Imputation and polygenic score performances of human genotyping arrays in diverse populations Article Swipe
 YOU?
·
· 2022
· Open Access
·
· DOI: https://doi.org/10.1101/2022.06.14.496059
YOU?
·
· 2022
· Open Access
·
· DOI: https://doi.org/10.1101/2022.06.14.496059
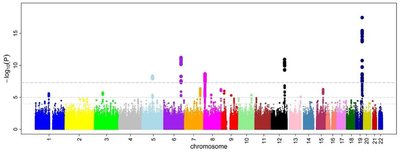
Regardless of the overwhelming use of next-generation sequencing technologies, microarray-based genotyping combined with the imputation of untyped variants remains a cost-effective means to interrogate genetic variations across the human genome. This technology is widely used in genome-wide association studies (GWAS) at bio-bank scales, and more recently, in polygenic score (PGS) analysis to predict and to stratify disease risk. Over the last decade, human genotyping arrays have undergone a tremendous growth in both number, and content making a comprehensive evaluation of their performances became more important. Here, we performed a comprehensive performance assessment for 23 available human genotyping arrays in 6 ancestry groups using diverse public, and in-house datasets. The analyses focus on performance estimation of derived imputation (in terms of accuracy and coverage) and PGS (in term of concordance to PGS estimated from whole genome sequencing data) in three different traits and diseases. We found that the arrays with a higher number of SNPs are not necessarily the ones with higher imputation performance, but the arrays that are well-optimized for the targeted population could provide very good imputation performance. In addition, PGS estimated by imputed SNP array data is highly correlated to PGS estimated by whole genome sequencing data in most of cases. When optimal arrays are used, the correlations of key PGS metrics between two types of data can be higher than 0.97, but interestingly, arrays with high density can result in lower PGS performance. Our results suggest the importance of properly selecting a suitable genotyping array for PGS applications. Finally, we developed a web tool that provide interactive analyses of tag SNP contents and imputation performance based on population and genomic regions of interest. This study would act as a practical guide for researchers to design their genotyping arrays-based studies. The tool is available at: https://genome.vinbigdata.org/tools/saa/
Related Topics
- Type
- preprint
- Language
- en
- Landing Page
- https://doi.org/10.1101/2022.06.14.496059
- https://www.biorxiv.org/content/biorxiv/early/2022/06/15/2022.06.14.496059.full.pdf
- OA Status
- green
- References
- 47
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4282918349
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4282918349Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1101/2022.06.14.496059Digital Object Identifier
- Title
-
Imputation and polygenic score performances of human genotyping arrays in diverse populationsWork title
- Type
-
preprintOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2022Year of publication
- Publication date
-
2022-06-15Full publication date if available
- Authors
-
Dat Nguyen, Trang Tran, Mai Tran, Khai Tran, Duy T. Pham, Nguyễn Thùy Dương, Quan Nguyen, Nam S. VoList of authors in order
- Landing page
-
https://doi.org/10.1101/2022.06.14.496059Publisher landing page
- PDF URL
-
https://www.biorxiv.org/content/biorxiv/early/2022/06/15/2022.06.14.496059.full.pdfDirect link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
greenOpen access status per OpenAlex
- OA URL
-
https://www.biorxiv.org/content/biorxiv/early/2022/06/15/2022.06.14.496059.full.pdfDirect OA link when available
- Concepts
-
Imputation (statistics), Genotyping, Genome-wide association study, Concordance, Biology, Single-nucleotide polymorphism, Computational biology, Genetic association, SNP genotyping, 1000 Genomes Project, Human genome, SNP, Genome, Genetics, Computer science, Statistics, Genotype, Missing data, Machine learning, Gene, MathematicsTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
0Total citation count in OpenAlex
- References (count)
-
47Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4282918349 |
|---|---|
| doi | https://doi.org/10.1101/2022.06.14.496059 |
| ids.doi | https://doi.org/10.1101/2022.06.14.496059 |
| ids.openalex | https://openalex.org/W4282918349 |
| fwci | |
| type | preprint |
| title | Imputation and polygenic score performances of human genotyping arrays in diverse populations |
| biblio.issue | |
