Genome-wide association and genomic prediction of resistance to viral nervous necrosis in European sea bass (Dicentrarchus labrax) using RAD sequencing Article Swipe
Related Concepts
Dicentrarchus
Biology
Sea bass
Genome
Genome-wide association study
DNA sequencing
Evolutionary biology
Genomic selection
Genetics
Computational biology
Fishery
Single-nucleotide polymorphism
Fish <Actinopterygii>
Gene
Genotype
Christos Palaiokostas
,
Sophie Cariou
,
Anastasia Bestin
,
Jean-Sébastien Bruant
,
Pierrick Haffray
,
Thierry Morin
,
Joëlle Cabon
,
François Allal
,
Marc Vandeputte
,
Ross D. Houston
·
 YOU?
·
· 2018
· Open Access
·
· DOI: https://doi.org/10.1186/s12711-018-0401-2
· OA: W2805462056
YOU?
·
· 2018
· Open Access
·
· DOI: https://doi.org/10.1186/s12711-018-0401-2
· OA: W2805462056
 YOU?
·
· 2018
· Open Access
·
· DOI: https://doi.org/10.1186/s12711-018-0401-2
· OA: W2805462056
YOU?
·
· 2018
· Open Access
·
· DOI: https://doi.org/10.1186/s12711-018-0401-2
· OA: W2805462056
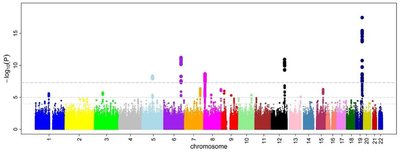
Genome-wide significant QTL were identified but each with relatively small effects on the trait. Tests of genomic prediction suggested that incorporating genome-wide SNP data is likely to result in higher accuracy of estimated breeding values for resistance to VNN. RAD sequencing is an effective method for generating such genome-wide SNPs, and our findings highlight the potential of genomic selection to breed farmed European sea bass with improved resistance to VNN.
Related Topics
Finding more related topics…