Hydroxychloroquine is Metabolized by Cytochrome P450 2D6, 3A4, and 2C8, and Inhibits Cytochrome P450 2D6, while its Metabolites also Inhibit Cytochrome P450 3A in vitro Article Swipe
 YOU?
·
· 2022
· Open Access
·
· DOI: https://doi.org/10.1124/dmd.122.001018
YOU?
·
· 2022
· Open Access
·
· DOI: https://doi.org/10.1124/dmd.122.001018
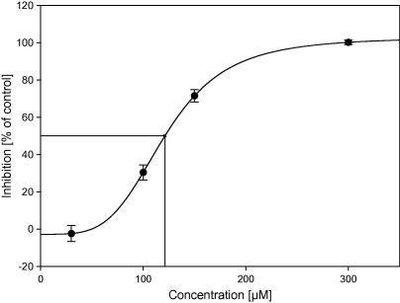
This study aimed to explore the cytochrome P450 (CYP) metabolic and inhibitory profile of hydroxychloroquine (HCQ). Hydroxychloroquine metabolism was studied using human liver microsomes (HLMs) and recombinant CYP enzymes. The inhibitory effects of HCQ and its metabolites on nine CYPs were also determined in HLMs, using an automated substrate cocktail method. Our metabolism data indicated that CYP3A4, CYP2D6, and CYP2C8 are the key enzymes involved in HCQ metabolism. All three CYPs formed the primary metabolites desethylchloroquine (DCQ) and desethylhydroxychloroquine (DHCQ) to various degrees. Although the intrinsic clearance (CLint) value of HCQ depletion by recombinant CYP2D6 was > 10-fold higher than that by CYP3A4 (0.87 versus 0.075 µl/min/pmol), scaling of recombinant CYP CLint to HLM level resulted in almost equal HLM CLint values for CYP2D6 and CYP3A4 (11 and 14 µl/min/mg, respectively). The scaled HLM CLint of CYP2C8 was 5.7 µl/min/mg. Data from HLM experiments with CYP-selective inhibitors also suggested relatively equal roles for CYP2D6 and CYP3A4 in HCQ metabolism, with a smaller contribution by CYP2C8. In CYP inhibition experiments, HCQ, DCQ, DHCQ, and the secondary metabolite didesethylchloroquine were direct CYP2D6 inhibitors, with 50% inhibitory concentration (IC50) values between 18 and 135 µM. HCQ did not inhibit other CYPs. Furthermore, all metabolites were time-dependent CYP3A inhibitors (IC50 shift 2.2-3.4). To conclude, HCQ is metabolized by CYP3A4, CYP2D6, and CYP2C8 in vitro. HCQ and its metabolites are reversible CYP2D6 inhibitors, and HCQ metabolites are time-dependent CYP3A inhibitors. These data can be used to improve physiologically-based pharmacokinetic models and update drug-drug interaction risk estimations for HCQ. SIGNIFICANCE STATEMENT: While CYP2D6, CYP3A4, and CYP2C8 have been shown to mediate chloroquine biotransformation, it appears that the role of CYP enzymes in hydroxychloroquine (HCQ) metabolism has not been studied. In addition, little is known about the CYP inhibitory effects of HCQ. Here, we demonstrate that CYP2D6, CYP3A4, and CYP2C8 are the key enzymes involved in HCQ metabolism. Furthermore, our findings show that HCQ and its metabolites are inhibitors of CYP2D6, which likely explains the previously observed interaction between HCQ and metoprolol.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.1124/dmd.122.001018
- https://dmd.aspetjournals.org/content/dmd/early/2022/11/29/dmd.122.001018.full.pdf
- OA Status
- hybrid
- Cited By
- 18
- References
- 47
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4310465622
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4310465622Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1124/dmd.122.001018Digital Object Identifier
- Title
-
Hydroxychloroquine is Metabolized by Cytochrome P450 2D6, 3A4, and 2C8, and Inhibits Cytochrome P450 2D6, while its Metabolites also Inhibit Cytochrome P450 3A in vitroWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2022Year of publication
- Publication date
-
2022-11-30Full publication date if available
- Authors
-
Marie‐Noëlle Paludetto, Mika Kurkela, Helinä Kahma, Janne T. Backman, Mikko Niemi, Anne M. FilppulaList of authors in order
- Landing page
-
https://doi.org/10.1124/dmd.122.001018Publisher landing page
- PDF URL
-
https://dmd.aspetjournals.org/content/dmd/early/2022/11/29/dmd.122.001018.full.pdfDirect link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
hybridOpen access status per OpenAlex
- OA URL
-
https://dmd.aspetjournals.org/content/dmd/early/2022/11/29/dmd.122.001018.full.pdfDirect OA link when available
- Concepts
-
CYP2D6, CYP3A4, Cytochrome P450, CYP2C8, Metabolite, Pharmacology, CYP3A, IC50, Chemistry, Microsome, CYP2C9, CYP2B6, Metabolism, Biochemistry, Enzyme, In vitro, BiologyTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
18Total citation count in OpenAlex
- Citations by year (recent)
-
2025: 8, 2024: 6, 2023: 4Per-year citation counts (last 5 years)
- References (count)
-
47Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4310465622 |
|---|---|
| doi | https://doi.org/10.1124/dmd.122.001018 |
| ids.doi | https://doi.org/10.1124/dmd.122.001018 |
| ids.pmid | https://pubmed.ncbi.nlm.nih.gov/36446607 |
| ids.openalex | https://openalex.org/W4310465622 |
| fwci | 4.19314069 |
| mesh[0].qualifier_ui | |
| mesh[0].descriptor_ui | D006801 |
| mesh[0].is_major_topic | False |
| mesh[0].qualifier_name | |
| mesh[0].descriptor_name | Humans |
| mesh[1].qualifier_ui | Q000378 |
| mesh[1].descriptor_ui | D019389 |
| mesh[1].is_major_topic | True |
| mesh[1].qualifier_name | metabolism |
| mesh[1].descriptor_name | Cytochrome P-450 CYP2D6 |
| mesh[2].qualifier_ui | Q000378 |
| mesh[2].descriptor_ui | D051544 |
| mesh[2].is_major_topic | True |
| mesh[2].qualifier_name | metabolism |
| mesh[2].descriptor_name | Cytochrome P-450 CYP3A |
| mesh[3].qualifier_ui | Q000378 |
| mesh[3].descriptor_ui | D006886 |
| mesh[3].is_major_topic | False |
| mesh[3].qualifier_name | metabolism |
| mesh[3].descriptor_name | Hydroxychloroquine |
| mesh[4].qualifier_ui | Q000494 |
| mesh[4].descriptor_ui | D006886 |
| mesh[4].is_major_topic | False |
| mesh[4].qualifier_name | pharmacology |
| mesh[4].descriptor_name | Hydroxychloroquine |
| mesh[5].qualifier_ui | Q000378 |
| mesh[5].descriptor_ui | D065727 |
| mesh[5].is_major_topic | False |
| mesh[5].qualifier_name | metabolism |
| mesh[5].descriptor_name | Cytochrome P-450 CYP2C8 |
| mesh[6].qualifier_ui | Q000494 |
| mesh[6].descriptor_ui | D065690 |
| mesh[6].is_major_topic | False |
| mesh[6].qualifier_name | pharmacology |
| mesh[6].descriptor_name | Cytochrome P-450 CYP2D6 Inhibitors |
| mesh[7].qualifier_ui | Q000494 |
| mesh[7].descriptor_ui | D065692 |
| mesh[7].is_major_topic | False |
| mesh[7].qualifier_name | pharmacology |
| mesh[7].descriptor_name | Cytochrome P-450 CYP3A Inhibitors |
| mesh[8].qualifier_ui | Q000378 |
| mesh[8].descriptor_ui | D003577 |
| mesh[8].is_major_topic | False |
| mesh[8].qualifier_name | metabolism |
| mesh[8].descriptor_name | Cytochrome P-450 Enzyme System |
| mesh[9].qualifier_ui | Q000494 |
| mesh[9].descriptor_ui | D065607 |
| mesh[9].is_major_topic | False |
| mesh[9].qualifier_name | pharmacology |
| mesh[9].descriptor_name | Cytochrome P-450 Enzyme Inhibitors |
| mesh[10].qualifier_ui | Q000378 |
| mesh[10].descriptor_ui | D008862 |
| mesh[10].is_major_topic | False |
| mesh[10].qualifier_name | metabolism |
| mesh[10].descriptor_name | Microsomes, Liver |
| mesh[11].qualifier_ui | |
| mesh[11].descriptor_ui | D006801 |
| mesh[11].is_major_topic | False |
| mesh[11].qualifier_name | |
| mesh[11].descriptor_name | Humans |
| mesh[12].qualifier_ui | Q000378 |
| mesh[12].descriptor_ui | D019389 |
| mesh[12].is_major_topic | True |
| mesh[12].qualifier_name | metabolism |
| mesh[12].descriptor_name | Cytochrome P-450 CYP2D6 |
| mesh[13].qualifier_ui | Q000378 |
| mesh[13].descriptor_ui | D051544 |
| mesh[13].is_major_topic | True |
| mesh[13].qualifier_name | metabolism |
| mesh[13].descriptor_name | Cytochrome P-450 CYP3A |
| mesh[14].qualifier_ui | Q000378 |
| mesh[14].descriptor_ui | D006886 |
| mesh[14].is_major_topic | False |
| mesh[14].qualifier_name | metabolism |
| mesh[14].descriptor_name | Hydroxychloroquine |
| mesh[15].qualifier_ui | Q000494 |
| mesh[15].descriptor_ui | D006886 |
| mesh[15].is_major_topic | False |
| mesh[15].qualifier_name | pharmacology |
| mesh[15].descriptor_name | Hydroxychloroquine |
| mesh[16].qualifier_ui | Q000378 |
| mesh[16].descriptor_ui | D065727 |
| mesh[16].is_major_topic | False |
| mesh[16].qualifier_name | metabolism |
| mesh[16].descriptor_name | Cytochrome P-450 CYP2C8 |
| mesh[17].qualifier_ui | Q000494 |
| mesh[17].descriptor_ui | D065690 |
| mesh[17].is_major_topic | False |
| mesh[17].qualifier_name | pharmacology |
| mesh[17].descriptor_name | Cytochrome P-450 CYP2D6 Inhibitors |
| mesh[18].qualifier_ui | Q000494 |
| mesh[18].descriptor_ui | D065692 |
| mesh[18].is_major_topic | False |
| mesh[18].qualifier_name | pharmacology |
| mesh[18].descriptor_name | Cytochrome P-450 CYP3A Inhibitors |
| mesh[19].qualifier_ui | Q000378 |
| mesh[19].descriptor_ui | D003577 |
| mesh[19].is_major_topic | False |
| mesh[19].qualifier_name | metabolism |
| mesh[19].descriptor_name | Cytochrome P-450 Enzyme System |
| mesh[20].qualifier_ui | Q000494 |
| mesh[20].descriptor_ui | D065607 |
| mesh[20].is_major_topic | False |
| mesh[20].qualifier_name | pharmacology |
| mesh[20].descriptor_name | Cytochrome P-450 Enzyme Inhibitors |
| mesh[21].qualifier_ui | Q000378 |
| mesh[21].descriptor_ui | D008862 |
| mesh[21].is_major_topic | False |
| mesh[21].qualifier_name | metabolism |
| mesh[21].descriptor_name | Microsomes, Liver |
| mesh[22].qualifier_ui | |
| mesh[22].descriptor_ui | D006801 |
| mesh[22].is_major_topic | False |
| mesh[22].qualifier_name | |
| mesh[22].descriptor_name | Humans |
| mesh[23].qualifier_ui | Q000378 |
| mesh[23].descriptor_ui | D019389 |
| mesh[23].is_major_topic | True |
| mesh[23].qualifier_name | metabolism |
| mesh[23].descriptor_name | Cytochrome P-450 CYP2D6 |
| mesh[24].qualifier_ui | Q000378 |
| mesh[24].descriptor_ui | D051544 |
| mesh[24].is_major_topic | True |
| mesh[24].qualifier_name | metabolism |
| mesh[24].descriptor_name | Cytochrome P-450 CYP3A |
| mesh[25].qualifier_ui | Q000378 |
| mesh[25].descriptor_ui | D006886 |
| mesh[25].is_major_topic | False |
| mesh[25].qualifier_name | metabolism |
| mesh[25].descriptor_name | Hydroxychloroquine |
| mesh[26].qualifier_ui | Q000494 |
| mesh[26].descriptor_ui | D006886 |
| mesh[26].is_major_topic | False |
| mesh[26].qualifier_name | pharmacology |
| mesh[26].descriptor_name | Hydroxychloroquine |
| mesh[27].qualifier_ui | Q000378 |
| mesh[27].descriptor_ui | D065727 |
| mesh[27].is_major_topic | False |
| mesh[27].qualifier_name | metabolism |
| mesh[27].descriptor_name | Cytochrome P-450 CYP2C8 |
| mesh[28].qualifier_ui | Q000494 |
| mesh[28].descriptor_ui | D065690 |
| mesh[28].is_major_topic | False |
| mesh[28].qualifier_name | pharmacology |
| mesh[28].descriptor_name | Cytochrome P-450 CYP2D6 Inhibitors |
| mesh[29].qualifier_ui | Q000494 |
| mesh[29].descriptor_ui | D065692 |
| mesh[29].is_major_topic | False |
| mesh[29].qualifier_name | pharmacology |
| mesh[29].descriptor_name | Cytochrome P-450 CYP3A Inhibitors |
| mesh[30].qualifier_ui | Q000378 |
| mesh[30].descriptor_ui | D003577 |
| mesh[30].is_major_topic | False |
| mesh[30].qualifier_name | metabolism |
| mesh[30].descriptor_name | Cytochrome P-450 Enzyme System |
| mesh[31].qualifier_ui | Q000494 |
| mesh[31].descriptor_ui | D065607 |
| mesh[31].is_major_topic | False |
| mesh[31].qualifier_name | pharmacology |
| mesh[31].descriptor_name | Cytochrome P-450 Enzyme Inhibitors |
| mesh[32].qualifier_ui | Q000378 |
| mesh[32].descriptor_ui | D008862 |
| mesh[32].is_major_topic | False |
| mesh[32].qualifier_name | metabolism |
| mesh[32].descriptor_name | Microsomes, Liver |
| type | article |
| title | Hydroxychloroquine is Metabolized by Cytochrome P450 2D6, 3A4, and 2C8, and Inhibits Cytochrome P450 2D6, while its Metabolites also Inhibit Cytochrome P450 3A in vitro |
| biblio.issue | 3 |
| biblio.volume | 51 |
| biblio.last_page | 305 |
| biblio.first_page | 293 |
| topics[0].id | https://openalex.org/T10375 |
| topics[0].field.id | https://openalex.org/fields/30 |
| topics[0].field.display_name | Pharmacology, Toxicology and Pharmaceutics |
| topics[0].score | 0.9994000196456909 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/3004 |
| topics[0].subfield.display_name | Pharmacology |
| topics[0].display_name | Pharmacogenetics and Drug Metabolism |
| topics[1].id | https://openalex.org/T13467 |
| topics[1].field.id | https://openalex.org/fields/27 |
| topics[1].field.display_name | Medicine |
| topics[1].score | 0.995199978351593 |
| topics[1].domain.id | https://openalex.org/domains/4 |
| topics[1].domain.display_name | Health Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/2731 |
| topics[1].subfield.display_name | Ophthalmology |
| topics[1].display_name | Drug-Induced Ocular Toxicity |
| topics[2].id | https://openalex.org/T10570 |
| topics[2].field.id | https://openalex.org/fields/27 |
| topics[2].field.display_name | Medicine |
| topics[2].score | 0.9945999979972839 |
| topics[2].domain.id | https://openalex.org/domains/4 |
| topics[2].domain.display_name | Health Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/2730 |
| topics[2].subfield.display_name | Oncology |
| topics[2].display_name | Drug Transport and Resistance Mechanisms |
| is_xpac | False |
| apc_list | |
| apc_paid.value | 1382 |
| apc_paid.currency | EUR |
| apc_paid.value_usd | 1490 |
| concepts[0].id | https://openalex.org/C33664856 |
| concepts[0].level | 4 |
| concepts[0].score | 0.8154451847076416 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q3271142 |
| concepts[0].display_name | CYP2D6 |
| concepts[1].id | https://openalex.org/C109650736 |
| concepts[1].level | 4 |
| concepts[1].score | 0.7907806634902954 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q424440 |
| concepts[1].display_name | CYP3A4 |
| concepts[2].id | https://openalex.org/C526171541 |
| concepts[2].level | 3 |
| concepts[2].score | 0.7439920902252197 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q407693 |
| concepts[2].display_name | Cytochrome P450 |
| concepts[3].id | https://openalex.org/C115368759 |
| concepts[3].level | 5 |
| concepts[3].score | 0.6804777383804321 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q14916190 |
| concepts[3].display_name | CYP2C8 |
| concepts[4].id | https://openalex.org/C2777477808 |
| concepts[4].level | 2 |
| concepts[4].score | 0.6528418660163879 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q407595 |
| concepts[4].display_name | Metabolite |
| concepts[5].id | https://openalex.org/C98274493 |
| concepts[5].level | 1 |
| concepts[5].score | 0.6417601108551025 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q128406 |
| concepts[5].display_name | Pharmacology |
| concepts[6].id | https://openalex.org/C11824378 |
| concepts[6].level | 4 |
| concepts[6].score | 0.6235172748565674 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q5014970 |
| concepts[6].display_name | CYP3A |
| concepts[7].id | https://openalex.org/C2777752497 |
| concepts[7].level | 3 |
| concepts[7].score | 0.6006848216056824 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q286779 |
| concepts[7].display_name | IC50 |
| concepts[8].id | https://openalex.org/C185592680 |
| concepts[8].level | 0 |
| concepts[8].score | 0.5958168506622314 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q2329 |
| concepts[8].display_name | Chemistry |
| concepts[9].id | https://openalex.org/C87644729 |
| concepts[9].level | 3 |
| concepts[9].score | 0.5539921522140503 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q547502 |
| concepts[9].display_name | Microsome |
| concepts[10].id | https://openalex.org/C3946865 |
| concepts[10].level | 4 |
| concepts[10].score | 0.46785905957221985 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q5014962 |
| concepts[10].display_name | CYP2C9 |
| concepts[11].id | https://openalex.org/C50605568 |
| concepts[11].level | 5 |
| concepts[11].score | 0.46189820766448975 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q14912267 |
| concepts[11].display_name | CYP2B6 |
| concepts[12].id | https://openalex.org/C62231903 |
| concepts[12].level | 2 |
| concepts[12].score | 0.4497029781341553 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q1057 |
| concepts[12].display_name | Metabolism |
| concepts[13].id | https://openalex.org/C55493867 |
| concepts[13].level | 1 |
| concepts[13].score | 0.36945661902427673 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q7094 |
| concepts[13].display_name | Biochemistry |
| concepts[14].id | https://openalex.org/C181199279 |
| concepts[14].level | 2 |
| concepts[14].score | 0.3649066388607025 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q8047 |
| concepts[14].display_name | Enzyme |
| concepts[15].id | https://openalex.org/C202751555 |
| concepts[15].level | 2 |
| concepts[15].score | 0.28561121225357056 |
| concepts[15].wikidata | https://www.wikidata.org/wiki/Q221681 |
| concepts[15].display_name | In vitro |
| concepts[16].id | https://openalex.org/C86803240 |
| concepts[16].level | 0 |
| concepts[16].score | 0.24302104115486145 |
| concepts[16].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[16].display_name | Biology |
| keywords[0].id | https://openalex.org/keywords/cyp2d6 |
| keywords[0].score | 0.8154451847076416 |
| keywords[0].display_name | CYP2D6 |
| keywords[1].id | https://openalex.org/keywords/cyp3a4 |
| keywords[1].score | 0.7907806634902954 |
| keywords[1].display_name | CYP3A4 |
| keywords[2].id | https://openalex.org/keywords/cytochrome-p450 |
| keywords[2].score | 0.7439920902252197 |
| keywords[2].display_name | Cytochrome P450 |
| keywords[3].id | https://openalex.org/keywords/cyp2c8 |
| keywords[3].score | 0.6804777383804321 |
| keywords[3].display_name | CYP2C8 |
| keywords[4].id | https://openalex.org/keywords/metabolite |
| keywords[4].score | 0.6528418660163879 |
| keywords[4].display_name | Metabolite |
| keywords[5].id | https://openalex.org/keywords/pharmacology |
| keywords[5].score | 0.6417601108551025 |
| keywords[5].display_name | Pharmacology |
| keywords[6].id | https://openalex.org/keywords/cyp3a |
| keywords[6].score | 0.6235172748565674 |
| keywords[6].display_name | CYP3A |
| keywords[7].id | https://openalex.org/keywords/ic50 |
| keywords[7].score | 0.6006848216056824 |
| keywords[7].display_name | IC50 |
| keywords[8].id | https://openalex.org/keywords/chemistry |
| keywords[8].score | 0.5958168506622314 |
| keywords[8].display_name | Chemistry |
| keywords[9].id | https://openalex.org/keywords/microsome |
| keywords[9].score | 0.5539921522140503 |
| keywords[9].display_name | Microsome |
| keywords[10].id | https://openalex.org/keywords/cyp2c9 |
| keywords[10].score | 0.46785905957221985 |
| keywords[10].display_name | CYP2C9 |
| keywords[11].id | https://openalex.org/keywords/cyp2b6 |
| keywords[11].score | 0.46189820766448975 |
| keywords[11].display_name | CYP2B6 |
| keywords[12].id | https://openalex.org/keywords/metabolism |
| keywords[12].score | 0.4497029781341553 |
| keywords[12].display_name | Metabolism |
| keywords[13].id | https://openalex.org/keywords/biochemistry |
| keywords[13].score | 0.36945661902427673 |
| keywords[13].display_name | Biochemistry |
| keywords[14].id | https://openalex.org/keywords/enzyme |
| keywords[14].score | 0.3649066388607025 |
| keywords[14].display_name | Enzyme |
| keywords[15].id | https://openalex.org/keywords/in-vitro |
| keywords[15].score | 0.28561121225357056 |
| keywords[15].display_name | In vitro |
| keywords[16].id | https://openalex.org/keywords/biology |
| keywords[16].score | 0.24302104115486145 |
| keywords[16].display_name | Biology |
| language | en |
| locations[0].id | doi:10.1124/dmd.122.001018 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S145039780 |
| locations[0].source.issn | 0090-9556, 1521-009X |
| locations[0].source.type | journal |
| locations[0].source.is_oa | False |
| locations[0].source.issn_l | 0090-9556 |
| locations[0].source.is_core | True |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | Drug Metabolism and Disposition |
| locations[0].source.host_organization | https://openalex.org/P4310316283 |
| locations[0].source.host_organization_name | American Society for Pharmacology and Experimental Therapeutics |
| locations[0].source.host_organization_lineage | https://openalex.org/P4310316283 |
| locations[0].source.host_organization_lineage_names | American Society for Pharmacology and Experimental Therapeutics |
| locations[0].license | cc-by-nc |
| locations[0].pdf_url | https://dmd.aspetjournals.org/content/dmd/early/2022/11/29/dmd.122.001018.full.pdf |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | https://openalex.org/licenses/cc-by-nc |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | Drug Metabolism and Disposition |
| locations[0].landing_page_url | https://doi.org/10.1124/dmd.122.001018 |
| locations[1].id | pmid:36446607 |
| locations[1].is_oa | False |
| locations[1].source.id | https://openalex.org/S4306525036 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | PubMed |
| locations[1].source.host_organization | https://openalex.org/I1299303238 |
| locations[1].source.host_organization_name | National Institutes of Health |
| locations[1].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[1].license | |
| locations[1].pdf_url | |
| locations[1].version | publishedVersion |
| locations[1].raw_type | |
| locations[1].license_id | |
| locations[1].is_accepted | True |
| locations[1].is_published | True |
| locations[1].raw_source_name | Drug metabolism and disposition: the biological fate of chemicals |
| locations[1].landing_page_url | https://pubmed.ncbi.nlm.nih.gov/36446607 |
| locations[2].id | pmh:oai:helda.helsinki.fi:10138/355282 |
| locations[2].is_oa | False |
| locations[2].source.id | https://openalex.org/S4306401476 |
| locations[2].source.issn | |
| locations[2].source.type | repository |
| locations[2].source.is_oa | False |
| locations[2].source.issn_l | |
| locations[2].source.is_core | False |
| locations[2].source.is_in_doaj | False |
| locations[2].source.display_name | Helda (University of Helsinki) |
| locations[2].source.host_organization | https://openalex.org/I133731052 |
| locations[2].source.host_organization_name | University of Helsinki |
| locations[2].source.host_organization_lineage | https://openalex.org/I133731052 |
| locations[2].license | |
| locations[2].pdf_url | |
| locations[2].version | submittedVersion |
| locations[2].raw_type | Article |
| locations[2].license_id | |
| locations[2].is_accepted | False |
| locations[2].is_published | False |
| locations[2].raw_source_name | |
| locations[2].landing_page_url | http://hdl.handle.net/10138/355282 |
| locations[3].id | pmh:oai:pure.atira.dk:publications/5337b0f4-1108-4ca7-9e0a-f38e07756afc |
| locations[3].is_oa | True |
| locations[3].source.id | https://openalex.org/S4306400216 |
| locations[3].source.issn | |
| locations[3].source.type | repository |
| locations[3].source.is_oa | False |
| locations[3].source.issn_l | |
| locations[3].source.is_core | False |
| locations[3].source.is_in_doaj | False |
| locations[3].source.display_name | Research Portal (King's College London) |
| locations[3].source.host_organization | https://openalex.org/I183935753 |
| locations[3].source.host_organization_name | King's College London |
| locations[3].source.host_organization_lineage | https://openalex.org/I183935753 |
| locations[3].license | cc-by-nc |
| locations[3].pdf_url | |
| locations[3].version | submittedVersion |
| locations[3].raw_type | article |
| locations[3].license_id | https://openalex.org/licenses/cc-by-nc |
| locations[3].is_accepted | False |
| locations[3].is_published | False |
| locations[3].raw_source_name | Paludetto , M-N , Kurkela , M , Kahma , H , Backman , J T , Niemi , M & Filppula , A M 2022 , ' Hydroxychloroquine is metabolized by CYP2D6, CYP3A4, and CYP2C8, and inhibits CYP2D6, while its metabolites also inhibit CYP3A in vitro ' , Drug Metabolism and Disposition . https://doi.org/10.1124/dmd.122.001018 |
| locations[3].landing_page_url | https://research.abo.fi/en/publications/5337b0f4-1108-4ca7-9e0a-f38e07756afc |
| locations[4].id | pmh:oai:pure.atira.dk:publications/f811d00e-4379-4217-9e1c-1d3a191892ea |
| locations[4].is_oa | False |
| locations[4].source.id | https://openalex.org/S4306400216 |
| locations[4].source.issn | |
| locations[4].source.type | repository |
| locations[4].source.is_oa | False |
| locations[4].source.issn_l | |
| locations[4].source.is_core | False |
| locations[4].source.is_in_doaj | False |
| locations[4].source.display_name | Research Portal (King's College London) |
| locations[4].source.host_organization | https://openalex.org/I183935753 |
| locations[4].source.host_organization_name | King's College London |
| locations[4].source.host_organization_lineage | https://openalex.org/I183935753 |
| locations[4].license | |
| locations[4].pdf_url | |
| locations[4].version | submittedVersion |
| locations[4].raw_type | article |
| locations[4].license_id | |
| locations[4].is_accepted | False |
| locations[4].is_published | False |
| locations[4].raw_source_name | Paludetto , M-N , Kurkela , M , Kahma , H , Backman , J T , Niemi , M & Filppula , A 2022 , ' Hydroxychloroquine is metabolized by CYP2D6, CYP3A4, and CYP2C8, and inhibits CYP2D6, while its metabolites also inhibit CYP3A in vitro ' , Drug Metabolism and Disposition . https://doi.org/10.1124/dmd.122.001018 |
| locations[4].landing_page_url | https://research.manchester.ac.uk/en/publications/f811d00e-4379-4217-9e1c-1d3a191892ea |
| indexed_in | crossref, pubmed |
| authorships[0].author.id | https://openalex.org/A5015557556 |
| authorships[0].author.orcid | https://orcid.org/0000-0002-8053-6889 |
| authorships[0].author.display_name | Marie‐Noëlle Paludetto |
| authorships[0].countries | FI |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I133731052 |
| authorships[0].affiliations[0].raw_affiliation_string | Department of Clinical Pharmacology, University of Helsinki, Finland |
| authorships[0].institutions[0].id | https://openalex.org/I133731052 |
| authorships[0].institutions[0].ror | https://ror.org/040af2s02 |
| authorships[0].institutions[0].type | education |
| authorships[0].institutions[0].lineage | https://openalex.org/I133731052 |
| authorships[0].institutions[0].country_code | FI |
| authorships[0].institutions[0].display_name | University of Helsinki |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Marie-Noëlle Paludetto |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Department of Clinical Pharmacology, University of Helsinki, Finland |
| authorships[1].author.id | https://openalex.org/A5000481959 |
| authorships[1].author.orcid | https://orcid.org/0000-0002-4153-4143 |
| authorships[1].author.display_name | Mika Kurkela |
| authorships[1].countries | FI |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I133731052 |
| authorships[1].affiliations[0].raw_affiliation_string | University of Helsinki, Finland |
| authorships[1].institutions[0].id | https://openalex.org/I133731052 |
| authorships[1].institutions[0].ror | https://ror.org/040af2s02 |
| authorships[1].institutions[0].type | education |
| authorships[1].institutions[0].lineage | https://openalex.org/I133731052 |
| authorships[1].institutions[0].country_code | FI |
| authorships[1].institutions[0].display_name | University of Helsinki |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Mika Kurkela |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | University of Helsinki, Finland |
| authorships[2].author.id | https://openalex.org/A5019961443 |
| authorships[2].author.orcid | https://orcid.org/0000-0003-1972-7368 |
| authorships[2].author.display_name | Helinä Kahma |
| authorships[2].countries | FI |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I133731052 |
| authorships[2].affiliations[0].raw_affiliation_string | Department of Clinical Pharmacology, University of Helsinki, Finland |
| authorships[2].institutions[0].id | https://openalex.org/I133731052 |
| authorships[2].institutions[0].ror | https://ror.org/040af2s02 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I133731052 |
| authorships[2].institutions[0].country_code | FI |
| authorships[2].institutions[0].display_name | University of Helsinki |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Helinä Kahma |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | Department of Clinical Pharmacology, University of Helsinki, Finland |
| authorships[3].author.id | https://openalex.org/A5017096947 |
| authorships[3].author.orcid | https://orcid.org/0000-0002-9577-2788 |
| authorships[3].author.display_name | Janne T. Backman |
| authorships[3].countries | FI |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I133731052 |
| authorships[3].affiliations[0].raw_affiliation_string | Department of Clinical Pharmacology, University of Helsinki, Finland |
| authorships[3].institutions[0].id | https://openalex.org/I133731052 |
| authorships[3].institutions[0].ror | https://ror.org/040af2s02 |
| authorships[3].institutions[0].type | education |
| authorships[3].institutions[0].lineage | https://openalex.org/I133731052 |
| authorships[3].institutions[0].country_code | FI |
| authorships[3].institutions[0].display_name | University of Helsinki |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Janne T. Backman |
| authorships[3].is_corresponding | False |
| authorships[3].raw_affiliation_strings | Department of Clinical Pharmacology, University of Helsinki, Finland |
| authorships[4].author.id | https://openalex.org/A5047851431 |
| authorships[4].author.orcid | https://orcid.org/0000-0003-4550-2189 |
| authorships[4].author.display_name | Mikko Niemi |
| authorships[4].countries | FI |
| authorships[4].affiliations[0].institution_ids | https://openalex.org/I133731052 |
| authorships[4].affiliations[0].raw_affiliation_string | Department of Clinical Pharmacology, University of Helsinki, Finland |
| authorships[4].institutions[0].id | https://openalex.org/I133731052 |
| authorships[4].institutions[0].ror | https://ror.org/040af2s02 |
| authorships[4].institutions[0].type | education |
| authorships[4].institutions[0].lineage | https://openalex.org/I133731052 |
| authorships[4].institutions[0].country_code | FI |
| authorships[4].institutions[0].display_name | University of Helsinki |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Mikko Niemi |
| authorships[4].is_corresponding | False |
| authorships[4].raw_affiliation_strings | Department of Clinical Pharmacology, University of Helsinki, Finland |
| authorships[5].author.id | https://openalex.org/A5032494672 |
| authorships[5].author.orcid | https://orcid.org/0000-0002-7932-8265 |
| authorships[5].author.display_name | Anne M. Filppula |
| authorships[5].countries | FI |
| authorships[5].affiliations[0].institution_ids | https://openalex.org/I133731052 |
| authorships[5].affiliations[0].raw_affiliation_string | Department of Clincial Pharmacology, University of Helsinki, Finland |
| authorships[5].institutions[0].id | https://openalex.org/I133731052 |
| authorships[5].institutions[0].ror | https://ror.org/040af2s02 |
| authorships[5].institutions[0].type | education |
| authorships[5].institutions[0].lineage | https://openalex.org/I133731052 |
| authorships[5].institutions[0].country_code | FI |
| authorships[5].institutions[0].display_name | University of Helsinki |
| authorships[5].author_position | last |
| authorships[5].raw_author_name | Anne M. Filppula |
| authorships[5].is_corresponding | False |
| authorships[5].raw_affiliation_strings | Department of Clincial Pharmacology, University of Helsinki, Finland |
| has_content.pdf | True |
| has_content.grobid_xml | True |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://dmd.aspetjournals.org/content/dmd/early/2022/11/29/dmd.122.001018.full.pdf |
| open_access.oa_status | hybrid |
| open_access.any_repository_has_fulltext | False |
| created_date | 2022-12-10T00:00:00 |
| display_name | Hydroxychloroquine is Metabolized by Cytochrome P450 2D6, 3A4, and 2C8, and Inhibits Cytochrome P450 2D6, while its Metabolites also Inhibit Cytochrome P450 3A in vitro |
| has_fulltext | True |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T10375 |
| primary_topic.field.id | https://openalex.org/fields/30 |
| primary_topic.field.display_name | Pharmacology, Toxicology and Pharmaceutics |
| primary_topic.score | 0.9994000196456909 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/3004 |
| primary_topic.subfield.display_name | Pharmacology |
| primary_topic.display_name | Pharmacogenetics and Drug Metabolism |
| related_works | https://openalex.org/W2082257800, https://openalex.org/W2378522772, https://openalex.org/W1797160017, https://openalex.org/W2567759360, https://openalex.org/W2804369996, https://openalex.org/W4206155619, https://openalex.org/W2068780263, https://openalex.org/W4213331117, https://openalex.org/W2115864561, https://openalex.org/W2985461840 |
| cited_by_count | 18 |
| counts_by_year[0].year | 2025 |
| counts_by_year[0].cited_by_count | 8 |
| counts_by_year[1].year | 2024 |
| counts_by_year[1].cited_by_count | 6 |
| counts_by_year[2].year | 2023 |
| counts_by_year[2].cited_by_count | 4 |
| locations_count | 5 |
| best_oa_location.id | doi:10.1124/dmd.122.001018 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S145039780 |
| best_oa_location.source.issn | 0090-9556, 1521-009X |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | False |
| best_oa_location.source.issn_l | 0090-9556 |
| best_oa_location.source.is_core | True |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | Drug Metabolism and Disposition |
| best_oa_location.source.host_organization | https://openalex.org/P4310316283 |
| best_oa_location.source.host_organization_name | American Society for Pharmacology and Experimental Therapeutics |
| best_oa_location.source.host_organization_lineage | https://openalex.org/P4310316283 |
| best_oa_location.source.host_organization_lineage_names | American Society for Pharmacology and Experimental Therapeutics |
| best_oa_location.license | cc-by-nc |
| best_oa_location.pdf_url | https://dmd.aspetjournals.org/content/dmd/early/2022/11/29/dmd.122.001018.full.pdf |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | journal-article |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by-nc |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | Drug Metabolism and Disposition |
| best_oa_location.landing_page_url | https://doi.org/10.1124/dmd.122.001018 |
| primary_location.id | doi:10.1124/dmd.122.001018 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S145039780 |
| primary_location.source.issn | 0090-9556, 1521-009X |
| primary_location.source.type | journal |
| primary_location.source.is_oa | False |
| primary_location.source.issn_l | 0090-9556 |
| primary_location.source.is_core | True |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | Drug Metabolism and Disposition |
| primary_location.source.host_organization | https://openalex.org/P4310316283 |
| primary_location.source.host_organization_name | American Society for Pharmacology and Experimental Therapeutics |
| primary_location.source.host_organization_lineage | https://openalex.org/P4310316283 |
| primary_location.source.host_organization_lineage_names | American Society for Pharmacology and Experimental Therapeutics |
| primary_location.license | cc-by-nc |
| primary_location.pdf_url | https://dmd.aspetjournals.org/content/dmd/early/2022/11/29/dmd.122.001018.full.pdf |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | https://openalex.org/licenses/cc-by-nc |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | Drug Metabolism and Disposition |
| primary_location.landing_page_url | https://doi.org/10.1124/dmd.122.001018 |
| publication_date | 2022-11-30 |
| publication_year | 2022 |
| referenced_works | https://openalex.org/W1985608865, https://openalex.org/W2799884779, https://openalex.org/W2770835683, https://openalex.org/W2790129692, https://openalex.org/W1593006094, https://openalex.org/W2116072674, https://openalex.org/W3091868035, https://openalex.org/W3115773889, https://openalex.org/W3135930892, https://openalex.org/W2121960436, https://openalex.org/W2007714746, https://openalex.org/W2253161284, https://openalex.org/W3036839244, https://openalex.org/W2121978955, https://openalex.org/W3182522906, https://openalex.org/W2071235764, https://openalex.org/W2013425106, https://openalex.org/W2970961444, https://openalex.org/W1968955854, https://openalex.org/W3110466220, https://openalex.org/W2006532982, https://openalex.org/W2802671657, https://openalex.org/W2061152607, https://openalex.org/W2133013045, https://openalex.org/W1959216194, https://openalex.org/W3004709581, https://openalex.org/W6843052851, https://openalex.org/W3043308024, https://openalex.org/W1929399969, https://openalex.org/W2762519124, https://openalex.org/W3118373206, https://openalex.org/W2529506256, https://openalex.org/W6690802718, https://openalex.org/W2005937589, https://openalex.org/W2031026401, https://openalex.org/W2137059909, https://openalex.org/W3088354768, https://openalex.org/W3159545476, https://openalex.org/W2003606483, https://openalex.org/W3089088198, https://openalex.org/W3082786135, https://openalex.org/W2106068866, https://openalex.org/W2152796502, https://openalex.org/W3136301451, https://openalex.org/W4296026703, https://openalex.org/W4242164740, https://openalex.org/W2246595845 |
| referenced_works_count | 47 |
| abstract_inverted_index.> | 96 |
| abstract_inverted_index.a | 160 |
| abstract_inverted_index.14 | 128 |
| abstract_inverted_index.18 | 188 |
| abstract_inverted_index.In | 165, 283 |
| abstract_inverted_index.To | 208 |
| abstract_inverted_index.an | 46 |
| abstract_inverted_index.be | 238 |
| abstract_inverted_index.by | 92, 101, 163, 213 |
| abstract_inverted_index.in | 43, 65, 116, 156, 218, 275, 308 |
| abstract_inverted_index.is | 211, 286 |
| abstract_inverted_index.it | 267 |
| abstract_inverted_index.of | 13, 32, 89, 108, 135, 272, 293, 322 |
| abstract_inverted_index.on | 37 |
| abstract_inverted_index.to | 3, 80, 112, 240, 263 |
| abstract_inverted_index.we | 296 |
| abstract_inverted_index.(11 | 126 |
| abstract_inverted_index.135 | 190 |
| abstract_inverted_index.5.7 | 138 |
| abstract_inverted_index.50% | 182 |
| abstract_inverted_index.All | 68 |
| abstract_inverted_index.CYP | 27, 110, 166, 273, 290 |
| abstract_inverted_index.HCQ | 33, 66, 90, 157, 192, 210, 220, 229, 309, 316, 332 |
| abstract_inverted_index.HLM | 113, 119, 133, 142 |
| abstract_inverted_index.Our | 51 |
| abstract_inverted_index.The | 29, 131 |
| abstract_inverted_index.all | 199 |
| abstract_inverted_index.and | 10, 25, 34, 58, 77, 124, 127, 154, 172, 189, 216, 221, 228, 245, 258, 301, 317, 333 |
| abstract_inverted_index.are | 60, 224, 231, 303, 320 |
| abstract_inverted_index.can | 237 |
| abstract_inverted_index.did | 193 |
| abstract_inverted_index.for | 122, 152, 251 |
| abstract_inverted_index.has | 279 |
| abstract_inverted_index.its | 35, 222, 318 |
| abstract_inverted_index.key | 62, 305 |
| abstract_inverted_index.not | 194, 280 |
| abstract_inverted_index.our | 312 |
| abstract_inverted_index.the | 5, 61, 72, 84, 173, 270, 289, 304, 327 |
| abstract_inverted_index.was | 18, 95, 137 |
| abstract_inverted_index.CYPs | 39, 70 |
| abstract_inverted_index.DCQ, | 170 |
| abstract_inverted_index.Data | 140 |
| abstract_inverted_index.HCQ, | 169 |
| abstract_inverted_index.HCQ. | 252, 294 |
| abstract_inverted_index.P450 | 7 |
| abstract_inverted_index.This | 0 |
| abstract_inverted_index.also | 41, 147 |
| abstract_inverted_index.been | 261, 281 |
| abstract_inverted_index.data | 53, 236 |
| abstract_inverted_index.from | 141 |
| abstract_inverted_index.have | 260 |
| abstract_inverted_index.nine | 38 |
| abstract_inverted_index.risk | 249 |
| abstract_inverted_index.role | 271 |
| abstract_inverted_index.show | 314 |
| abstract_inverted_index.than | 99 |
| abstract_inverted_index.that | 55, 100, 269, 298, 315 |
| abstract_inverted_index.used | 239 |
| abstract_inverted_index.were | 40, 177, 201 |
| abstract_inverted_index.with | 144, 159, 181 |
| abstract_inverted_index.(0.87 | 103 |
| abstract_inverted_index.(CYP) | 8 |
| abstract_inverted_index.(DCQ) | 76 |
| abstract_inverted_index.(HCQ) | 277 |
| abstract_inverted_index.0.075 | 105 |
| abstract_inverted_index.CYP3A | 203, 233 |
| abstract_inverted_index.CYPs. | 197 |
| abstract_inverted_index.DHCQ, | 171 |
| abstract_inverted_index.HLMs, | 44 |
| abstract_inverted_index.Here, | 295 |
| abstract_inverted_index.These | 235 |
| abstract_inverted_index.While | 255 |
| abstract_inverted_index.about | 288 |
| abstract_inverted_index.aimed | 2 |
| abstract_inverted_index.equal | 118, 150 |
| abstract_inverted_index.human | 21 |
| abstract_inverted_index.known | 287 |
| abstract_inverted_index.level | 114 |
| abstract_inverted_index.liver | 22 |
| abstract_inverted_index.other | 196 |
| abstract_inverted_index.roles | 151 |
| abstract_inverted_index.shift | 206 |
| abstract_inverted_index.shown | 262 |
| abstract_inverted_index.study | 1 |
| abstract_inverted_index.three | 69 |
| abstract_inverted_index.using | 20, 45 |
| abstract_inverted_index.value | 88 |
| abstract_inverted_index.which | 324 |
| abstract_inverted_index.(DHCQ) | 79 |
| abstract_inverted_index.(HCQ). | 15 |
| abstract_inverted_index.(HLMs) | 24 |
| abstract_inverted_index.CYP2C8 | 59, 136, 217, 259, 302 |
| abstract_inverted_index.CYP2D6 | 94, 123, 153, 179, 226 |
| abstract_inverted_index.CYP3A4 | 102, 125, 155 |
| abstract_inverted_index.almost | 117 |
| abstract_inverted_index.direct | 178 |
| abstract_inverted_index.formed | 71 |
| abstract_inverted_index.higher | 98 |
| abstract_inverted_index.likely | 325 |
| abstract_inverted_index.little | 285 |
| abstract_inverted_index.models | 244 |
| abstract_inverted_index.scaled | 132 |
| abstract_inverted_index.update | 246 |
| abstract_inverted_index.values | 121, 186 |
| abstract_inverted_index.vitro. | 219 |
| abstract_inverted_index.10-fold | 97 |
| abstract_inverted_index.CYP2C8. | 164 |
| abstract_inverted_index.CYP2D6, | 57, 215, 256, 299, 323 |
| abstract_inverted_index.CYP3A4, | 56, 214, 257, 300 |
| abstract_inverted_index.appears | 268 |
| abstract_inverted_index.between | 187, 331 |
| abstract_inverted_index.effects | 31, 292 |
| abstract_inverted_index.enzymes | 63, 274, 306 |
| abstract_inverted_index.explore | 4 |
| abstract_inverted_index.improve | 241 |
| abstract_inverted_index.inhibit | 195 |
| abstract_inverted_index.mediate | 264 |
| abstract_inverted_index.method. | 50 |
| abstract_inverted_index.primary | 73 |
| abstract_inverted_index.profile | 12 |
| abstract_inverted_index.scaling | 107 |
| abstract_inverted_index.smaller | 161 |
| abstract_inverted_index.studied | 19 |
| abstract_inverted_index.various | 81 |
| abstract_inverted_index.Although | 83 |
| abstract_inverted_index.cocktail | 49 |
| abstract_inverted_index.degrees. | 82 |
| abstract_inverted_index.enzymes. | 28 |
| abstract_inverted_index.explains | 326 |
| abstract_inverted_index.findings | 313 |
| abstract_inverted_index.involved | 64, 307 |
| abstract_inverted_index.observed | 329 |
| abstract_inverted_index.resulted | 115 |
| abstract_inverted_index.studied. | 282 |
| abstract_inverted_index.2.2-3.4). | 207 |
| abstract_inverted_index.addition, | 284 |
| abstract_inverted_index.automated | 47 |
| abstract_inverted_index.clearance | 86 |
| abstract_inverted_index.conclude, | 209 |
| abstract_inverted_index.depletion | 91 |
| abstract_inverted_index.drug-drug | 247 |
| abstract_inverted_index.indicated | 54 |
| abstract_inverted_index.intrinsic | 85 |
| abstract_inverted_index.metabolic | 9 |
| abstract_inverted_index.secondary | 174 |
| abstract_inverted_index.substrate | 48 |
| abstract_inverted_index.suggested | 148 |
| abstract_inverted_index.STATEMENT: | 254 |
| abstract_inverted_index.cytochrome | 6 |
| abstract_inverted_index.determined | 42 |
| abstract_inverted_index.inhibition | 167 |
| abstract_inverted_index.inhibitors | 146, 204, 321 |
| abstract_inverted_index.inhibitory | 11, 30, 183, 291 |
| abstract_inverted_index.metabolism | 17, 52, 278 |
| abstract_inverted_index.metabolite | 175 |
| abstract_inverted_index.microsomes | 23 |
| abstract_inverted_index.previously | 328 |
| abstract_inverted_index.relatively | 149 |
| abstract_inverted_index.reversible | 225 |
| abstract_inverted_index.<i>µ</i>M. | 191 |
| abstract_inverted_index.chloroquine | 265 |
| abstract_inverted_index.demonstrate | 297 |
| abstract_inverted_index.estimations | 250 |
| abstract_inverted_index.experiments | 143 |
| abstract_inverted_index.inhibitors, | 180, 227 |
| abstract_inverted_index.inhibitors. | 234 |
| abstract_inverted_index.interaction | 248, 330 |
| abstract_inverted_index.metabolism, | 158 |
| abstract_inverted_index.metabolism. | 67, 310 |
| abstract_inverted_index.metabolites | 36, 74, 200, 223, 230, 319 |
| abstract_inverted_index.metabolized | 212 |
| abstract_inverted_index.metoprolol. | 334 |
| abstract_inverted_index.recombinant | 26, 93, 109 |
| abstract_inverted_index.Furthermore, | 198, 311 |
| abstract_inverted_index.SIGNIFICANCE | 253 |
| abstract_inverted_index.contribution | 162 |
| abstract_inverted_index.experiments, | 168 |
| abstract_inverted_index.<i>versus</i> | 104 |
| abstract_inverted_index.CYP-selective | 145 |
| abstract_inverted_index.concentration | 184 |
| abstract_inverted_index.respectively). | 130 |
| abstract_inverted_index.time-dependent | 202, 232 |
| abstract_inverted_index.pharmacokinetic | 243 |
| abstract_inverted_index.(IC<sub>50</sub> | 205 |
| abstract_inverted_index.CL<sub>int</sub> | 111, 120, 134 |
| abstract_inverted_index.(IC<sub>50</sub>) | 185 |
| abstract_inverted_index.(CL<sub>int</sub>) | 87 |
| abstract_inverted_index.<i>µ</i>l/min/mg, | 129 |
| abstract_inverted_index.<i>µ</i>l/min/mg. | 139 |
| abstract_inverted_index.Hydroxychloroquine | 16 |
| abstract_inverted_index.biotransformation, | 266 |
| abstract_inverted_index.hydroxychloroquine | 14, 276 |
| abstract_inverted_index.desethylchloroquine | 75 |
| abstract_inverted_index.<i>µ</i>l/min/pmol), | 106 |
| abstract_inverted_index.didesethylchloroquine | 176 |
| abstract_inverted_index.physiologically-based | 242 |
| abstract_inverted_index.desethylhydroxychloroquine | 78 |
| cited_by_percentile_year.max | 99 |
| cited_by_percentile_year.min | 97 |
| countries_distinct_count | 1 |
| institutions_distinct_count | 6 |
| citation_normalized_percentile.value | 0.91736456 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | True |