| biblio.volume | |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T10261 |
| topics[0].field.id | https://openalex.org/fields/13 |
| topics[0].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[0].score | 0.9998999834060669 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1311 |
| topics[0].subfield.display_name | Genetics |
| topics[0].display_name | Genetic Associations and Epidemiology |
| topics[1].id | https://openalex.org/T11468 |
| topics[1].field.id | https://openalex.org/fields/13 |
| topics[1].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[1].score | 0.9994000196456909 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1311 |
| topics[1].subfield.display_name | Genetics |
| topics[1].display_name | Genetic Mapping and Diversity in Plants and Animals |
| topics[2].id | https://openalex.org/T10594 |
| topics[2].field.id | https://openalex.org/fields/13 |
| topics[2].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[2].score | 0.9988999962806702 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1311 |
| topics[2].subfield.display_name | Genetics |
| topics[2].display_name | Genetic and phenotypic traits in livestock |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C58041806 |
| concepts[0].level | 3 |
| concepts[0].score | 0.8741490244865417 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q1660484 |
| concepts[0].display_name | Imputation (statistics) |
| concepts[1].id | https://openalex.org/C31467283 |
| concepts[1].level | 4 |
| concepts[1].score | 0.8323341608047485 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q912147 |
| concepts[1].display_name | Genotyping |
| concepts[2].id | https://openalex.org/C106208931 |
| concepts[2].level | 5 |
| concepts[2].score | 0.6342840790748596 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q1098876 |
| concepts[2].display_name | Genome-wide association study |
| concepts[3].id | https://openalex.org/C160798450 |
| concepts[3].level | 2 |
| concepts[3].score | 0.5968185067176819 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q4230870 |
| concepts[3].display_name | Concordance |
| concepts[4].id | https://openalex.org/C86803240 |
| concepts[4].level | 0 |
| concepts[4].score | 0.5523102879524231 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[4].display_name | Biology |
| concepts[5].id | https://openalex.org/C153209595 |
| concepts[5].level | 4 |
| concepts[5].score | 0.549034059047699 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q501128 |
| concepts[5].display_name | Single-nucleotide polymorphism |
| concepts[6].id | https://openalex.org/C70721500 |
| concepts[6].level | 1 |
| concepts[6].score | 0.4772919714450836 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q177005 |
| concepts[6].display_name | Computational biology |
| concepts[7].id | https://openalex.org/C186413461 |
| concepts[7].level | 5 |
| concepts[7].score | 0.466608464717865 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q744727 |
| concepts[7].display_name | Genetic association |
| concepts[8].id | https://openalex.org/C163691529 |
| concepts[8].level | 5 |
| concepts[8].score | 0.4646090567111969 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q7391886 |
| concepts[8].display_name | SNP genotyping |
| concepts[9].id | https://openalex.org/C97425143 |
| concepts[9].level | 5 |
| concepts[9].score | 0.4518839120864868 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q164786 |
| concepts[9].display_name | 1000 Genomes Project |
| concepts[10].id | https://openalex.org/C197077220 |
| concepts[10].level | 4 |
| concepts[10].score | 0.4515777826309204 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q720988 |
| concepts[10].display_name | Human genome |
| concepts[11].id | https://openalex.org/C139275648 |
| concepts[11].level | 5 |
| concepts[11].score | 0.4117887020111084 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q17134011 |
| concepts[11].display_name | SNP |
| concepts[12].id | https://openalex.org/C141231307 |
| concepts[12].level | 3 |
| concepts[12].score | 0.3667636215686798 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q7020 |
| concepts[12].display_name | Genome |
| concepts[13].id | https://openalex.org/C54355233 |
| concepts[13].level | 1 |
| concepts[13].score | 0.36420056223869324 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[13].display_name | Genetics |
| concepts[14].id | https://openalex.org/C41008148 |
| concepts[14].level | 0 |
| concepts[14].score | 0.34779298305511475 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q21198 |
| concepts[14].display_name | Computer science |
| concepts[15].id | https://openalex.org/C105795698 |
| concepts[15].level | 1 |
| concepts[15].score | 0.3339438736438751 |
| concepts[15].wikidata | https://www.wikidata.org/wiki/Q12483 |
| concepts[15].display_name | Statistics |
| concepts[16].id | https://openalex.org/C135763542 |
| concepts[16].level | 3 |
| concepts[16].score | 0.27554911375045776 |
| concepts[16].wikidata | https://www.wikidata.org/wiki/Q106016 |
| concepts[16].display_name | Genotype |
| concepts[17].id | https://openalex.org/C9357733 |
| concepts[17].level | 2 |
| concepts[17].score | 0.22671827673912048 |
| concepts[17].wikidata | https://www.wikidata.org/wiki/Q6878417 |
| concepts[17].display_name | Missing data |
| concepts[18].id | https://openalex.org/C119857082 |
| concepts[18].level | 1 |
| concepts[18].score | 0.15525564551353455 |
| concepts[18].wikidata | https://www.wikidata.org/wiki/Q2539 |
| concepts[18].display_name | Machine learning |
| concepts[19].id | https://openalex.org/C104317684 |
| concepts[19].level | 2 |
| concepts[19].score | 0.15042021870613098 |
| concepts[19].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[19].display_name | Gene |
| concepts[20].id | https://openalex.org/C33923547 |
| concepts[20].level | 0 |
| concepts[20].score | 0.12392506003379822 |
| concepts[20].wikidata | https://www.wikidata.org/wiki/Q395 |
| concepts[20].display_name | Mathematics |
| keywords[0].id | https://openalex.org/keywords/imputation |
| keywords[0].score | 0.8741490244865417 |
| keywords[0].display_name | Imputation (statistics) |
| keywords[1].id | https://openalex.org/keywords/genotyping |
| keywords[1].score | 0.8323341608047485 |
| keywords[1].display_name | Genotyping |
| keywords[2].id | https://openalex.org/keywords/genome-wide-association-study |
| keywords[2].score | 0.6342840790748596 |
| keywords[2].display_name | Genome-wide association study |
| keywords[3].id | https://openalex.org/keywords/concordance |
| keywords[3].score | 0.5968185067176819 |
| keywords[3].display_name | Concordance |
| keywords[4].id | https://openalex.org/keywords/biology |
| keywords[4].score | 0.5523102879524231 |
| keywords[4].display_name | Biology |
| keywords[5].id | https://openalex.org/keywords/single-nucleotide-polymorphism |
| keywords[5].score | 0.549034059047699 |
| keywords[5].display_name | Single-nucleotide polymorphism |
| keywords[6].id | https://openalex.org/keywords/computational-biology |
| keywords[6].score | 0.4772919714450836 |
| keywords[6].display_name | Computational biology |
| keywords[7].id | https://openalex.org/keywords/genetic-association |
| keywords[7].score | 0.466608464717865 |
| keywords[7].display_name | Genetic association |
| keywords[8].id | https://openalex.org/keywords/snp-genotyping |
| keywords[8].score | 0.4646090567111969 |
| keywords[8].display_name | SNP genotyping |
| keywords[9].id | https://openalex.org/keywords/1000-genomes-project |
| keywords[9].score | 0.4518839120864868 |
| keywords[9].display_name | 1000 Genomes Project |
| keywords[10].id | https://openalex.org/keywords/human-genome |
| keywords[10].score | 0.4515777826309204 |
| keywords[10].display_name | Human genome |
| keywords[11].id | https://openalex.org/keywords/snp |
| keywords[11].score | 0.4117887020111084 |
| keywords[11].display_name | SNP |
| keywords[12].id | https://openalex.org/keywords/genome |
| keywords[12].score | 0.3667636215686798 |
| keywords[12].display_name | Genome |
| keywords[13].id | https://openalex.org/keywords/genetics |
| keywords[13].score | 0.36420056223869324 |
| keywords[13].display_name | Genetics |
| keywords[14].id | https://openalex.org/keywords/computer-science |
| keywords[14].score | 0.34779298305511475 |
| keywords[14].display_name | Computer science |
| keywords[15].id | https://openalex.org/keywords/statistics |
| keywords[15].score | 0.3339438736438751 |
| keywords[15].display_name | Statistics |
| keywords[16].id | https://openalex.org/keywords/genotype |
| keywords[16].score | 0.27554911375045776 |
| keywords[16].display_name | Genotype |
| keywords[17].id | https://openalex.org/keywords/missing-data |
| keywords[17].score | 0.22671827673912048 |
| keywords[17].display_name | Missing data |
| keywords[18].id | https://openalex.org/keywords/machine-learning |
| keywords[18].score | 0.15525564551353455 |
| keywords[18].display_name | Machine learning |
| keywords[19].id | https://openalex.org/keywords/gene |
| keywords[19].score | 0.15042021870613098 |
| keywords[19].display_name | Gene |
| keywords[20].id | https://openalex.org/keywords/mathematics |
| keywords[20].score | 0.12392506003379822 |
| keywords[20].display_name | Mathematics |
| language | en |
| locations[0].id | doi:10.1101/2022.06.14.496059 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4306402567 |
| locations[0].source.issn | |
| locations[0].source.type | repository |
| locations[0].source.is_oa | False |
| locations[0].source.issn_l | |
| locations[0].source.is_core | False |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| locations[0].source.host_organization | https://openalex.org/I2750212522 |
| locations[0].source.host_organization_name | Cold Spring Harbor Laboratory |
| locations[0].source.host_organization_lineage | https://openalex.org/I2750212522 |
| locations[0].license | cc-by-nd |
| locations[0].pdf_url | https://www.biorxiv.org/content/biorxiv/early/2022/06/15/2022.06.14.496059.full.pdf |
| locations[0].version | acceptedVersion |
| locations[0].raw_type | posted-content |
| locations[0].license_id | https://openalex.org/licenses/cc-by-nd |
| locations[0].is_accepted | True |
| locations[0].is_published | False |
| locations[0].raw_source_name | |
| locations[0].landing_page_url | https://doi.org/10.1101/2022.06.14.496059 |
| indexed_in | crossref |
| authorships[0].author.id | https://openalex.org/A5020547925 |
| authorships[0].author.orcid | https://orcid.org/0000-0003-3852-9578 |
| authorships[0].author.display_name | Dat Nguyen |
| authorships[0].affiliations[0].raw_affiliation_string | Center for Biomedical Informatics, Vingroup Big Data Institute, Hanoi, Vietnam |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Dat Thanh Nguyen |
| authorships[0].is_corresponding | True |
| authorships[0].raw_affiliation_strings | Center for Biomedical Informatics, Vingroup Big Data Institute, Hanoi, Vietnam |
| authorships[1].author.id | https://openalex.org/A5077184620 |
| authorships[1].author.orcid | https://orcid.org/0000-0003-3674-9286 |
| authorships[1].author.display_name | Trang Tran |
| authorships[1].affiliations[0].raw_affiliation_string | Center for Biomedical Informatics, Vingroup Big Data Institute, Hanoi, Vietnam |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Trang Tran |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | Center for Biomedical Informatics, Vingroup Big Data Institute, Hanoi, Vietnam |
| authorships[2].author.id | https://openalex.org/A5077334388 |
| authorships[2].author.orcid | https://orcid.org/0000-0002-0294-2558 |
| authorships[2].author.display_name | Mai Tran |
| authorships[2].affiliations[0].raw_affiliation_string | Center for Biomedical Informatics, Vingroup Big Data Institute, Hanoi, Vietnam |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Mai Tran |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | Center for Biomedical Informatics, Vingroup Big Data Institute, Hanoi, Vietnam |
| authorships[3].author.id | https://openalex.org/A5034366374 |
| authorships[3].author.orcid | https://orcid.org/0000-0002-2923-3823 |
| authorships[3].author.display_name | Khai Tran |
| authorships[3].affiliations[0].raw_affiliation_string | Center for Biomedical Informatics, Vingroup Big Data Institute, Hanoi, Vietnam |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Khai Tran |
| authorships[3].is_corresponding | False |
| authorships[3].raw_affiliation_strings | Center for Biomedical Informatics, Vingroup Big Data Institute, Hanoi, Vietnam |
| authorships[4].author.id | https://openalex.org/A5101420627 |
| authorships[4].author.orcid | https://orcid.org/0009-0007-2586-2087 |
| authorships[4].author.display_name | Duy T. Pham |
| authorships[4].countries | AU |
| authorships[4].affiliations[0].institution_ids | https://openalex.org/I165143802 |
| authorships[4].affiliations[0].raw_affiliation_string | Institute for Molecular Bioscience, University of Queensland, Brisbane, Australia |
| authorships[4].institutions[0].id | https://openalex.org/I165143802 |
| authorships[4].institutions[0].ror | https://ror.org/00rqy9422 |
| authorships[4].institutions[0].type | education |
| authorships[4].institutions[0].lineage | https://openalex.org/I165143802 |
| authorships[4].institutions[0].country_code | AU |
| authorships[4].institutions[0].display_name | The University of Queensland |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Duy Pham |
| authorships[4].is_corresponding | False |
| authorships[4].raw_affiliation_strings | Institute for Molecular Bioscience, University of Queensland, Brisbane, Australia |
| authorships[5].author.id | https://openalex.org/A5025792398 |
| authorships[5].author.orcid | https://orcid.org/0000-0001-8691-9138 |
| authorships[5].author.display_name | Nguyễn Thùy Dương |
| authorships[5].countries | VN |
| authorships[5].affiliations[0].institution_ids | https://openalex.org/I70349855 |
| authorships[5].affiliations[0].raw_affiliation_string | Institute of Genome Research, Vietnam Academy of Science and Technology, Hanoi, Vietnam |
| authorships[5].affiliations[1].raw_affiliation_string | Center for Biomedical Informatics, Vingroup Big Data Institute, Hanoi, Vietnam |
| authorships[5].institutions[0].id | https://openalex.org/I70349855 |
| authorships[5].institutions[0].ror | https://ror.org/02wsd5p50 |
| authorships[5].institutions[0].type | government |
| authorships[5].institutions[0].lineage | https://openalex.org/I70349855 |
| authorships[5].institutions[0].country_code | VN |
| authorships[5].institutions[0].display_name | Vietnam Academy of Science and Technology |
| authorships[5].author_position | middle |
| authorships[5].raw_author_name | Nguyen Thuy Duong |
| authorships[5].is_corresponding | False |
| authorships[5].raw_affiliation_strings | Center for Biomedical Informatics, Vingroup Big Data Institute, Hanoi, Vietnam, Institute of Genome Research, Vietnam Academy of Science and Technology, Hanoi, Vietnam |
| authorships[6].author.id | https://openalex.org/A5034599952 |
| authorships[6].author.orcid | https://orcid.org/0000-0001-7870-5703 |
| authorships[6].author.display_name | Quan Nguyen |
| authorships[6].countries | AU |
| authorships[6].affiliations[0].institution_ids | https://openalex.org/I165143802 |
| authorships[6].affiliations[0].raw_affiliation_string | Institute for Molecular Bioscience, University of Queensland, Brisbane, Australia |
| authorships[6].institutions[0].id | https://openalex.org/I165143802 |
| authorships[6].institutions[0].ror | https://ror.org/00rqy9422 |
| authorships[6].institutions[0].type | education |
| authorships[6].institutions[0].lineage | https://openalex.org/I165143802 |
| authorships[6].institutions[0].country_code | AU |
| authorships[6].institutions[0].display_name | The University of Queensland |
| authorships[6].author_position | middle |
| authorships[6].raw_author_name | Quan Nguyen |
| authorships[6].is_corresponding | True |
| authorships[6].raw_affiliation_strings | Institute for Molecular Bioscience, University of Queensland, Brisbane, Australia |
| authorships[7].author.id | https://openalex.org/A5043745628 |
| authorships[7].author.orcid | https://orcid.org/0000-0002-5454-9176 |
| authorships[7].author.display_name | Nam S. Vo |
| authorships[7].countries | VN |
| authorships[7].affiliations[0].institution_ids | https://openalex.org/I4210142044 |
| authorships[7].affiliations[0].raw_affiliation_string | College of Engineering and Computer Science, VinUniversity, Hanoi, Vietnam |
| authorships[7].affiliations[1].raw_affiliation_string | Center for Biomedical Informatics, Vingroup Big Data Institute, Hanoi, Vietnam |
| authorships[7].institutions[0].id | https://openalex.org/I4210142044 |
| authorships[7].institutions[0].ror | https://ror.org/052dmdr17 |
| authorships[7].institutions[0].type | education |
| authorships[7].institutions[0].lineage | https://openalex.org/I4210142044 |
| authorships[7].institutions[0].country_code | VN |
| authorships[7].institutions[0].display_name | VinUniversity |
| authorships[7].author_position | last |
| authorships[7].raw_author_name | Nam S. Vo |
| authorships[7].is_corresponding | True |
| authorships[7].raw_affiliation_strings | Center for Biomedical Informatics, Vingroup Big Data Institute, Hanoi, Vietnam, College of Engineering and Computer Science, VinUniversity, Hanoi, Vietnam |
| has_content.pdf | True |
| has_content.grobid_xml | True |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://www.biorxiv.org/content/biorxiv/early/2022/06/15/2022.06.14.496059.full.pdf |
| open_access.oa_status | green |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Imputation and polygenic score performances of human genotyping arrays in diverse populations |
| has_fulltext | True |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T10261 |
| primary_topic.field.id | https://openalex.org/fields/13 |
| primary_topic.field.display_name | Biochemistry, Genetics and Molecular Biology |
| primary_topic.score | 0.9998999834060669 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1311 |
| primary_topic.subfield.display_name | Genetics |
| primary_topic.display_name | Genetic Associations and Epidemiology |
| related_works | https://openalex.org/W4286002085, https://openalex.org/W2178628622, https://openalex.org/W2122946544, https://openalex.org/W4381248205, https://openalex.org/W4387141664, https://openalex.org/W4317801964, https://openalex.org/W1968654496, https://openalex.org/W2126214064, https://openalex.org/W2216458865, https://openalex.org/W1989743336 |
| cited_by_count | 0 |
| locations_count | 1 |
| best_oa_location.id | doi:10.1101/2022.06.14.496059 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4306402567 |
| best_oa_location.source.issn | |
| best_oa_location.source.type | repository |
| best_oa_location.source.is_oa | False |
| best_oa_location.source.issn_l | |
| best_oa_location.source.is_core | False |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| best_oa_location.source.host_organization | https://openalex.org/I2750212522 |
| best_oa_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| best_oa_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| best_oa_location.license | cc-by-nd |
| best_oa_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2022/06/15/2022.06.14.496059.full.pdf |
| best_oa_location.version | acceptedVersion |
| best_oa_location.raw_type | posted-content |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by-nd |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | False |
| best_oa_location.raw_source_name | |
| best_oa_location.landing_page_url | https://doi.org/10.1101/2022.06.14.496059 |
| primary_location.id | doi:10.1101/2022.06.14.496059 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4306402567 |
| primary_location.source.issn | |
| primary_location.source.type | repository |
| primary_location.source.is_oa | False |
| primary_location.source.issn_l | |
| primary_location.source.is_core | False |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| primary_location.source.host_organization | https://openalex.org/I2750212522 |
| primary_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| primary_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| primary_location.license | cc-by-nd |
| primary_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2022/06/15/2022.06.14.496059.full.pdf |
| primary_location.version | acceptedVersion |
| primary_location.raw_type | posted-content |
| primary_location.license_id | https://openalex.org/licenses/cc-by-nd |
| primary_location.is_accepted | True |
| primary_location.is_published | False |
| primary_location.raw_source_name | |
| primary_location.landing_page_url | https://doi.org/10.1101/2022.06.14.496059 |
| publication_date | 2022-06-15 |
| publication_year | 2022 |
| referenced_works | https://openalex.org/W2018823764, https://openalex.org/W2901303766, https://openalex.org/W2895486342, https://openalex.org/W3128153948, https://openalex.org/W2015408724, https://openalex.org/W1971771243, https://openalex.org/W3108355566, https://openalex.org/W3045441625, https://openalex.org/W2958615822, https://openalex.org/W2104549677, https://openalex.org/W2119279196, https://openalex.org/W2149992227, https://openalex.org/W2510973425, https://openalex.org/W2924041486, https://openalex.org/W2550933502, https://openalex.org/W3038990418, https://openalex.org/W6959219552, https://openalex.org/W1997775525, https://openalex.org/W2081067541, https://openalex.org/W2107121611, https://openalex.org/W2111307685, https://openalex.org/W2216458865, https://openalex.org/W1487785114, https://openalex.org/W2980086570, https://openalex.org/W3028329270, https://openalex.org/W3126861453, https://openalex.org/W2126361119, https://openalex.org/W2971867024, https://openalex.org/W2087036932, https://openalex.org/W2794134966, https://openalex.org/W2932671560, https://openalex.org/W2511515754, https://openalex.org/W2920792207, https://openalex.org/W2112869109, https://openalex.org/W4205690135, https://openalex.org/W4220726561, https://openalex.org/W2005935256, https://openalex.org/W4254359144, https://openalex.org/W2922534626, https://openalex.org/W2944540884, https://openalex.org/W2803332630, https://openalex.org/W3165929960, https://openalex.org/W2725988230, https://openalex.org/W4220827000, https://openalex.org/W2883622734, https://openalex.org/W2952883732, https://openalex.org/W2101071606 |
| referenced_works_count | 47 |
| abstract_inverted_index.6 | 100 |
| abstract_inverted_index.a | 20, 68, 77, 89, 150, 245, 255, 282 |
| abstract_inverted_index.23 | 94 |
| abstract_inverted_index.In | 180 |
| abstract_inverted_index.We | 144 |
| abstract_inverted_index.as | 281 |
| abstract_inverted_index.at | 41 |
| abstract_inverted_index.be | 221 |
| abstract_inverted_index.by | 184, 195 |
| abstract_inverted_index.in | 36, 47, 71, 99, 138, 200, 233 |
| abstract_inverted_index.is | 33, 189, 295 |
| abstract_inverted_index.of | 2, 6, 16, 80, 115, 120, 128, 153, 202, 211, 218, 242, 262, 275 |
| abstract_inverted_index.on | 112, 270 |
| abstract_inverted_index.to | 23, 52, 55, 130, 192, 287 |
| abstract_inverted_index.we | 87, 253 |
| abstract_inverted_index.(in | 118, 126 |
| abstract_inverted_index.Our | 237 |
| abstract_inverted_index.PGS | 125, 131, 182, 193, 213, 235, 250 |
| abstract_inverted_index.SNP | 186, 264 |
| abstract_inverted_index.The | 109, 293 |
| abstract_inverted_index.act | 280 |
| abstract_inverted_index.and | 44, 54, 74, 106, 122, 124, 142, 266, 272 |
| abstract_inverted_index.are | 155, 168, 207 |
| abstract_inverted_index.at: | 297 |
| abstract_inverted_index.but | 164, 225 |
| abstract_inverted_index.can | 220, 231 |
| abstract_inverted_index.for | 93, 170, 249, 285 |
| abstract_inverted_index.key | 212 |
| abstract_inverted_index.not | 156 |
| abstract_inverted_index.tag | 263 |
| abstract_inverted_index.the | 3, 14, 28, 60, 147, 158, 165, 171, 209, 240 |
| abstract_inverted_index.two | 216 |
| abstract_inverted_index.use | 5 |
| abstract_inverted_index.web | 256 |
| abstract_inverted_index.Over | 59 |
| abstract_inverted_index.SNPs | 154 |
| abstract_inverted_index.This | 31, 277 |
| abstract_inverted_index.When | 204 |
| abstract_inverted_index.both | 72 |
| abstract_inverted_index.data | 188, 199, 219 |
| abstract_inverted_index.from | 133 |
| abstract_inverted_index.good | 177 |
| abstract_inverted_index.have | 66 |
| abstract_inverted_index.high | 229 |
| abstract_inverted_index.last | 61 |
| abstract_inverted_index.more | 45, 84 |
| abstract_inverted_index.most | 201 |
| abstract_inverted_index.ones | 159 |
| abstract_inverted_index.term | 127 |
| abstract_inverted_index.than | 223 |
| abstract_inverted_index.that | 146, 167, 258 |
| abstract_inverted_index.tool | 257, 294 |
| abstract_inverted_index.used | 35 |
| abstract_inverted_index.very | 176 |
| abstract_inverted_index.with | 13, 149, 160, 228 |
| abstract_inverted_index.(PGS) | 50 |
| abstract_inverted_index.0.97, | 224 |
| abstract_inverted_index.Here, | 86 |
| abstract_inverted_index.array | 187, 248 |
| abstract_inverted_index.based | 269 |
| abstract_inverted_index.could | 174 |
| abstract_inverted_index.data) | 137 |
| abstract_inverted_index.focus | 111 |
| abstract_inverted_index.found | 145 |
| abstract_inverted_index.guide | 284 |
| abstract_inverted_index.human | 29, 63, 96 |
| abstract_inverted_index.lower | 234 |
| abstract_inverted_index.means | 22 |
| abstract_inverted_index.risk. | 58 |
| abstract_inverted_index.score | 49 |
| abstract_inverted_index.study | 278 |
| abstract_inverted_index.terms | 119 |
| abstract_inverted_index.their | 81, 289 |
| abstract_inverted_index.three | 139 |
| abstract_inverted_index.types | 217 |
| abstract_inverted_index.used, | 208 |
| abstract_inverted_index.using | 103 |
| abstract_inverted_index.whole | 134, 196 |
| abstract_inverted_index.would | 279 |
| abstract_inverted_index.(GWAS) | 40 |
| abstract_inverted_index.across | 27 |
| abstract_inverted_index.arrays | 65, 98, 148, 166, 206, 227 |
| abstract_inverted_index.became | 83 |
| abstract_inverted_index.cases. | 203 |
| abstract_inverted_index.design | 288 |
| abstract_inverted_index.genome | 135, 197 |
| abstract_inverted_index.groups | 102 |
| abstract_inverted_index.growth | 70 |
| abstract_inverted_index.higher | 151, 161, 222 |
| abstract_inverted_index.highly | 190 |
| abstract_inverted_index.making | 76 |
| abstract_inverted_index.number | 152 |
| abstract_inverted_index.result | 232 |
| abstract_inverted_index.traits | 141 |
| abstract_inverted_index.widely | 34 |
| abstract_inverted_index.between | 215 |
| abstract_inverted_index.content | 75 |
| abstract_inverted_index.decade, | 62 |
| abstract_inverted_index.density | 230 |
| abstract_inverted_index.derived | 116 |
| abstract_inverted_index.disease | 57 |
| abstract_inverted_index.diverse | 104 |
| abstract_inverted_index.genetic | 25 |
| abstract_inverted_index.genome. | 30 |
| abstract_inverted_index.genomic | 273 |
| abstract_inverted_index.imputed | 185 |
| abstract_inverted_index.metrics | 214 |
| abstract_inverted_index.number, | 73 |
| abstract_inverted_index.optimal | 205 |
| abstract_inverted_index.predict | 53 |
| abstract_inverted_index.provide | 175, 259 |
| abstract_inverted_index.public, | 105 |
| abstract_inverted_index.regions | 274 |
| abstract_inverted_index.remains | 19 |
| abstract_inverted_index.results | 238 |
| abstract_inverted_index.scales, | 43 |
| abstract_inverted_index.studies | 39 |
| abstract_inverted_index.suggest | 239 |
| abstract_inverted_index.untyped | 17 |
| abstract_inverted_index.ABSTRACT | 0 |
| abstract_inverted_index.Finally, | 252 |
| abstract_inverted_index.accuracy | 121 |
| abstract_inverted_index.analyses | 110, 261 |
| abstract_inverted_index.analysis | 51 |
| abstract_inverted_index.ancestry | 101 |
| abstract_inverted_index.bio-bank | 42 |
| abstract_inverted_index.combined | 12 |
| abstract_inverted_index.contents | 265 |
| abstract_inverted_index.in-house | 107 |
| abstract_inverted_index.properly | 243 |
| abstract_inverted_index.stratify | 56 |
| abstract_inverted_index.studies. | 292 |
| abstract_inverted_index.suitable | 246 |
| abstract_inverted_index.targeted | 172 |
| abstract_inverted_index.variants | 18 |
| abstract_inverted_index.addition, | 181 |
| abstract_inverted_index.available | 95, 296 |
| abstract_inverted_index.coverage) | 123 |
| abstract_inverted_index.datasets. | 108 |
| abstract_inverted_index.developed | 254 |
| abstract_inverted_index.different | 140 |
| abstract_inverted_index.diseases. | 143 |
| abstract_inverted_index.estimated | 132, 183, 194 |
| abstract_inverted_index.interest. | 276 |
| abstract_inverted_index.performed | 88 |
| abstract_inverted_index.polygenic | 48 |
| abstract_inverted_index.practical | 283 |
| abstract_inverted_index.recently, | 46 |
| abstract_inverted_index.selecting | 244 |
| abstract_inverted_index.undergone | 67 |
| abstract_inverted_index.Regardless | 1 |
| abstract_inverted_index.assessment | 92 |
| abstract_inverted_index.correlated | 191 |
| abstract_inverted_index.estimation | 114 |
| abstract_inverted_index.evaluation | 79 |
| abstract_inverted_index.genotyping | 11, 64, 97, 247, 290 |
| abstract_inverted_index.importance | 241 |
| abstract_inverted_index.important. | 85 |
| abstract_inverted_index.imputation | 15, 117, 162, 178, 267 |
| abstract_inverted_index.population | 173, 271 |
| abstract_inverted_index.sequencing | 8, 136, 198 |
| abstract_inverted_index.technology | 32 |
| abstract_inverted_index.tremendous | 69 |
| abstract_inverted_index.variations | 26 |
| abstract_inverted_index.association | 38 |
| abstract_inverted_index.concordance | 129 |
| abstract_inverted_index.genome-wide | 37 |
| abstract_inverted_index.interactive | 260 |
| abstract_inverted_index.interrogate | 24 |
| abstract_inverted_index.necessarily | 157 |
| abstract_inverted_index.performance | 91, 113, 268 |
| abstract_inverted_index.researchers | 286 |
| abstract_inverted_index.arrays-based | 291 |
| abstract_inverted_index.correlations | 210 |
| abstract_inverted_index.overwhelming | 4 |
| abstract_inverted_index.performance, | 163 |
| abstract_inverted_index.performance. | 179, 236 |
| abstract_inverted_index.performances | 82 |
| abstract_inverted_index.applications. | 251 |
| abstract_inverted_index.comprehensive | 78, 90 |
| abstract_inverted_index.technologies, | 9 |
| abstract_inverted_index.cost-effective | 21 |
| abstract_inverted_index.interestingly, | 226 |
| abstract_inverted_index.well-optimized | 169 |
| abstract_inverted_index.next-generation | 7 |
| abstract_inverted_index.microarray-based | 10 |
| abstract_inverted_index.https://genome.vinbigdata.org/tools/saa/ | 298 |
| cited_by_percentile_year | |
| corresponding_author_ids | https://openalex.org/A5043745628, https://openalex.org/A5020547925, https://openalex.org/A5034599952 |
| countries_distinct_count | 2 |
| institutions_distinct_count | 8 |
| corresponding_institution_ids | https://openalex.org/I165143802, https://openalex.org/I4210142044 |
| sustainable_development_goals[0].id | https://metadata.un.org/sdg/17 |
| sustainable_development_goals[0].score | 0.4099999964237213 |
| sustainable_development_goals[0].display_name | Partnerships for the goals |
| citation_normalized_percentile.value | 0.08214647 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |