[4].institutions[0].type | facility |
| authorships[4].institutions[0].lineage | https://openalex.org/I1286583668, https://openalex.org/I58422712 |
| authorships[4].institutions[0].country_code | KE |
| authorships[4].institutions[0].display_name | International Livestock Research Institute |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Bernard Oduor |
| authorships[4].is_corresponding | True |
| authorships[4].raw_affiliation_strings | Animal and Human Health Program, International Livestock Research Institute, Nairobi, Kenya |
| authorships[5].author.id | https://openalex.org/A5069678363 |
| authorships[5].author.orcid | https://orcid.org/0000-0002-5594-4418 |
| authorships[5].author.display_name | Hussein M. Abkallo |
| authorships[5].countries | KE |
| authorships[5].affiliations[0].institution_ids | https://openalex.org/I58422712 |
| authorships[5].affiliations[0].raw_affiliation_string | Animal and Human Health Program, International Livestock Research Institute, Nairobi, Kenya |
| authorships[5].institutions[0].id | https://openalex.org/I58422712 |
| authorships[5].institutions[0].ror | https://ror.org/01jxjwb74 |
| authorships[5].institutions[0].type | facility |
| authorships[5].institutions[0].lineage | https://openalex.org/I1286583668, https://openalex.org/I58422712 |
| authorships[5].institutions[0].country_code | KE |
| authorships[5].institutions[0].display_name | International Livestock Research Institute |
| authorships[5].author_position | middle |
| authorships[5].raw_author_name | Hussein M. Abkallo |
| authorships[5].is_corresponding | True |
| authorships[5].raw_affiliation_strings | Animal and Human Health Program, International Livestock Research Institute, Nairobi, Kenya |
| authorships[6].author.id | https://openalex.org/A5011382763 |
| authorships[6].author.orcid | https://orcid.org/0000-0002-9711-7584 |
| authorships[6].author.display_name | Michael Maina |
| authorships[6].countries | KE |
| authorships[6].affiliations[0].institution_ids | https://openalex.org/I250207469 |
| authorships[6].affiliations[0].raw_affiliation_string | Centre for Research in Infectious Diseases, College of Graduate Studies and Research, Mount Kenya University, Thika, Kenya |
| authorships[6].institutions[0].id | https://openalex.org/I250207469 |
| authorships[6].institutions[0].ror | https://ror.org/04kq7tf63 |
| authorships[6].institutions[0].type | education |
| authorships[6].institutions[0].lineage | https://openalex.org/I250207469 |
| authorships[6].institutions[0].country_code | KE |
| authorships[6].institutions[0].display_name | Mount Kenya University |
| authorships[6].author_position | middle |
| authorships[6].raw_author_name | Michael Maina |
| authorships[6].is_corresponding | True |
| authorships[6].raw_affiliation_strings | Centre for Research in Infectious Diseases, College of Graduate Studies and Research, Mount Kenya University, Thika, Kenya |
| authorships[7].author.id | https://openalex.org/A5056665544 |
| authorships[7].author.orcid | https://orcid.org/0000-0001-8132-4151 |
| authorships[7].author.display_name | Harrison Waweru |
| authorships[7].countries | KE |
| authorships[7].affiliations[0].institution_ids | https://openalex.org/I250207469 |
| authorships[7].affiliations[0].raw_affiliation_string | Centre for Research in Infectious Diseases, College of Graduate Studies and Research, Mount Kenya University, Thika, Kenya |
| authorships[7].institutions[0].id | https://openalex.org/I250207469 |
| authorships[7].institutions[0].ror | https://ror.org/04kq7tf63 |
| authorships[7].institutions[0].type | education |
| authorships[7].institutions[0].lineage | https://openalex.org/I250207469 |
| authorships[7].institutions[0].country_code | KE |
| authorships[7].institutions[0].display_name | Mount Kenya University |
| authorships[7].author_position | middle |
| authorships[7].raw_author_name | Harrison Waweru |
| authorships[7].is_corresponding | True |
| authorships[7].raw_affiliation_strings | Centre for Research in Infectious Diseases, College of Graduate Studies and Research, Mount Kenya University, Thika, Kenya |
| authorships[8].author.id | https://openalex.org/A5068914872 |
| authorships[8].author.orcid | https://orcid.org/0000-0002-1756-932X |
| authorships[8].author.display_name | Moses Kamita |
| authorships[8].countries | KE |
| authorships[8].affiliations[0].institution_ids | https://openalex.org/I250207469 |
| authorships[8].affiliations[0].raw_affiliation_string | Centre for Research in Infectious Diseases, College of Graduate Studies and Research, Mount Kenya University, Thika, Kenya |
| authorships[8].institutions[0].id | https://openalex.org/I250207469 |
| authorships[8].institutions[0].ror | https://ror.org/04kq7tf63 |
| authorships[8].institutions[0].type | education |
| authorships[8].institutions[0].lineage | https://openalex.org/I250207469 |
| authorships[8].institutions[0].country_code | KE |
| authorships[8].institutions[0].display_name | Mount Kenya University |
| authorships[8].author_position | middle |
| authorships[8].raw_author_name | Moses Kamita |
| authorships[8].is_corresponding | True |
| authorships[8].raw_affiliation_strings | Centre for Research in Infectious Diseases, College of Graduate Studies and Research, Mount Kenya University, Thika, Kenya |
| authorships[9].author.id | https://openalex.org/A5043061476 |
| authorships[9].author.orcid | https://orcid.org/0000-0002-9391-9387 |
| authorships[9].author.display_name | Nicole Pamme |
| authorships[9].countries | SE |
| authorships[9].affiliations[0].institution_ids | https://openalex.org/I161593684 |
| authorships[9].affiliations[0].raw_affiliation_string | Department of Materials and Environmental Chemistry, Stockholm University, Stockholm, Sweden |
| authorships[9].institutions[0].id | https://openalex.org/I161593684 |
| authorships[9].institutions[0].ror | https://ror.org/05f0yaq80 |
| authorships[9].institutions[0].type | education |
| authorships[9].institutions[0].lineage | https://openalex.org/I161593684 |
| authorships[9].institutions[0].country_code | SE |
| authorships[9].institutions[0].display_name | Stockholm University |
| authorships[9].author_position | middle |
| authorships[9].raw_author_name | Nicole Pamme |
| authorships[9].is_corresponding | True |
| authorships[9].raw_affiliation_strings | Department of Materials and Environmental Chemistry, Stockholm University, Stockholm, Sweden |
| authorships[10].author.id | https://openalex.org/A5092524816 |
| authorships[10].author.orcid | |
| authorships[10].author.display_name | Joshua Dupaty |
| authorships[10].countries | US |
| authorships[10].affiliations[0].institution_ids | https://openalex.org/I111088046 |
| authorships[10].affiliations[0].raw_affiliation_string | Department of Biomedical Engineering, Boston University, Boston, Massachusetts, United States of America |
| authorships[10].institutions[0].id | https://openalex.org/I111088046 |
| authorships[10].institutions[0].ror | https://ror.org/05qwgg493 |
| authorships[10].institutions[0].type | education |
| authorships[10].institutions[0].lineage | https://openalex.org/I111088046 |
| authorships[10].institutions[0].country_code | US |
| authorships[10].institutions[0].display_name | Boston University |
| authorships[10].author_position | middle |
| authorships[10].raw_author_name | Joshua Dupaty |
| authorships[10].is_corresponding | True |
| authorships[10].raw_affiliation_strings | Department of Biomedical Engineering, Boston University, Boston, Massachusetts, United States of America |
| authorships[11].author.id | https://openalex.org/A5005255971 |
| authorships[11].author.orcid | https://orcid.org/0000-0001-6103-849X |
| authorships[11].author.display_name | Catherine M. Klapperich |
| authorships[11].countries | US |
| authorships[11].affiliations[0].institution_ids | https://openalex.org/I111088046 |
| authorships[11].affiliations[0].raw_affiliation_string | Department of Biomedical Engineering, Boston University, Boston, Massachusetts, United States of America |
| authorships[11].institutions[0].id | https://openalex.org/I111088046 |
| authorships[11].institutions[0].ror | https://ror.org/05qwgg493 |
| authorships[11].institutions[0].type | education |
| authorships[11].institutions[0].lineage | https://openalex.org/I111088046 |
| authorships[11].institutions[0].country_code | US |
| authorships[11].institutions[0].display_name | Boston University |
| authorships[11].author_position | middle |
| authorships[11].raw_author_name | Catherine M. Klapperich |
| authorships[11].is_corresponding | True |
| authorships[11].raw_affiliation_strings | Department of Biomedical Engineering, Boston University, Boston, Massachusetts, United States of America |
| authorships[12].author.id | https://openalex.org/A5027238632 |
| authorships[12].author.orcid | https://orcid.org/0000-0002-5505-9320 |
| authorships[12].author.display_name | Srinivasa Raju Lolabattu |
| authorships[12].affiliations[0].raw_affiliation_string | Division of Research and Development, Jigsaw Bio Solutions Private Limited, Bangalore, India |
| authorships[12].author_position | middle |
| authorships[12].raw_author_name | Srinivasa Raju Lolabattu |
| authorships[12].is_corresponding | True |
| authorships[12].raw_affiliation_strings | Division of Research and Development, Jigsaw Bio Solutions Private Limited, Bangalore, India |
| authorships[13].author.id | https://openalex.org/A5013867593 |
| authorships[13].author.orcid | https://orcid.org/0000-0001-7516-522X |
| authorships[13].author.display_name | Jesse Gitaka |
| authorships[13].countries | KE |
| authorships[13].affiliations[0].institution_ids | https://openalex.org/I250207469 |
| authorships[13].affiliations[0].raw_affiliation_string | Centre for Research in Infectious Diseases, College of Graduate Studies and Research, Mount Kenya University, Thika, Kenya |
| authorships[13].institutions[0].id | https://openalex.org/I250207469 |
| authorships[13].institutions[0].ror | https://ror.org/04kq7tf63 |
| authorships[13].institutions[0].type | education |
| authorships[13].institutions[0].lineage | https://openalex.org/I250207469 |
| authorships[13].institutions[0].country_code | KE |
| authorships[13].institutions[0].display_name | Mount Kenya University |
| authorships[13].author_position | last |
| authorships[13].raw_author_name | Jesse Gitaka |
| authorships[13].is_corresponding | True |
| authorships[13].raw_affiliation_strings | Centre for Research in Infectious Diseases, College of Graduate Studies and Research, Mount Kenya University, Thika, Kenya |
| has_content.pdf | False |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://doi.org/10.1371/journal.pone.0317958 |
| open_access.oa_status | gold |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Highly sensitive molecular assay based on Identical Multi-Repeat Sequence (IMRS) algorithm for the detection of Trichomonas vaginalis infection |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T10625 |
| primary_topic.field.id | https://openalex.org/fields/27 |
| primary_topic.field.display_name | Medicine |
| primary_topic.score | 0.9986000061035156 |
| primary_topic.domain.id | https://openalex.org/domains/4 |
| primary_topic.domain.display_name | Health Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/2713 |
| primary_topic.subfield.display_name | Epidemiology |
| primary_topic.display_name | Herpesvirus Infections and Treatments |
| related_works | https://openalex.org/W2091919795, https://openalex.org/W4248960193, https://openalex.org/W2138177734, https://openalex.org/W4246838622, https://openalex.org/W2033447691, https://openalex.org/W1837566555, https://openalex.org/W2405735505, https://openalex.org/W2065241456, https://openalex.org/W1975476769, https://openalex.org/W1982163012 |
| cited_by_count | 0 |
| locations_count | 4 |
| best_oa_location.id | doi:10.1371/journal.pone.0317958 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S202381698 |
| best_oa_location.source.issn | 1932-6203 |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | True |
| best_oa_location.source.issn_l | 1932-6203 |
| best_oa_location.source.is_core | True |
| best_oa_location.source.is_in_doaj | True |
| best_oa_location.source.display_name | PLoS ONE |
| best_oa_location.source.host_organization | https://openalex.org/P4310315706 |
| best_oa_location.source.host_organization_name | Public Library of Science |
| best_oa_location.source.host_organization_lineage | https://openalex.org/P4310315706 |
| best_oa_location.source.host_organization_lineage_names | Public Library of Science |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | journal-article |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | PLOS ONE |
| best_oa_location.landing_page_url | https://doi.org/10.1371/journal.pone.0317958 |
| primary_location.id | doi:10.1371/journal.pone.0317958 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S202381698 |
| primary_location.source.issn | 1932-6203 |
| primary_location.source.type | journal |
| primary_location.source.is_oa | True |
| primary_location.source.issn_l | 1932-6203 |
| primary_location.source.is_core | True |
| primary_location.source.is_in_doaj | True |
| primary_location.source.display_name | PLoS ONE |
| primary_location.source.host_organization | https://openalex.org/P4310315706 |
| primary_location.source.host_organization_name | Public Library of Science |
| primary_location.source.host_organization_lineage | https://openalex.org/P4310315706 |
| primary_location.source.host_organization_lineage_names | Public Library of Science |
| primary_location.license | cc-by |
| primary_location.pdf_url | |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | PLOS ONE |
| primary_location.landing_page_url | https://doi.org/10.1371/journal.pone.0317958 |
| publication_date | 2025-02-07 |
| publication_year | 2025 |
| referenced_works | https://openalex.org/W4388773910, https://openalex.org/W4311433910, https://openalex.org/W4385225298, https://openalex.org/W70460679, https://openalex.org/W2102875285, https://openalex.org/W2258841189, https://openalex.org/W2168047762, https://openalex.org/W2147119980, https://openalex.org/W2162720994, https://openalex.org/W2767687501, https://openalex.org/W2947693962, https://openalex.org/W4214883834, https://openalex.org/W2789791939, https://openalex.org/W2732332911, https://openalex.org/W2792142692, https://openalex.org/W4388206213, https://openalex.org/W2057514444, https://openalex.org/W3075820966, https://openalex.org/W1981979522, https://openalex.org/W2160770318, https://openalex.org/W2084042629, https://openalex.org/W2010973136, https://openalex.org/W1978783962, https://openalex.org/W2206240473, https://openalex.org/W4220855209, https://openalex.org/W2089135077, https://openalex.org/W2139982241, https://openalex.org/W2105275554, https://openalex.org/W2057876450, https://openalex.org/W2065098811, https://openalex.org/W2770752141, https://openalex.org/W2121212030, https://openalex.org/W4210582501, https://openalex.org/W4210451305 |
| referenced_works_count | 34 |
| abstract_inverted_index.( | 12 |
| abstract_inverted_index.) | 16 |
| abstract_inverted_index.. | 14, 28, 72, 154, 156, 176, 194, 231, 276, 297 |
| abstract_inverted_index.2 | 201 |
| abstract_inverted_index.L | 244 |
| abstract_inverted_index.T | 13, 27, 71, 153, 175, 193, 230, 275, 296 |
| abstract_inverted_index.a | 94, 181, 186, 212, 236, 305, 314 |
| abstract_inverted_index.l | 208 |
| abstract_inverted_index.69 | 189 |
| abstract_inverted_index.De | 291 |
| abstract_inverted_index.In | 48 |
| abstract_inverted_index.We | 158 |
| abstract_inverted_index.as | 36, 84, 106, 119, 145, 211, 300, 304 |
| abstract_inverted_index.by | 9 |
| abstract_inverted_index.in | 23, 46, 214 |
| abstract_inverted_index.is | 74 |
| abstract_inverted_index.of | 19, 70, 92, 102, 140, 188, 240, 257, 273, 295, 317 |
| abstract_inverted_index.on | 163, 173 |
| abstract_inverted_index.to | 65, 136, 169, 179, 202, 246, 284 |
| abstract_inverted_index.μ | 207, 243, 253 |
| abstract_inverted_index.174 | 3 |
| abstract_inverted_index.18S | 222, 248 |
| abstract_inverted_index.DNA | 196 |
| abstract_inverted_index.For | 219 |
| abstract_inverted_index.L). | 254 |
| abstract_inverted_index.PCR | 224, 250 |
| abstract_inverted_index.The | 229, 255 |
| abstract_inverted_index.and | 40, 78, 98, 108, 125, 128, 209, 313 |
| abstract_inverted_index.are | 7, 31 |
| abstract_inverted_index.can | 63 |
| abstract_inverted_index.fg/ | 242 |
| abstract_inverted_index.for | 57, 76, 148, 152, 259, 307, 311 |
| abstract_inverted_index.one | 287 |
| abstract_inverted_index.pg/ | 252 |
| abstract_inverted_index.the | 51, 90, 100, 138, 174, 215, 247, 260, 270, 274 |
| abstract_inverted_index.use | 101, 139 |
| abstract_inverted_index.was | 197, 226, 263, 280 |
| abstract_inverted_index.0.03 | 241 |
| abstract_inverted_index.0.58 | 264 |
| abstract_inverted_index.Half | 18 |
| abstract_inverted_index.IMRS | 299 |
| abstract_inverted_index.PCR, | 269 |
| abstract_inverted_index.This | 133 |
| abstract_inverted_index.case | 59 |
| abstract_inverted_index.from | 199 |
| abstract_inverted_index.have | 89, 115 |
| abstract_inverted_index.high | 120 |
| abstract_inverted_index.lead | 64 |
| abstract_inverted_index.less | 285 |
| abstract_inverted_index.novo | 292 |
| abstract_inverted_index.pair | 183 |
| abstract_inverted_index.rRNA | 223, 249 |
| abstract_inverted_index.seen | 45 |
| abstract_inverted_index.such | 35, 83, 105, 118 |
| abstract_inverted_index.test | 238 |
| abstract_inverted_index.than | 286 |
| abstract_inverted_index.that | 184 |
| abstract_inverted_index.this | 62 |
| abstract_inverted_index.time | 97 |
| abstract_inverted_index.used | 159, 210 |
| abstract_inverted_index.wide | 315 |
| abstract_inverted_index.with | 33 |
| abstract_inverted_index.−4 | 204 |
| abstract_inverted_index.(PCR) | 88 |
| abstract_inverted_index.(WHO) | 55 |
| abstract_inverted_index.-IMRS | 233, 278 |
| abstract_inverted_index.Chain | 86 |
| abstract_inverted_index.Using | 267 |
| abstract_inverted_index.World | 52 |
| abstract_inverted_index.aimed | 135 |
| abstract_inverted_index.allow | 99 |
| abstract_inverted_index.among | 131 |
| abstract_inverted_index.assay | 225, 262 |
| abstract_inverted_index.based | 162 |
| abstract_inverted_index.false | 123 |
| abstract_inverted_index.limit | 256 |
| abstract_inverted_index.occur | 22 |
| abstract_inverted_index.range | 316 |
| abstract_inverted_index.short | 95 |
| abstract_inverted_index.study | 134 |
| abstract_inverted_index.tests | 82 |
| abstract_inverted_index.these | 20, 112 |
| abstract_inverted_index.total | 187 |
| abstract_inverted_index.urine | 107 |
| abstract_inverted_index.(0.714 | 251 |
| abstract_inverted_index.(IMRS) | 144, 167 |
| abstract_inverted_index.Health | 53 |
| abstract_inverted_index.assay. | 218 |
| abstract_inverted_index.costs, | 122 |
| abstract_inverted_index.design | 180 |
| abstract_inverted_index.genome | 178, 205, 265, 288, 293 |
| abstract_inverted_index.having | 93 |
| abstract_inverted_index.higher | 237 |
| abstract_inverted_index.mining | 294 |
| abstract_inverted_index.mostly | 44 |
| abstract_inverted_index.pelvic | 37 |
| abstract_inverted_index.people | 5 |
| abstract_inverted_index.primer | 182 |
| abstract_inverted_index.prompt | 79 |
| abstract_inverted_index.repeat | 190 |
| abstract_inverted_index.serves | 303 |
| abstract_inverted_index.swabs. | 110 |
| abstract_inverted_index.women. | 47 |
| abstract_inverted_index.5.8×10 | 200, 203 |
| abstract_inverted_index.Genomic | 192 |
| abstract_inverted_index.Methods | 157 |
| abstract_inverted_index.Results | 228 |
| abstract_inverted_index.adverse | 41 |
| abstract_inverted_index.copies/ | 206 |
| abstract_inverted_index.diluted | 198 |
| abstract_inverted_index.disease | 39 |
| abstract_inverted_index.million | 4 |
| abstract_inverted_index.offered | 235 |
| abstract_inverted_index.others. | 132 |
| abstract_inverted_index.pg/μL, | 282 |
| abstract_inverted_index.primers | 147, 234, 279, 302 |
| abstract_inverted_index.targets | 185 |
| abstract_inverted_index.vaginal | 109 |
| abstract_inverted_index.<0.01 | 281 |
| abstract_inverted_index.Accurate | 68 |
| abstract_inverted_index.However, | 61, 111 |
| abstract_inverted_index.Reaction | 87 |
| abstract_inverted_index.affected | 8 |
| abstract_inverted_index.compared | 245 |
| abstract_inverted_index.evaluate | 137 |
| abstract_inverted_index.globally | 6 |
| abstract_inverted_index.identify | 170 |
| abstract_inverted_index.negative | 124 |
| abstract_inverted_index.numerous | 116 |
| abstract_inverted_index.outcomes | 43 |
| abstract_inverted_index.platform | 306 |
| abstract_inverted_index.positive | 126 |
| abstract_inverted_index.regions, | 50 |
| abstract_inverted_index.regions. | 25 |
| abstract_inverted_index.required | 75 |
| abstract_inverted_index.results, | 127 |
| abstract_inverted_index.template | 213 |
| abstract_inverted_index.Annually, | 1 |
| abstract_inverted_index.Molecular | 81 |
| abstract_inverted_index.Untreated | 26 |
| abstract_inverted_index.advantage | 91 |
| abstract_inverted_index.advocates | 56 |
| abstract_inverted_index.algorithm | 168 |
| abstract_inverted_index.copy/μL. | 289 |
| abstract_inverted_index.detection | 258 |
| abstract_inverted_index.diagnosis | 69 |
| abstract_inverted_index.effective | 77 |
| abstract_inverted_index.employed. | 227 |
| abstract_inverted_index.identical | 141, 164 |
| abstract_inverted_index.pregnancy | 42 |
| abstract_inverted_index.real-time | 268 |
| abstract_inverted_index.sequences | 143, 166, 171 |
| abstract_inverted_index.specimens | 104 |
| abstract_inverted_index.syndromic | 58 |
| abstract_inverted_index.vaginalis | 11, 15, 29, 73, 155, 177, 195, 232, 277, 298 |
| abstract_inverted_index.variation | 130 |
| abstract_inverted_index.Conclusion | 290 |
| abstract_inverted_index.IMRS-based | 216 |
| abstract_inverted_index.Isothermal | 261 |
| abstract_inverted_index.Polymerase | 85 |
| abstract_inverted_index.analytical | 271 |
| abstract_inverted_index.approaches | 161 |
| abstract_inverted_index.associated | 32 |
| abstract_inverted_index.copies/mL. | 266 |
| abstract_inverted_index.developing | 149, 308 |
| abstract_inverted_index.diagnostic | 113, 151 |
| abstract_inverted_index.equivalent | 283 |
| abstract_inverted_index.infection. | 17 |
| abstract_inverted_index.infections | 21, 30 |
| abstract_inverted_index.infectious | 318 |
| abstract_inverted_index.pathogens. | 319 |
| abstract_inverted_index.sequences. | 191 |
| abstract_inverted_index.techniques | 114 |
| abstract_inverted_index.treatment. | 67, 80 |
| abstract_inverted_index.Trichomonas | 10 |
| abstract_inverted_index.comparison, | 221 |
| abstract_inverted_index.diagnostics | 310 |
| abstract_inverted_index.distributed | 172 |
| abstract_inverted_index.interstrain | 129 |
| abstract_inverted_index.management. | 60 |
| abstract_inverted_index.performance | 220 |
| abstract_inverted_index.sensitivity | 239, 272 |
| abstract_inverted_index.turn-around | 96 |
| abstract_inverted_index.unnecessary | 66 |
| abstract_inverted_index.Introduction | 0 |
| abstract_inverted_index.Organization | 54 |
| abstract_inverted_index.inflammatory | 38 |
| abstract_inverted_index.multi-repeat | 142, 165 |
| abstract_inverted_index.non-invasive | 103 |
| abstract_inverted_index.amplification | 146, 217, 301 |
| abstract_inverted_index.approximately | 2 |
| abstract_inverted_index.complications | 34 |
| abstract_inverted_index.disadvantages | 117 |
| abstract_inverted_index.genome-mining | 160 |
| abstract_inverted_index.Trichomoniasis | 312 |
| abstract_inverted_index.infrastructure | 121 |
| abstract_inverted_index.ultrasensitive | 150, 309 |
| abstract_inverted_index.resource-limited | 24, 49 |
| cited_by_percentile_year | |
| corresponding_author_ids | https://openalex.org/A5063699921, https://openalex.org/A5011382763, https://openalex.org/A5056665544, https://openalex.org/A5092524816, https://openalex.org/A5068914872, https://openalex.org/A5005255971, https://openalex.org/A5104147491, https://openalex.org/A5069678363, https://openalex.org/A5000772668, https://openalex.org/A5023621728, https://openalex.org/A5043061476, https://openalex.org/A5013867593, https://openalex.org/A5027238632, https://openalex.org/A5023267458 |
| countries_distinct_count | 3 |
| institutions_distinct_count | 14 |
| corresponding_institution_ids | https://openalex.org/I111088046, https://openalex.org/I161593684, https://openalex.org/I250207469, https://openalex.org/I58422712 |
| sustainable_development_goals[0].id | https://metadata.un.org/sdg/9 |
| sustainable_development_goals[0].score | 0.5899999737739563 |
| sustainable_development_goals[0].display_name | Industry, innovation and infrastructure |
| citation_normalized_percentile.value | 0.05730184 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |