Identification of gene-sex hormone interactions associated with type 2 diabetes among men and women Article Swipe
 YOU?
·
· 2025
· Open Access
·
· DOI: https://doi.org/10.1371/journal.pgen.1011470
· OA: W4413924685
YOU?
·
· 2025
· Open Access
·
· DOI: https://doi.org/10.1371/journal.pgen.1011470
· OA: W4413924685
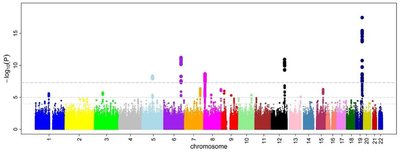
Background and Objectives Type 2 diabetes affects an increasing number of people worldwide. Although genome-wide association studies (GWAS) of type 2 diabetes have identified hundreds of loci, their interactions with other risk factors aren’t well understood. We investigated genetic interactions with three sex hormones (total testosterone, bioavailable testosterone, and sex hormone binding globulin (SHBG)) to identify additional type 2 diabetes-related loci that were undetected in traditional GWAS. Methods The study population consisted of white European UK Biobank participants. Individuals with type 1 diabetes were excluded. We examined sex-stratified interactions of polygenic risk score (PRS) for type 2 diabetes with sex hormone levels. We analyzed sex-stratified, genome-wide SNP × sex hormone interactions, adjusting for age and the top ten principal ancestry components. Results We found significant (P < 0.05) interactions for each of the sex hormones with PRS in both men and women, with the most significant being between SHBG and PRS in women (OR 0.88; 95% CI: 0.85-0.90; P = 1.09E-18). We identified 3 SNP × sex hormone interactions in men and 14 in women that achieved genome-wide significance (GWS; P < 5 × 10 -8 ). Applying a 2-degree of freedom test, we identified GWS loci (10 in men and 23 in women) that were not GWS when testing marginal genetic effects alone. Conclusion Including interaction terms in GWAS may identify additional risk loci and improve the understanding of genetic architecture for type 2 diabetes. Different genetic interactions with sex hormones in men and women emphasize the importance of sex-stratified analysis in sex differential diseases.