Hospital and urban wastewaters shape the structure and active resistome of environmental biofilms Article Swipe
 YOU?
·
· 2023
· Open Access
·
· DOI: https://doi.org/10.1101/2023.01.19.524754
YOU?
·
· 2023
· Open Access
·
· DOI: https://doi.org/10.1101/2023.01.19.524754
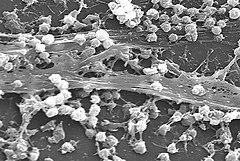
Background Demonstration of the transfer, dynamics, and regulation of antibiotic resistance genes (ARGs) and mobile genetic elements (MGEs) in a complex environmental matrix is yet experimentally challenging, with many essential open questions such as how and where transfer and dissemination of ARGs happens in nature. The extent and conditions of MGEs transfer that carry ARGs is still largely unexplored in natural environments and microbial communities. Biofilms are structures that include high density multi-species bacterial communities embedded in self-produced extracellular polymeric substances (EPS) constituting a matrix that facilitates gene transfer and where bacteria exhibit high tolerance to stress and to antibiotics. In this study we implemented a sampling and analysis approach that allows phenotypic and genomic analyses of in situ and reconstituted in vitro hospital and urban wastewater (WW) biofilms. To assess the potential of hospital and urban WW biofilms to efficiently disseminate ARGs in the WW system, we explored the EPS within the biofilm matrix and assessed the expression of the resistome (ARGs) and mobilome (MGEs) by metatranscriptomics. Results We first showed that a) the composition of EPS differs depending on their growth environment ( in situ and in vitro ) and their sampling origin (hospital vs urban WW) and that b) a low amount of ciprofloxacin impacted the composition of the EPS. Next, the metatranscriptomic approach showed that a) expression of ARGs and MGEs increase upon adding a low amount of ciprofloxacin for biofilms from hospital WW but not for those from urban WW and b) that expression of specific plasmids that carry individual or multiple ARGs varies depending on the WW origins of the biofilms. When the same plasmids were expressed in both, urban and hospital WW biofilms, they carried and expressed different ARGs. Conclusion We show that hospital and urban wastewaters shape the structure and active resistome of environmental biofilms, and we confirmed that hospital WW is an important hot spot for the dissemination and selection of AMR. The different responses to antibiotic pressure in hospital vs urban biofilms, coupled with differences in biofilm structure helps delineate distinct characteristics of hospital and urban WW biofilms highlighting the relationships between the resistome and its expression in environmental biofilms and their surrounding ecosystems.
Related Topics
- Type
- preprint
- Language
- en
- Landing Page
- https://doi.org/10.1101/2023.01.19.524754
- https://www.biorxiv.org/content/biorxiv/early/2023/02/10/2023.01.19.524754.full.pdf
- OA Status
- green
- References
- 67
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4317597452
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4317597452Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1101/2023.01.19.524754Digital Object Identifier
- Title
-
Hospital and urban wastewaters shape the structure and active resistome of environmental biofilmsWork title
- Type
-
preprintOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2023Year of publication
- Publication date
-
2023-01-20Full publication date if available
- Authors
-
Elena Buelow, Catherine Dauga, Claire Carrion, Hugo Mathé‐Hubert, Sophia Achaibou, Margaux Gaschet, Thomas Jové, O. Chesneau, Sean Kennedy, Marie-Cécile Ploy, Sandra Da Re, Christophe DagotList of authors in order
- Landing page
-
https://doi.org/10.1101/2023.01.19.524754Publisher landing page
- PDF URL
-
https://www.biorxiv.org/content/biorxiv/early/2023/02/10/2023.01.19.524754.full.pdfDirect link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
greenOpen access status per OpenAlex
- OA URL
-
https://www.biorxiv.org/content/biorxiv/early/2023/02/10/2023.01.19.524754.full.pdfDirect OA link when available
- Concepts
-
Biofilm, Resistome, Mobile genetic elements, Biology, Plasmid, Horizontal gene transfer, Microbiology, Bacteria, Gene, Genetics, GenomeTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
0Total citation count in OpenAlex
- References (count)
-
67Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4317597452 |
|---|---|
| doi | https://doi.org/10.1101/2023.01.19.524754 |
| ids.doi | https://doi.org/10.1101/2023.01.19.524754 |
| ids.openalex | https://openalex.org/W4317597452 |
| fwci | 0.0 |
| type | preprint |
| title | Hospital and urban wastewaters shape the structure and active resistome of environmental biofilms |
| biblio.issue | |
| biblio.volume | |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T10419 |
| topics[0].field.id | https://openalex.org/fields/23 |
| topics[0].field.display_name | Environmental Science |
| topics[0].score | 0.9995999932289124 |
| topics[0].domain.id | https://openalex.org/domains/3 |
| topics[0].domain.display_name | Physical Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/2310 |
| topics[0].subfield.display_name | Pollution |
| topics[0].display_name | Pharmaceutical and Antibiotic Environmental Impacts |
| topics[1].id | https://openalex.org/T10593 |
| topics[1].field.id | https://openalex.org/fields/13 |
| topics[1].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[1].score | 0.9901000261306763 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1312 |
| topics[1].subfield.display_name | Molecular Biology |
| topics[1].display_name | Bacterial biofilms and quorum sensing |
| topics[2].id | https://openalex.org/T10147 |
| topics[2].field.id | https://openalex.org/fields/13 |
| topics[2].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[2].score | 0.9894000291824341 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1313 |
| topics[2].subfield.display_name | Molecular Medicine |
| topics[2].display_name | Antibiotic Resistance in Bacteria |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C58123911 |
| concepts[0].level | 3 |
| concepts[0].score | 0.8803766965866089 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q467410 |
| concepts[0].display_name | Biofilm |
| concepts[1].id | https://openalex.org/C2776666601 |
| concepts[1].level | 5 |
| concepts[1].score | 0.7737840414047241 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q3457855 |
| concepts[1].display_name | Resistome |
| concepts[2].id | https://openalex.org/C132533263 |
| concepts[2].level | 4 |
| concepts[2].score | 0.6030710339546204 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q2347362 |
| concepts[2].display_name | Mobile genetic elements |
| concepts[3].id | https://openalex.org/C86803240 |
| concepts[3].level | 0 |
| concepts[3].score | 0.5027596950531006 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[3].display_name | Biology |
| concepts[4].id | https://openalex.org/C22744801 |
| concepts[4].level | 3 |
| concepts[4].score | 0.4982573986053467 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q172778 |
| concepts[4].display_name | Plasmid |
| concepts[5].id | https://openalex.org/C92938381 |
| concepts[5].level | 4 |
| concepts[5].score | 0.46948155760765076 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q83185 |
| concepts[5].display_name | Horizontal gene transfer |
| concepts[6].id | https://openalex.org/C89423630 |
| concepts[6].level | 1 |
| concepts[6].score | 0.4075823128223419 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q7193 |
| concepts[6].display_name | Microbiology |
| concepts[7].id | https://openalex.org/C523546767 |
| concepts[7].level | 2 |
| concepts[7].score | 0.4024217128753662 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q10876 |
| concepts[7].display_name | Bacteria |
| concepts[8].id | https://openalex.org/C104317684 |
| concepts[8].level | 2 |
| concepts[8].score | 0.2926425039768219 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[8].display_name | Gene |
| concepts[9].id | https://openalex.org/C54355233 |
| concepts[9].level | 1 |
| concepts[9].score | 0.25646042823791504 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[9].display_name | Genetics |
| concepts[10].id | https://openalex.org/C141231307 |
| concepts[10].level | 3 |
| concepts[10].score | 0.19347724318504333 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q7020 |
| concepts[10].display_name | Genome |
| keywords[0].id | https://openalex.org/keywords/biofilm |
| keywords[0].score | 0.8803766965866089 |
| keywords[0].display_name | Biofilm |
| keywords[1].id | https://openalex.org/keywords/resistome |
| keywords[1].score | 0.7737840414047241 |
| keywords[1].display_name | Resistome |
| keywords[2].id | https://openalex.org/keywords/mobile-genetic-elements |
| keywords[2].score | 0.6030710339546204 |
| keywords[2].display_name | Mobile genetic elements |
| keywords[3].id | https://openalex.org/keywords/biology |
| keywords[3].score | 0.5027596950531006 |
| keywords[3].display_name | Biology |
| keywords[4].id | https://openalex.org/keywords/plasmid |
| keywords[4].score | 0.4982573986053467 |
| keywords[4].display_name | Plasmid |
| keywords[5].id | https://openalex.org/keywords/horizontal-gene-transfer |
| keywords[5].score | 0.46948155760765076 |
| keywords[5].display_name | Horizontal gene transfer |
| keywords[6].id | https://openalex.org/keywords/microbiology |
| keywords[6].score | 0.4075823128223419 |
| keywords[6].display_name | Microbiology |
| keywords[7].id | https://openalex.org/keywords/bacteria |
| keywords[7].score | 0.4024217128753662 |
| keywords[7].display_name | Bacteria |
| keywords[8].id | https://openalex.org/keywords/gene |
| keywords[8].score | 0.2926425039768219 |
| keywords[8].display_name | Gene |
| keywords[9].id | https://openalex.org/keywords/genetics |
| keywords[9].score | 0.25646042823791504 |
| keywords[9].display_name | Genetics |
| keywords[10].id | https://openalex.org/keywords/genome |
| keywords[10].score | 0.19347724318504333 |
| keywords[10].display_name | Genome |
| language | en |
| locations[0].id | doi:10.1101/2023.01.19.524754 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4306402567 |
| locations[0].source.issn | |
| locations[0].source.type | repository |
| locations[0].source.is_oa | False |
| locations[0].source.issn_l | |
| locations[0].source.is_core | False |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| locations[0].source.host_organization | https://openalex.org/I2750212522 |
| locations[0].source.host_organization_name | Cold Spring Harbor Laboratory |
| locations[0].source.host_organization_lineage | https://openalex.org/I2750212522 |
| locations[0].license | cc-by-nc-nd |
| locations[0].pdf_url | https://www.biorxiv.org/content/biorxiv/early/2023/02/10/2023.01.19.524754.full.pdf |
| locations[0].version | acceptedVersion |
| locations[0].raw_type | posted-content |
| locations[0].license_id | https://openalex.org/licenses/cc-by-nc-nd |
| locations[0].is_accepted | True |
| locations[0].is_published | False |
| locations[0].raw_source_name | |
| locations[0].landing_page_url | https://doi.org/10.1101/2023.01.19.524754 |
| indexed_in | crossref |
| authorships[0].author.id | https://openalex.org/A5032228618 |
| authorships[0].author.orcid | https://orcid.org/0000-0003-1458-4791 |
| authorships[0].author.display_name | Elena Buelow |
| authorships[0].countries | FR |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I154526488, https://openalex.org/I65806277 |
| authorships[0].affiliations[0].raw_affiliation_string | INSERM, CHU Limoges, RESINFIT, UMR109, University of Limoges, Limoges, France |
| authorships[0].affiliations[1].institution_ids | https://openalex.org/I106785703, https://openalex.org/I1294671590, https://openalex.org/I4210104684, https://openalex.org/I899635006 |
| authorships[0].affiliations[1].raw_affiliation_string | Univ. Grenoble Alpes, CNRS, UMR 5525, VetAgro Sup, Grenoble INP, TIMC, 38000 Grenoble, France |
| authorships[0].institutions[0].id | https://openalex.org/I1294671590 |
| authorships[0].institutions[0].ror | https://ror.org/02feahw73 |
| authorships[0].institutions[0].type | government |
| authorships[0].institutions[0].lineage | https://openalex.org/I1294671590 |
| authorships[0].institutions[0].country_code | FR |
| authorships[0].institutions[0].display_name | Centre National de la Recherche Scientifique |
| authorships[0].institutions[1].id | https://openalex.org/I154526488 |
| authorships[0].institutions[1].ror | https://ror.org/02vjkv261 |
| authorships[0].institutions[1].type | government |
| authorships[0].institutions[1].lineage | https://openalex.org/I154526488 |
| authorships[0].institutions[1].country_code | FR |
| authorships[0].institutions[1].display_name | Inserm |
| authorships[0].institutions[2].id | https://openalex.org/I106785703 |
| authorships[0].institutions[2].ror | https://ror.org/05sbt2524 |
| authorships[0].institutions[2].type | education |
| authorships[0].institutions[2].lineage | https://openalex.org/I106785703, https://openalex.org/I899635006 |
| authorships[0].institutions[2].country_code | FR |
| authorships[0].institutions[2].display_name | Institut polytechnique de Grenoble |
| authorships[0].institutions[3].id | https://openalex.org/I899635006 |
| authorships[0].institutions[3].ror | https://ror.org/02rx3b187 |
| authorships[0].institutions[3].type | education |
| authorships[0].institutions[3].lineage | https://openalex.org/I899635006 |
| authorships[0].institutions[3].country_code | FR |
| authorships[0].institutions[3].display_name | Université Grenoble Alpes |
| authorships[0].institutions[4].id | https://openalex.org/I65806277 |
| authorships[0].institutions[4].ror | https://ror.org/02cp04407 |
| authorships[0].institutions[4].type | education |
| authorships[0].institutions[4].lineage | https://openalex.org/I65806277 |
| authorships[0].institutions[4].country_code | FR |
| authorships[0].institutions[4].display_name | Université de Limoges |
| authorships[0].institutions[5].id | https://openalex.org/I4210104684 |
| authorships[0].institutions[5].ror | https://ror.org/01c7wz417 |
| authorships[0].institutions[5].type | education |
| authorships[0].institutions[5].lineage | https://openalex.org/I4210104684 |
| authorships[0].institutions[5].country_code | FR |
| authorships[0].institutions[5].display_name | VetAgro Sup |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Elena Buelow |
| authorships[0].is_corresponding | True |
| authorships[0].raw_affiliation_strings | INSERM, CHU Limoges, RESINFIT, UMR109, University of Limoges, Limoges, France, Univ. Grenoble Alpes, CNRS, UMR 5525, VetAgro Sup, Grenoble INP, TIMC, 38000 Grenoble, France |
| authorships[1].author.id | https://openalex.org/A5064644109 |
| authorships[1].author.orcid | https://orcid.org/0000-0002-1794-4871 |
| authorships[1].author.display_name | Catherine Dauga |
| authorships[1].countries | FR |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I157536573, https://openalex.org/I4210150352 |
| authorships[1].affiliations[0].raw_affiliation_string | Biomics Pole, CITECH, Institut Pasteur, F-75015 Paris, France |
| authorships[1].affiliations[1].institution_ids | https://openalex.org/I157536573, https://openalex.org/I204730241 |
| authorships[1].affiliations[1].raw_affiliation_string | Institut Pasteur, Université Paris Cité, Département Biologie Computationnelle, F-75015 Paris, France |
| authorships[1].institutions[0].id | https://openalex.org/I157536573 |
| authorships[1].institutions[0].ror | https://ror.org/0495fxg12 |
| authorships[1].institutions[0].type | nonprofit |
| authorships[1].institutions[0].lineage | https://openalex.org/I157536573 |
| authorships[1].institutions[0].country_code | FR |
| authorships[1].institutions[0].display_name | Institut Pasteur |
| authorships[1].institutions[1].id | https://openalex.org/I4210150352 |
| authorships[1].institutions[1].ror | https://ror.org/05mv80p58 |
| authorships[1].institutions[1].type | education |
| authorships[1].institutions[1].lineage | https://openalex.org/I4210150352 |
| authorships[1].institutions[1].country_code | FR |
| authorships[1].institutions[1].display_name | Institut Textile et Chimique |
| authorships[1].institutions[2].id | https://openalex.org/I204730241 |
| authorships[1].institutions[2].ror | https://ror.org/05f82e368 |
| authorships[1].institutions[2].type | education |
| authorships[1].institutions[2].lineage | https://openalex.org/I204730241 |
| authorships[1].institutions[2].country_code | FR |
| authorships[1].institutions[2].display_name | Université Paris Cité |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Catherine Dauga |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | Biomics Pole, CITECH, Institut Pasteur, F-75015 Paris, France, Institut Pasteur, Université Paris Cité, Département Biologie Computationnelle, F-75015 Paris, France |
| authorships[2].author.id | https://openalex.org/A5037574721 |
| authorships[2].author.orcid | https://orcid.org/0000-0003-3221-6319 |
| authorships[2].author.display_name | Claire Carrion |
| authorships[2].countries | FR |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I1294671590 |
| authorships[2].affiliations[0].raw_affiliation_string | CHU Limoges, UMR CNRS 7276, F-87000 Limoges, France |
| authorships[2].affiliations[1].institution_ids | https://openalex.org/I154526488, https://openalex.org/I65806277 |
| authorships[2].affiliations[1].raw_affiliation_string | University of Limoges, INSERM |
| authorships[2].institutions[0].id | https://openalex.org/I1294671590 |
| authorships[2].institutions[0].ror | https://ror.org/02feahw73 |
| authorships[2].institutions[0].type | government |
| authorships[2].institutions[0].lineage | https://openalex.org/I1294671590 |
| authorships[2].institutions[0].country_code | FR |
| authorships[2].institutions[0].display_name | Centre National de la Recherche Scientifique |
| authorships[2].institutions[1].id | https://openalex.org/I154526488 |
| authorships[2].institutions[1].ror | https://ror.org/02vjkv261 |
| authorships[2].institutions[1].type | government |
| authorships[2].institutions[1].lineage | https://openalex.org/I154526488 |
| authorships[2].institutions[1].country_code | FR |
| authorships[2].institutions[1].display_name | Inserm |
| authorships[2].institutions[2].id | https://openalex.org/I65806277 |
| authorships[2].institutions[2].ror | https://ror.org/02cp04407 |
| authorships[2].institutions[2].type | education |
| authorships[2].institutions[2].lineage | https://openalex.org/I65806277 |
| authorships[2].institutions[2].country_code | FR |
| authorships[2].institutions[2].display_name | Université de Limoges |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Claire Carrion |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | CHU Limoges, UMR CNRS 7276, F-87000 Limoges, France, University of Limoges, INSERM |
| authorships[3].author.id | https://openalex.org/A5074549878 |
| authorships[3].author.orcid | https://orcid.org/0000-0003-4785-433X |
| authorships[3].author.display_name | Hugo Mathé‐Hubert |
| authorships[3].countries | FR |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I106785703, https://openalex.org/I1294671590, https://openalex.org/I4210104684, https://openalex.org/I899635006 |
| authorships[3].affiliations[0].raw_affiliation_string | Univ. Grenoble Alpes, CNRS, UMR 5525, VetAgro Sup, Grenoble INP, TIMC, 38000 Grenoble, France |
| authorships[3].institutions[0].id | https://openalex.org/I1294671590 |
| authorships[3].institutions[0].ror | https://ror.org/02feahw73 |
| authorships[3].institutions[0].type | government |
| authorships[3].institutions[0].lineage | https://openalex.org/I1294671590 |
| authorships[3].institutions[0].country_code | FR |
| authorships[3].institutions[0].display_name | Centre National de la Recherche Scientifique |
| authorships[3].institutions[1].id | https://openalex.org/I106785703 |
| authorships[3].institutions[1].ror | https://ror.org/05sbt2524 |
| authorships[3].institutions[1].type | education |
| authorships[3].institutions[1].lineage | https://openalex.org/I106785703, https://openalex.org/I899635006 |
| authorships[3].institutions[1].country_code | FR |
| authorships[3].institutions[1].display_name | Institut polytechnique de Grenoble |
| authorships[3].institutions[2].id | https://openalex.org/I899635006 |
| authorships[3].institutions[2].ror | https://ror.org/02rx3b187 |
| authorships[3].institutions[2].type | education |
| authorships[3].institutions[2].lineage | https://openalex.org/I899635006 |
| authorships[3].institutions[2].country_code | FR |
| authorships[3].institutions[2].display_name | Université Grenoble Alpes |
| authorships[3].institutions[3].id | https://openalex.org/I4210104684 |
| authorships[3].institutions[3].ror | https://ror.org/01c7wz417 |
| authorships[3].institutions[3].type | education |
| authorships[3].institutions[3].lineage | https://openalex.org/I4210104684 |
| authorships[3].institutions[3].country_code | FR |
| authorships[3].institutions[3].display_name | VetAgro Sup |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Hugo Mathé-Hubert |
| authorships[3].is_corresponding | False |
| authorships[3].raw_affiliation_strings | Univ. Grenoble Alpes, CNRS, UMR 5525, VetAgro Sup, Grenoble INP, TIMC, 38000 Grenoble, France |
| authorships[4].author.id | https://openalex.org/A5069702517 |
| authorships[4].author.orcid | |
| authorships[4].author.display_name | Sophia Achaibou |
| authorships[4].countries | FR |
| authorships[4].affiliations[0].institution_ids | https://openalex.org/I157536573, https://openalex.org/I4210150352 |
| authorships[4].affiliations[0].raw_affiliation_string | Biomics Pole, CITECH, Institut Pasteur, F-75015 Paris, France |
| authorships[4].institutions[0].id | https://openalex.org/I157536573 |
| authorships[4].institutions[0].ror | https://ror.org/0495fxg12 |
| authorships[4].institutions[0].type | nonprofit |
| authorships[4].institutions[0].lineage | https://openalex.org/I157536573 |
| authorships[4].institutions[0].country_code | FR |
| authorships[4].institutions[0].display_name | Institut Pasteur |
| authorships[4].institutions[1].id | https://openalex.org/I4210150352 |
| authorships[4].institutions[1].ror | https://ror.org/05mv80p58 |
| authorships[4].institutions[1].type | education |
| authorships[4].institutions[1].lineage | https://openalex.org/I4210150352 |
| authorships[4].institutions[1].country_code | FR |
| authorships[4].institutions[1].display_name | Institut Textile et Chimique |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Sophia Achaibou |
| authorships[4].is_corresponding | False |
| authorships[4].raw_affiliation_strings | Biomics Pole, CITECH, Institut Pasteur, F-75015 Paris, France |
| authorships[5].author.id | https://openalex.org/A5015831463 |
| authorships[5].author.orcid | |
| authorships[5].author.display_name | Margaux Gaschet |
| authorships[5].countries | FR |
| authorships[5].affiliations[0].institution_ids | https://openalex.org/I154526488, https://openalex.org/I65806277 |
| authorships[5].affiliations[0].raw_affiliation_string | INSERM, CHU Limoges, RESINFIT, UMR109, University of Limoges, Limoges, France |
| authorships[5].institutions[0].id | https://openalex.org/I154526488 |
| authorships[5].institutions[0].ror | https://ror.org/02vjkv261 |
| authorships[5].institutions[0].type | government |
| authorships[5].institutions[0].lineage | https://openalex.org/I154526488 |
| authorships[5].institutions[0].country_code | FR |
| authorships[5].institutions[0].display_name | Inserm |
| authorships[5].institutions[1].id | https://openalex.org/I65806277 |
| authorships[5].institutions[1].ror | https://ror.org/02cp04407 |
| authorships[5].institutions[1].type | education |
| authorships[5].institutions[1].lineage | https://openalex.org/I65806277 |
| authorships[5].institutions[1].country_code | FR |
| authorships[5].institutions[1].display_name | Université de Limoges |
| authorships[5].author_position | middle |
| authorships[5].raw_author_name | Margaux Gaschet |
| authorships[5].is_corresponding | False |
| authorships[5].raw_affiliation_strings | INSERM, CHU Limoges, RESINFIT, UMR109, University of Limoges, Limoges, France |
| authorships[6].author.id | https://openalex.org/A5085367142 |
| authorships[6].author.orcid | https://orcid.org/0000-0002-9983-2880 |
| authorships[6].author.display_name | Thomas Jové |
| authorships[6].countries | FR |
| authorships[6].affiliations[0].institution_ids | https://openalex.org/I154526488, https://openalex.org/I65806277 |
| authorships[6].affiliations[0].raw_affiliation_string | INSERM, CHU Limoges, RESINFIT, UMR109, University of Limoges, Limoges, France |
| authorships[6].institutions[0].id | https://openalex.org/I154526488 |
| authorships[6].institutions[0].ror | https://ror.org/02vjkv261 |
| authorships[6].institutions[0].type | government |
| authorships[6].institutions[0].lineage | https://openalex.org/I154526488 |
| authorships[6].institutions[0].country_code | FR |
| authorships[6].institutions[0].display_name | Inserm |
| authorships[6].institutions[1].id | https://openalex.org/I65806277 |
| authorships[6].institutions[1].ror | https://ror.org/02cp04407 |
| authorships[6].institutions[1].type | education |
| authorships[6].institutions[1].lineage | https://openalex.org/I65806277 |
| authorships[6].institutions[1].country_code | FR |
| authorships[6].institutions[1].display_name | Université de Limoges |
| authorships[6].author_position | middle |
| authorships[6].raw_author_name | Thomas Jové |
| authorships[6].is_corresponding | False |
| authorships[6].raw_affiliation_strings | INSERM, CHU Limoges, RESINFIT, UMR109, University of Limoges, Limoges, France |
| authorships[7].author.id | https://openalex.org/A5015544930 |
| authorships[7].author.orcid | https://orcid.org/0000-0002-9718-0444 |
| authorships[7].author.display_name | O. Chesneau |
| authorships[7].countries | FR |
| authorships[7].affiliations[0].institution_ids | https://openalex.org/I157536573 |
| authorships[7].affiliations[0].raw_affiliation_string | Collection de l'Institut Pasteur (CIP), Microbiology Department, Institut Pasteur, Paris 75015, France |
| authorships[7].institutions[0].id | https://openalex.org/I157536573 |
| authorships[7].institutions[0].ror | https://ror.org/0495fxg12 |
| authorships[7].institutions[0].type | nonprofit |
| authorships[7].institutions[0].lineage | https://openalex.org/I157536573 |
| authorships[7].institutions[0].country_code | FR |
| authorships[7].institutions[0].display_name | Institut Pasteur |
| authorships[7].author_position | middle |
| authorships[7].raw_author_name | Olivier Chesneau |
| authorships[7].is_corresponding | False |
| authorships[7].raw_affiliation_strings | Collection de l'Institut Pasteur (CIP), Microbiology Department, Institut Pasteur, Paris 75015, France |
| authorships[8].author.id | https://openalex.org/A5039721313 |
| authorships[8].author.orcid | https://orcid.org/0000-0002-3932-8922 |
| authorships[8].author.display_name | Sean Kennedy |
| authorships[8].countries | FR |
| authorships[8].affiliations[0].institution_ids | https://openalex.org/I157536573, https://openalex.org/I204730241 |
| authorships[8].affiliations[0].raw_affiliation_string | Institut Pasteur, Université Paris Cité, Département Biologie Computationnelle, F-75015 Paris, France |
| authorships[8].institutions[0].id | https://openalex.org/I157536573 |
| authorships[8].institutions[0].ror | https://ror.org/0495fxg12 |
| authorships[8].institutions[0].type | nonprofit |
| authorships[8].institutions[0].lineage | https://openalex.org/I157536573 |
| authorships[8].institutions[0].country_code | FR |
| authorships[8].institutions[0].display_name | Institut Pasteur |
| authorships[8].institutions[1].id | https://openalex.org/I204730241 |
| authorships[8].institutions[1].ror | https://ror.org/05f82e368 |
| authorships[8].institutions[1].type | education |
| authorships[8].institutions[1].lineage | https://openalex.org/I204730241 |
| authorships[8].institutions[1].country_code | FR |
| authorships[8].institutions[1].display_name | Université Paris Cité |
| authorships[8].author_position | middle |
| authorships[8].raw_author_name | Sean P. Kennedy |
| authorships[8].is_corresponding | False |
| authorships[8].raw_affiliation_strings | Institut Pasteur, Université Paris Cité, Département Biologie Computationnelle, F-75015 Paris, France |
| authorships[9].author.id | https://openalex.org/A5011238769 |
| authorships[9].author.orcid | https://orcid.org/0000-0002-0718-4979 |
| authorships[9].author.display_name | Marie-Cécile Ploy |
| authorships[9].countries | FR |
| authorships[9].affiliations[0].institution_ids | https://openalex.org/I154526488, https://openalex.org/I65806277 |
| authorships[9].affiliations[0].raw_affiliation_string | INSERM, CHU Limoges, RESINFIT, UMR109, University of Limoges, Limoges, France |
| authorships[9].institutions[0].id | https://openalex.org/I154526488 |
| authorships[9].institutions[0].ror | https://ror.org/02vjkv261 |
| authorships[9].institutions[0].type | government |
| authorships[9].institutions[0].lineage | https://openalex.org/I154526488 |
| authorships[9].institutions[0].country_code | FR |
| authorships[9].institutions[0].display_name | Inserm |
| authorships[9].institutions[1].id | https://openalex.org/I65806277 |
| authorships[9].institutions[1].ror | https://ror.org/02cp04407 |
| authorships[9].institutions[1].type | education |
| authorships[9].institutions[1].lineage | https://openalex.org/I65806277 |
| authorships[9].institutions[1].country_code | FR |
| authorships[9].institutions[1].display_name | Université de Limoges |
| authorships[9].author_position | middle |
| authorships[9].raw_author_name | Marie-Cecile Ploy |
| authorships[9].is_corresponding | False |
| authorships[9].raw_affiliation_strings | INSERM, CHU Limoges, RESINFIT, UMR109, University of Limoges, Limoges, France |
| authorships[10].author.id | https://openalex.org/A5010714436 |
| authorships[10].author.orcid | https://orcid.org/0000-0003-3579-9185 |
| authorships[10].author.display_name | Sandra Da Re |
| authorships[10].countries | FR |
| authorships[10].affiliations[0].institution_ids | https://openalex.org/I154526488, https://openalex.org/I65806277 |
| authorships[10].affiliations[0].raw_affiliation_string | INSERM, CHU Limoges, RESINFIT, UMR109, University of Limoges, Limoges, France |
| authorships[10].institutions[0].id | https://openalex.org/I154526488 |
| authorships[10].institutions[0].ror | https://ror.org/02vjkv261 |
| authorships[10].institutions[0].type | government |
| authorships[10].institutions[0].lineage | https://openalex.org/I154526488 |
| authorships[10].institutions[0].country_code | FR |
| authorships[10].institutions[0].display_name | Inserm |
| authorships[10].institutions[1].id | https://openalex.org/I65806277 |
| authorships[10].institutions[1].ror | https://ror.org/02cp04407 |
| authorships[10].institutions[1].type | education |
| authorships[10].institutions[1].lineage | https://openalex.org/I65806277 |
| authorships[10].institutions[1].country_code | FR |
| authorships[10].institutions[1].display_name | Université de Limoges |
| authorships[10].author_position | middle |
| authorships[10].raw_author_name | Sandra Da Re |
| authorships[10].is_corresponding | False |
| authorships[10].raw_affiliation_strings | INSERM, CHU Limoges, RESINFIT, UMR109, University of Limoges, Limoges, France |
| authorships[11].author.id | https://openalex.org/A5007669705 |
| authorships[11].author.orcid | https://orcid.org/0000-0003-3632-0420 |
| authorships[11].author.display_name | Christophe Dagot |
| authorships[11].countries | FR |
| authorships[11].affiliations[0].institution_ids | https://openalex.org/I154526488, https://openalex.org/I65806277 |
| authorships[11].affiliations[0].raw_affiliation_string | INSERM, CHU Limoges, RESINFIT, UMR109, University of Limoges, Limoges, France |
| authorships[11].institutions[0].id | https://openalex.org/I154526488 |
| authorships[11].institutions[0].ror | https://ror.org/02vjkv261 |
| authorships[11].institutions[0].type | government |
| authorships[11].institutions[0].lineage | https://openalex.org/I154526488 |
| authorships[11].institutions[0].country_code | FR |
| authorships[11].institutions[0].display_name | Inserm |
| authorships[11].institutions[1].id | https://openalex.org/I65806277 |
| authorships[11].institutions[1].ror | https://ror.org/02cp04407 |
| authorships[11].institutions[1].type | education |
| authorships[11].institutions[1].lineage | https://openalex.org/I65806277 |
| authorships[11].institutions[1].country_code | FR |
| authorships[11].institutions[1].display_name | Université de Limoges |
| authorships[11].author_position | last |
| authorships[11].raw_author_name | Christophe Dagot |
| authorships[11].is_corresponding | False |
| authorships[11].raw_affiliation_strings | INSERM, CHU Limoges, RESINFIT, UMR109, University of Limoges, Limoges, France |
| has_content.pdf | True |
| has_content.grobid_xml | True |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://www.biorxiv.org/content/biorxiv/early/2023/02/10/2023.01.19.524754.full.pdf |
| open_access.oa_status | green |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Hospital and urban wastewaters shape the structure and active resistome of environmental biofilms |
| has_fulltext | True |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T10419 |
| primary_topic.field.id | https://openalex.org/fields/23 |
| primary_topic.field.display_name | Environmental Science |
| primary_topic.score | 0.9995999932289124 |
| primary_topic.domain.id | https://openalex.org/domains/3 |
| primary_topic.domain.display_name | Physical Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/2310 |
| primary_topic.subfield.display_name | Pollution |
| primary_topic.display_name | Pharmaceutical and Antibiotic Environmental Impacts |
| related_works | https://openalex.org/W2305704688, https://openalex.org/W1989898630, https://openalex.org/W4412076945, https://openalex.org/W2945663299, https://openalex.org/W2006478187, https://openalex.org/W4390909895, https://openalex.org/W1891155455, https://openalex.org/W2788164399, https://openalex.org/W3185032264, https://openalex.org/W2543813668 |
| cited_by_count | 0 |
| locations_count | 1 |
| best_oa_location.id | doi:10.1101/2023.01.19.524754 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4306402567 |
| best_oa_location.source.issn | |
| best_oa_location.source.type | repository |
| best_oa_location.source.is_oa | False |
| best_oa_location.source.issn_l | |
| best_oa_location.source.is_core | False |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| best_oa_location.source.host_organization | https://openalex.org/I2750212522 |
| best_oa_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| best_oa_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| best_oa_location.license | cc-by-nc-nd |
| best_oa_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2023/02/10/2023.01.19.524754.full.pdf |
| best_oa_location.version | acceptedVersion |
| best_oa_location.raw_type | posted-content |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by-nc-nd |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | False |
| best_oa_location.raw_source_name | |
| best_oa_location.landing_page_url | https://doi.org/10.1101/2023.01.19.524754 |
| primary_location.id | doi:10.1101/2023.01.19.524754 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4306402567 |
| primary_location.source.issn | |
| primary_location.source.type | repository |
| primary_location.source.is_oa | False |
| primary_location.source.issn_l | |
| primary_location.source.is_core | False |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| primary_location.source.host_organization | https://openalex.org/I2750212522 |
| primary_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| primary_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| primary_location.license | cc-by-nc-nd |
| primary_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2023/02/10/2023.01.19.524754.full.pdf |
| primary_location.version | acceptedVersion |
| primary_location.raw_type | posted-content |
| primary_location.license_id | https://openalex.org/licenses/cc-by-nc-nd |
| primary_location.is_accepted | True |
| primary_location.is_published | False |
| primary_location.raw_source_name | |
| primary_location.landing_page_url | https://doi.org/10.1101/2023.01.19.524754 |
| publication_date | 2023-01-20 |
| publication_year | 2023 |
| referenced_works | https://openalex.org/W2181033092, https://openalex.org/W2134604195, https://openalex.org/W1982021484, https://openalex.org/W3007319283, https://openalex.org/W1985716049, https://openalex.org/W2793090722, https://openalex.org/W3004510819, https://openalex.org/W2039353072, https://openalex.org/W2069431280, https://openalex.org/W4294170467, https://openalex.org/W2969037787, https://openalex.org/W2158298888, https://openalex.org/W2008088946, https://openalex.org/W2009690857, https://openalex.org/W2027951608, https://openalex.org/W1825642470, https://openalex.org/W1992298603, https://openalex.org/W2949973543, https://openalex.org/W2123299336, https://openalex.org/W2547337573, https://openalex.org/W2796460172, https://openalex.org/W2048693902, https://openalex.org/W4200420863, https://openalex.org/W3116222147, https://openalex.org/W2199204051, https://openalex.org/W2913795513, https://openalex.org/W2986199620, https://openalex.org/W2804144344, https://openalex.org/W2725896266, https://openalex.org/W2197156841, https://openalex.org/W2739852532, https://openalex.org/W2049312611, https://openalex.org/W2018345882, https://openalex.org/W2050116248, https://openalex.org/W2159954944, https://openalex.org/W2091922579, https://openalex.org/W2399343623, https://openalex.org/W2982531601, https://openalex.org/W2149559438, https://openalex.org/W2946191791, https://openalex.org/W1963539735, https://openalex.org/W2159267296, https://openalex.org/W2163861300, https://openalex.org/W2029950408, https://openalex.org/W2126330768, https://openalex.org/W2014408727, https://openalex.org/W2021369951, https://openalex.org/W2005717348, https://openalex.org/W2995538658, https://openalex.org/W3093507631, https://openalex.org/W2042160058, https://openalex.org/W2065031948, https://openalex.org/W2151952137, https://openalex.org/W2172205368, https://openalex.org/W1988657382, https://openalex.org/W1971757487, https://openalex.org/W2142104073, https://openalex.org/W2180484294, https://openalex.org/W3019884735, https://openalex.org/W2604889568, https://openalex.org/W2047893579, https://openalex.org/W3205833625, https://openalex.org/W2027438571, https://openalex.org/W2070399793, https://openalex.org/W2158398230, https://openalex.org/W3128039816, https://openalex.org/W3136475225 |
| referenced_works_count | 67 |
| abstract_inverted_index.( | 185 |
| abstract_inverted_index.) | 191 |
| abstract_inverted_index.a | 20, 84, 106, 203, 229 |
| abstract_inverted_index.In | 101 |
| abstract_inverted_index.To | 130 |
| abstract_inverted_index.WW | 138, 146, 238, 245, 263, 279, 309, 347 |
| abstract_inverted_index.We | 170, 288 |
| abstract_inverted_index.a) | 174, 220 |
| abstract_inverted_index.an | 311 |
| abstract_inverted_index.as | 34 |
| abstract_inverted_index.b) | 202, 247 |
| abstract_inverted_index.by | 167 |
| abstract_inverted_index.in | 19, 44, 60, 77, 118, 122, 144, 186, 189, 274, 328, 336, 358 |
| abstract_inverted_index.is | 24, 56, 310 |
| abstract_inverted_index.of | 3, 9, 41, 50, 117, 134, 160, 177, 206, 211, 222, 232, 250, 265, 301, 320, 343 |
| abstract_inverted_index.on | 181, 261 |
| abstract_inverted_index.or | 256 |
| abstract_inverted_index.to | 96, 99, 140, 325 |
| abstract_inverted_index.vs | 197, 330 |
| abstract_inverted_index.we | 104, 148, 305 |
| abstract_inverted_index.EPS | 151, 178 |
| abstract_inverted_index.The | 46, 322 |
| abstract_inverted_index.WW) | 199 |
| abstract_inverted_index.and | 7, 14, 36, 39, 48, 63, 90, 98, 108, 114, 120, 125, 136, 156, 164, 188, 192, 200, 224, 246, 277, 283, 292, 298, 304, 318, 345, 355, 361 |
| abstract_inverted_index.are | 67 |
| abstract_inverted_index.but | 239 |
| abstract_inverted_index.for | 234, 241, 315 |
| abstract_inverted_index.hot | 313 |
| abstract_inverted_index.how | 35 |
| abstract_inverted_index.its | 356 |
| abstract_inverted_index.low | 204, 230 |
| abstract_inverted_index.not | 240 |
| abstract_inverted_index.the | 4, 132, 145, 150, 153, 158, 161, 175, 209, 212, 215, 262, 266, 269, 296, 316, 350, 353 |
| abstract_inverted_index.yet | 25 |
| abstract_inverted_index.(WW) | 128 |
| abstract_inverted_index.AMR. | 321 |
| abstract_inverted_index.ARGs | 42, 55, 143, 223, 258 |
| abstract_inverted_index.EPS. | 213 |
| abstract_inverted_index.MGEs | 51, 225 |
| abstract_inverted_index.When | 268 |
| abstract_inverted_index.from | 236, 243 |
| abstract_inverted_index.gene | 88 |
| abstract_inverted_index.high | 71, 94 |
| abstract_inverted_index.many | 29 |
| abstract_inverted_index.open | 31 |
| abstract_inverted_index.same | 270 |
| abstract_inverted_index.show | 289 |
| abstract_inverted_index.situ | 119, 187 |
| abstract_inverted_index.spot | 314 |
| abstract_inverted_index.such | 33 |
| abstract_inverted_index.that | 53, 69, 86, 111, 173, 201, 219, 248, 253, 290, 307 |
| abstract_inverted_index.they | 281 |
| abstract_inverted_index.this | 102 |
| abstract_inverted_index.upon | 227 |
| abstract_inverted_index.were | 272 |
| abstract_inverted_index.with | 28, 334 |
| abstract_inverted_index.(EPS) | 82 |
| abstract_inverted_index.ARGs. | 286 |
| abstract_inverted_index.Next, | 214 |
| abstract_inverted_index.both, | 275 |
| abstract_inverted_index.carry | 54, 254 |
| abstract_inverted_index.first | 171 |
| abstract_inverted_index.genes | 12 |
| abstract_inverted_index.helps | 339 |
| abstract_inverted_index.shape | 295 |
| abstract_inverted_index.still | 57 |
| abstract_inverted_index.study | 103 |
| abstract_inverted_index.their | 182, 193, 362 |
| abstract_inverted_index.those | 242 |
| abstract_inverted_index.urban | 126, 137, 198, 244, 276, 293, 331, 346 |
| abstract_inverted_index.vitro | 123, 190 |
| abstract_inverted_index.where | 37, 91 |
| abstract_inverted_index.(ARGs) | 13, 163 |
| abstract_inverted_index.(MGEs) | 18, 166 |
| abstract_inverted_index.active | 299 |
| abstract_inverted_index.adding | 228 |
| abstract_inverted_index.allows | 112 |
| abstract_inverted_index.amount | 205, 231 |
| abstract_inverted_index.assess | 131 |
| abstract_inverted_index.extent | 47 |
| abstract_inverted_index.growth | 183 |
| abstract_inverted_index.matrix | 23, 85, 155 |
| abstract_inverted_index.mobile | 15 |
| abstract_inverted_index.origin | 195 |
| abstract_inverted_index.showed | 172, 218 |
| abstract_inverted_index.stress | 97 |
| abstract_inverted_index.varies | 259 |
| abstract_inverted_index.within | 152 |
| abstract_inverted_index.Results | 169 |
| abstract_inverted_index.between | 352 |
| abstract_inverted_index.biofilm | 154, 337 |
| abstract_inverted_index.carried | 282 |
| abstract_inverted_index.complex | 21 |
| abstract_inverted_index.coupled | 333 |
| abstract_inverted_index.density | 72 |
| abstract_inverted_index.differs | 179 |
| abstract_inverted_index.exhibit | 93 |
| abstract_inverted_index.genetic | 16 |
| abstract_inverted_index.genomic | 115 |
| abstract_inverted_index.happens | 43 |
| abstract_inverted_index.include | 70 |
| abstract_inverted_index.largely | 58 |
| abstract_inverted_index.natural | 61 |
| abstract_inverted_index.nature. | 45 |
| abstract_inverted_index.origins | 264 |
| abstract_inverted_index.system, | 147 |
| abstract_inverted_index.Abstract | 0 |
| abstract_inverted_index.Biofilms | 66 |
| abstract_inverted_index.analyses | 116 |
| abstract_inverted_index.analysis | 109 |
| abstract_inverted_index.approach | 110, 217 |
| abstract_inverted_index.assessed | 157 |
| abstract_inverted_index.bacteria | 92 |
| abstract_inverted_index.biofilms | 139, 235, 348, 360 |
| abstract_inverted_index.distinct | 341 |
| abstract_inverted_index.elements | 17 |
| abstract_inverted_index.embedded | 76 |
| abstract_inverted_index.explored | 149 |
| abstract_inverted_index.hospital | 124, 135, 237, 278, 291, 308, 329, 344 |
| abstract_inverted_index.impacted | 208 |
| abstract_inverted_index.increase | 226 |
| abstract_inverted_index.mobilome | 165 |
| abstract_inverted_index.multiple | 257 |
| abstract_inverted_index.plasmids | 252, 271 |
| abstract_inverted_index.pressure | 327 |
| abstract_inverted_index.sampling | 107, 194 |
| abstract_inverted_index.specific | 251 |
| abstract_inverted_index.transfer | 38, 52, 89 |
| abstract_inverted_index.(hospital | 196 |
| abstract_inverted_index.bacterial | 74 |
| abstract_inverted_index.biofilms, | 280, 303, 332 |
| abstract_inverted_index.biofilms. | 129, 267 |
| abstract_inverted_index.confirmed | 306 |
| abstract_inverted_index.delineate | 340 |
| abstract_inverted_index.depending | 180, 260 |
| abstract_inverted_index.different | 285, 323 |
| abstract_inverted_index.dynamics, | 6 |
| abstract_inverted_index.essential | 30 |
| abstract_inverted_index.expressed | 273, 284 |
| abstract_inverted_index.important | 312 |
| abstract_inverted_index.microbial | 64 |
| abstract_inverted_index.polymeric | 80 |
| abstract_inverted_index.potential | 133 |
| abstract_inverted_index.questions | 32 |
| abstract_inverted_index.resistome | 162, 300, 354 |
| abstract_inverted_index.responses | 324 |
| abstract_inverted_index.selection | 319 |
| abstract_inverted_index.structure | 297, 338 |
| abstract_inverted_index.tolerance | 95 |
| abstract_inverted_index.transfer, | 5 |
| abstract_inverted_index.Background | 1 |
| abstract_inverted_index.Conclusion | 287 |
| abstract_inverted_index.antibiotic | 10, 326 |
| abstract_inverted_index.conditions | 49 |
| abstract_inverted_index.expression | 159, 221, 249, 357 |
| abstract_inverted_index.individual | 255 |
| abstract_inverted_index.phenotypic | 113 |
| abstract_inverted_index.regulation | 8 |
| abstract_inverted_index.resistance | 11 |
| abstract_inverted_index.structures | 68 |
| abstract_inverted_index.substances | 81 |
| abstract_inverted_index.unexplored | 59 |
| abstract_inverted_index.wastewater | 127 |
| abstract_inverted_index.communities | 75 |
| abstract_inverted_index.composition | 176, 210 |
| abstract_inverted_index.differences | 335 |
| abstract_inverted_index.disseminate | 142 |
| abstract_inverted_index.ecosystems. | 364 |
| abstract_inverted_index.efficiently | 141 |
| abstract_inverted_index.environment | 184 |
| abstract_inverted_index.facilitates | 87 |
| abstract_inverted_index.implemented | 105 |
| abstract_inverted_index.surrounding | 363 |
| abstract_inverted_index.wastewaters | 294 |
| abstract_inverted_index.antibiotics. | 100 |
| abstract_inverted_index.challenging, | 27 |
| abstract_inverted_index.communities. | 65 |
| abstract_inverted_index.constituting | 83 |
| abstract_inverted_index.environments | 62 |
| abstract_inverted_index.highlighting | 349 |
| abstract_inverted_index.Demonstration | 2 |
| abstract_inverted_index.ciprofloxacin | 207, 233 |
| abstract_inverted_index.dissemination | 40, 317 |
| abstract_inverted_index.environmental | 22, 302, 359 |
| abstract_inverted_index.extracellular | 79 |
| abstract_inverted_index.multi-species | 73 |
| abstract_inverted_index.reconstituted | 121 |
| abstract_inverted_index.relationships | 351 |
| abstract_inverted_index.self-produced | 78 |
| abstract_inverted_index.experimentally | 26 |
| abstract_inverted_index.characteristics | 342 |
| abstract_inverted_index.metatranscriptomic | 216 |
| abstract_inverted_index.metatranscriptomics. | 168 |
| cited_by_percentile_year | |
| corresponding_author_ids | https://openalex.org/A5032228618 |
| countries_distinct_count | 1 |
| institutions_distinct_count | 12 |
| corresponding_institution_ids | https://openalex.org/I106785703, https://openalex.org/I1294671590, https://openalex.org/I154526488, https://openalex.org/I4210104684, https://openalex.org/I65806277, https://openalex.org/I899635006 |
| sustainable_development_goals[0].id | https://metadata.un.org/sdg/6 |
| sustainable_development_goals[0].score | 0.75 |
| sustainable_development_goals[0].display_name | Clean water and sanitation |
| citation_normalized_percentile.value | 0.00506398 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |