Influenza A virus segments five and six can harbor artificial introns allowing expanded coding capacity Article Swipe
 YOU?
·
· 2021
· Open Access
·
· DOI: https://doi.org/10.1371/journal.ppat.1009951
YOU?
·
· 2021
· Open Access
·
· DOI: https://doi.org/10.1371/journal.ppat.1009951
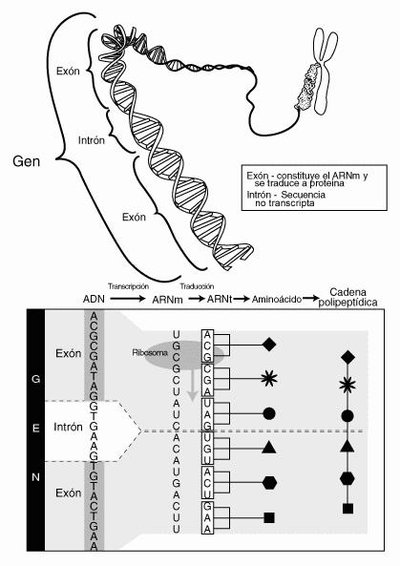
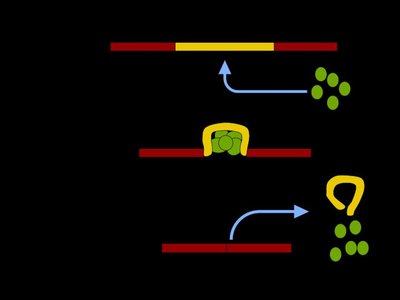
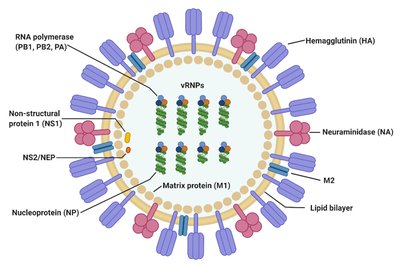
Influenza A viruses encode their genomes across eight, negative sense RNA segments. The six largest segments produce mRNA transcripts that do not generally splice; however, the two smallest segments are actively spliced to produce the essential viral proteins NEP and M2. Thus, viral utilization of RNA splicing effectively expands the viral coding capacity without increasing the number of genomic segments. As a first step towards understanding why splicing is not more broadly utilized across genomic segments, we designed and inserted an artificial intron into the normally nonsplicing NA segment. This insertion was tolerated and, although viral mRNAs were incompletely spliced, we observed only minor effects on viral fitness. To take advantage of the unspliced viral RNAs, we encoded a reporter luciferase gene in frame with the viral ORF such that when the intron was not removed the reporter protein would be produced. This approach, which we also show can be applied to the NP encoding segment and in different viral genetic backgrounds, led to high levels of reporter protein expression with minimal effects on the kinetics of viral replication or the ability to cause disease in experimentally infected animals. These data together show that the influenza viral genome is more tolerant of splicing than previously appreciated and this knowledge can be leveraged to develop viral genetic platforms with utility for biotechnology applications.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.1371/journal.ppat.1009951
- https://journals.plos.org/plospathogens/article/file?id=10.1371/journal.ppat.1009951&type=printable
- OA Status
- gold
- Cited By
- 7
- References
- 51
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W3204471398
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W3204471398Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1371/journal.ppat.1009951Digital Object Identifier
- Title
-
Influenza A virus segments five and six can harbor artificial introns allowing expanded coding capacityWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2021Year of publication
- Publication date
-
2021-09-27Full publication date if available
- Authors
-
Heather M. Froggatt, Kaitlyn N. Burke, Ryan R. Chaparian, Hector A. Miranda, Xinyu Zhu, Benjamin S. Chambers, Nicholas S. HeatonList of authors in order
- Landing page
-
https://doi.org/10.1371/journal.ppat.1009951Publisher landing page
- PDF URL
-
https://journals.plos.org/plospathogens/article/file?id=10.1371/journal.ppat.1009951&type=printableDirect link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
goldOpen access status per OpenAlex
- OA URL
-
https://journals.plos.org/plospathogens/article/file?id=10.1371/journal.ppat.1009951&type=printableDirect OA link when available
- Concepts
-
Intron, Biology, RNA splicing, Gene, Genome, Genetics, Coding region, RNA, Computational biology, Influenza A virus, Viral replication, Reporter gene, Virus, Virology, Gene expressionTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
7Total citation count in OpenAlex
- Citations by year (recent)
-
2025: 2, 2023: 3, 2022: 2Per-year citation counts (last 5 years)
- References (count)
-
51Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W3204471398 |
|---|---|
| doi | https://doi.org/10.1371/journal.ppat.1009951 |
| ids.doi | https://doi.org/10.1371/journal.ppat.1009951 |
| ids.mag | 3204471398 |
| ids.pmid | https://pubmed.ncbi.nlm.nih.gov/34570829 |
| ids.openalex | https://openalex.org/W3204471398 |
| fwci | 0.93339385 |
| mesh[0].qualifier_ui | |
| mesh[0].descriptor_ui | D000818 |
| mesh[0].is_major_topic | False |
| mesh[0].qualifier_name | |
| mesh[0].descriptor_name | Animals |
| mesh[1].qualifier_ui | |
| mesh[1].descriptor_ui | D006801 |
| mesh[1].is_major_topic | False |
| mesh[1].qualifier_name | |
| mesh[1].descriptor_name | Humans |
| mesh[2].qualifier_ui | Q000235 |
| mesh[2].descriptor_ui | D009980 |
| mesh[2].is_major_topic | False |
| mesh[2].qualifier_name | genetics |
| mesh[2].descriptor_name | Influenza A virus |
| mesh[3].qualifier_ui | Q000235 |
| mesh[3].descriptor_ui | D007438 |
| mesh[3].is_major_topic | False |
| mesh[3].qualifier_name | genetics |
| mesh[3].descriptor_name | Introns |
| mesh[4].qualifier_ui | Q000235 |
| mesh[4].descriptor_ui | D012326 |
| mesh[4].is_major_topic | False |
| mesh[4].qualifier_name | genetics |
| mesh[4].descriptor_name | RNA Splicing |
| mesh[5].qualifier_ui | Q000235 |
| mesh[5].descriptor_ui | D012367 |
| mesh[5].is_major_topic | False |
| mesh[5].qualifier_name | genetics |
| mesh[5].descriptor_name | RNA, Viral |
| mesh[6].qualifier_ui | |
| mesh[6].descriptor_ui | D000818 |
| mesh[6].is_major_topic | False |
| mesh[6].qualifier_name | |
| mesh[6].descriptor_name | Animals |
| mesh[7].qualifier_ui | |
| mesh[7].descriptor_ui | D006801 |
| mesh[7].is_major_topic | False |
| mesh[7].qualifier_name | |
| mesh[7].descriptor_name | Humans |
| mesh[8].qualifier_ui | Q000235 |
| mesh[8].descriptor_ui | D009980 |
| mesh[8].is_major_topic | False |
| mesh[8].qualifier_name | genetics |
| mesh[8].descriptor_name | Influenza A virus |
| mesh[9].qualifier_ui | Q000235 |
| mesh[9].descriptor_ui | D007438 |
| mesh[9].is_major_topic | False |
| mesh[9].qualifier_name | genetics |
| mesh[9].descriptor_name | Introns |
| mesh[10].qualifier_ui | Q000235 |
| mesh[10].descriptor_ui | D012326 |
| mesh[10].is_major_topic | False |
| mesh[10].qualifier_name | genetics |
| mesh[10].descriptor_name | RNA Splicing |
| mesh[11].qualifier_ui | Q000235 |
| mesh[11].descriptor_ui | D012367 |
| mesh[11].is_major_topic | False |
| mesh[11].qualifier_name | genetics |
| mesh[11].descriptor_name | RNA, Viral |
| type | article |
| title | Influenza A virus segments five and six can harbor artificial introns allowing expanded coding capacity |
| awards[0].id | https://openalex.org/G4435362199 |
| awards[0].funder_id | https://openalex.org/F4320337355 |
| awards[0].display_name | |
| awards[0].funder_award_id | 75N93019C00050 |
| awards[0].funder_display_name | National Institute of Allergy and Infectious Diseases |
| awards[1].id | https://openalex.org/G7068991450 |
| awards[1].funder_id | https://openalex.org/F4320337338 |
| awards[1].display_name | |
| awards[1].funder_award_id | R01-AI137031 |
| awards[1].funder_display_name | National Heart, Lung, and Blood Institute |
| biblio.issue | 9 |
| biblio.volume | 17 |
| biblio.last_page | e1009951 |
| biblio.first_page | e1009951 |
| topics[0].id | https://openalex.org/T10167 |
| topics[0].field.id | https://openalex.org/fields/27 |
| topics[0].field.display_name | Medicine |
| topics[0].score | 0.9998000264167786 |
| topics[0].domain.id | https://openalex.org/domains/4 |
| topics[0].domain.display_name | Health Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/2713 |
| topics[0].subfield.display_name | Epidemiology |
| topics[0].display_name | Influenza Virus Research Studies |
| topics[1].id | https://openalex.org/T10521 |
| topics[1].field.id | https://openalex.org/fields/13 |
| topics[1].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[1].score | 0.9994000196456909 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1312 |
| topics[1].subfield.display_name | Molecular Biology |
| topics[1].display_name | RNA and protein synthesis mechanisms |
| topics[2].id | https://openalex.org/T11243 |
| topics[2].field.id | https://openalex.org/fields/27 |
| topics[2].field.display_name | Medicine |
| topics[2].score | 0.9975000023841858 |
| topics[2].domain.id | https://openalex.org/domains/4 |
| topics[2].domain.display_name | Health Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/2713 |
| topics[2].subfield.display_name | Epidemiology |
| topics[2].display_name | Respiratory viral infections research |
| funders[0].id | https://openalex.org/F4320337338 |
| funders[0].ror | https://ror.org/012pb6c26 |
| funders[0].display_name | National Heart, Lung, and Blood Institute |
| funders[1].id | https://openalex.org/F4320337355 |
| funders[1].ror | https://ror.org/043z4tv69 |
| funders[1].display_name | National Institute of Allergy and Infectious Diseases |
| is_xpac | False |
| apc_list.value | 2655 |
| apc_list.currency | USD |
| apc_list.value_usd | 2655 |
| apc_paid.value | 2655 |
| apc_paid.currency | USD |
| apc_paid.value_usd | 2655 |
| concepts[0].id | https://openalex.org/C94671646 |
| concepts[0].level | 3 |
| concepts[0].score | 0.7516258955001831 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q207551 |
| concepts[0].display_name | Intron |
| concepts[1].id | https://openalex.org/C86803240 |
| concepts[1].level | 0 |
| concepts[1].score | 0.7444047331809998 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[1].display_name | Biology |
| concepts[2].id | https://openalex.org/C54458228 |
| concepts[2].level | 4 |
| concepts[2].score | 0.7167929410934448 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q237218 |
| concepts[2].display_name | RNA splicing |
| concepts[3].id | https://openalex.org/C104317684 |
| concepts[3].level | 2 |
| concepts[3].score | 0.5861406922340393 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[3].display_name | Gene |
| concepts[4].id | https://openalex.org/C141231307 |
| concepts[4].level | 3 |
| concepts[4].score | 0.5374979972839355 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q7020 |
| concepts[4].display_name | Genome |
| concepts[5].id | https://openalex.org/C54355233 |
| concepts[5].level | 1 |
| concepts[5].score | 0.5322494506835938 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[5].display_name | Genetics |
| concepts[6].id | https://openalex.org/C91779695 |
| concepts[6].level | 3 |
| concepts[6].score | 0.5295520424842834 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q3780824 |
| concepts[6].display_name | Coding region |
| concepts[7].id | https://openalex.org/C67705224 |
| concepts[7].level | 3 |
| concepts[7].score | 0.5233924984931946 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q11053 |
| concepts[7].display_name | RNA |
| concepts[8].id | https://openalex.org/C70721500 |
| concepts[8].level | 1 |
| concepts[8].score | 0.514819324016571 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q177005 |
| concepts[8].display_name | Computational biology |
| concepts[9].id | https://openalex.org/C2777546802 |
| concepts[9].level | 3 |
| concepts[9].score | 0.4932491183280945 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q834390 |
| concepts[9].display_name | Influenza A virus |
| concepts[10].id | https://openalex.org/C140704245 |
| concepts[10].level | 3 |
| concepts[10].score | 0.4714404344558716 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q3933202 |
| concepts[10].display_name | Viral replication |
| concepts[11].id | https://openalex.org/C161733203 |
| concepts[11].level | 4 |
| concepts[11].score | 0.452894389629364 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q911828 |
| concepts[11].display_name | Reporter gene |
| concepts[12].id | https://openalex.org/C2522874641 |
| concepts[12].level | 2 |
| concepts[12].score | 0.3902927041053772 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q808 |
| concepts[12].display_name | Virus |
| concepts[13].id | https://openalex.org/C159047783 |
| concepts[13].level | 1 |
| concepts[13].score | 0.3479119837284088 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q7215 |
| concepts[13].display_name | Virology |
| concepts[14].id | https://openalex.org/C150194340 |
| concepts[14].level | 3 |
| concepts[14].score | 0.26869064569473267 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q26972 |
| concepts[14].display_name | Gene expression |
| keywords[0].id | https://openalex.org/keywords/intron |
| keywords[0].score | 0.7516258955001831 |
| keywords[0].display_name | Intron |
| keywords[1].id | https://openalex.org/keywords/biology |
| keywords[1].score | 0.7444047331809998 |
| keywords[1].display_name | Biology |
| keywords[2].id | https://openalex.org/keywords/rna-splicing |
| keywords[2].score | 0.7167929410934448 |
| keywords[2].display_name | RNA splicing |
| keywords[3].id | https://openalex.org/keywords/gene |
| keywords[3].score | 0.5861406922340393 |
| keywords[3].display_name | Gene |
| keywords[4].id | https://openalex.org/keywords/genome |
| keywords[4].score | 0.5374979972839355 |
| keywords[4].display_name | Genome |
| keywords[5].id | https://openalex.org/keywords/genetics |
| keywords[5].score | 0.5322494506835938 |
| keywords[5].display_name | Genetics |
| keywords[6].id | https://openalex.org/keywords/coding-region |
| keywords[6].score | 0.5295520424842834 |
| keywords[6].display_name | Coding region |
| keywords[7].id | https://openalex.org/keywords/rna |
| keywords[7].score | 0.5233924984931946 |
| keywords[7].display_name | RNA |
| keywords[8].id | https://openalex.org/keywords/computational-biology |
| keywords[8].score | 0.514819324016571 |
| keywords[8].display_name | Computational biology |
| keywords[9].id | https://openalex.org/keywords/influenza-a-virus |
| keywords[9].score | 0.4932491183280945 |
| keywords[9].display_name | Influenza A virus |
| keywords[10].id | https://openalex.org/keywords/viral-replication |
| keywords[10].score | 0.4714404344558716 |
| keywords[10].display_name | Viral replication |
| keywords[11].id | https://openalex.org/keywords/reporter-gene |
| keywords[11].score | 0.452894389629364 |
| keywords[11].display_name | Reporter gene |
| keywords[12].id | https://openalex.org/keywords/virus |
| keywords[12].score | 0.3902927041053772 |
| keywords[12].display_name | Virus |
| keywords[13].id | https://openalex.org/keywords/virology |
| keywords[13].score | 0.3479119837284088 |
| keywords[13].display_name | Virology |
| keywords[14].id | https://openalex.org/keywords/gene-expression |
| keywords[14].score | 0.26869064569473267 |
| keywords[14].display_name | Gene expression |
| language | en |
| locations[0].id | doi:10.1371/journal.ppat.1009951 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S2004986 |
| locations[0].source.issn | 1553-7366, 1553-7374 |
| locations[0].source.type | journal |
| locations[0].source.is_oa | True |
| locations[0].source.issn_l | 1553-7366 |
| locations[0].source.is_core | True |
| locations[0].source.is_in_doaj | True |
| locations[0].source.display_name | PLoS Pathogens |
| locations[0].source.host_organization | https://openalex.org/P4310315706 |
| locations[0].source.host_organization_name | Public Library of Science |
| locations[0].source.host_organization_lineage | https://openalex.org/P4310315706 |
| locations[0].source.host_organization_lineage_names | Public Library of Science |
| locations[0].license | cc-by |
| locations[0].pdf_url | https://journals.plos.org/plospathogens/article/file?id=10.1371/journal.ppat.1009951&type=printable |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | PLOS Pathogens |
| locations[0].landing_page_url | https://doi.org/10.1371/journal.ppat.1009951 |
| locations[1].id | pmid:34570829 |
| locations[1].is_oa | False |
| locations[1].source.id | https://openalex.org/S4306525036 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | PubMed |
| locations[1].source.host_organization | https://openalex.org/I1299303238 |
| locations[1].source.host_organization_name | National Institutes of Health |
| locations[1].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[1].license | |
| locations[1].pdf_url | |
| locations[1].version | publishedVersion |
| locations[1].raw_type | |
| locations[1].license_id | |
| locations[1].is_accepted | True |
| locations[1].is_published | True |
| locations[1].raw_source_name | PLoS pathogens |
| locations[1].landing_page_url | https://pubmed.ncbi.nlm.nih.gov/34570829 |
| locations[2].id | pmh:oai:doaj.org/article:5f9db5129dae43e99d5e7c7d6e8b2e01 |
| locations[2].is_oa | True |
| locations[2].source.id | https://openalex.org/S4306401280 |
| locations[2].source.issn | |
| locations[2].source.type | repository |
| locations[2].source.is_oa | False |
| locations[2].source.issn_l | |
| locations[2].source.is_core | False |
| locations[2].source.is_in_doaj | False |
| locations[2].source.display_name | DOAJ (DOAJ: Directory of Open Access Journals) |
| locations[2].source.host_organization | |
| locations[2].source.host_organization_name | |
| locations[2].license | cc-by-sa |
| locations[2].pdf_url | |
| locations[2].version | submittedVersion |
| locations[2].raw_type | article |
| locations[2].license_id | https://openalex.org/licenses/cc-by-sa |
| locations[2].is_accepted | False |
| locations[2].is_published | False |
| locations[2].raw_source_name | PLoS Pathogens, Vol 17, Iss 9, p e1009951 (2021) |
| locations[2].landing_page_url | https://doaj.org/article/5f9db5129dae43e99d5e7c7d6e8b2e01 |
| locations[3].id | pmh:oai:europepmc.org:7389041 |
| locations[3].is_oa | True |
| locations[3].source.id | https://openalex.org/S4306400806 |
| locations[3].source.issn | |
| locations[3].source.type | repository |
| locations[3].source.is_oa | False |
| locations[3].source.issn_l | |
| locations[3].source.is_core | False |
| locations[3].source.is_in_doaj | False |
| locations[3].source.display_name | Europe PMC (PubMed Central) |
| locations[3].source.host_organization | https://openalex.org/I1303153112 |
| locations[3].source.host_organization_name | European Bioinformatics Institute |
| locations[3].source.host_organization_lineage | https://openalex.org/I1303153112 |
| locations[3].license | other-oa |
| locations[3].pdf_url | |
| locations[3].version | submittedVersion |
| locations[3].raw_type | Text |
| locations[3].license_id | https://openalex.org/licenses/other-oa |
| locations[3].is_accepted | False |
| locations[3].is_published | False |
| locations[3].raw_source_name | |
| locations[3].landing_page_url | http://europepmc.org/pmc/articles/PMC8496794 |
| locations[4].id | pmh:oai:pubmedcentral.nih.gov:8496794 |
| locations[4].is_oa | True |
| locations[4].source.id | https://openalex.org/S2764455111 |
| locations[4].source.issn | |
| locations[4].source.type | repository |
| locations[4].source.is_oa | False |
| locations[4].source.issn_l | |
| locations[4].source.is_core | False |
| locations[4].source.is_in_doaj | False |
| locations[4].source.display_name | PubMed Central |
| locations[4].source.host_organization | https://openalex.org/I1299303238 |
| locations[4].source.host_organization_name | National Institutes of Health |
| locations[4].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[4].license | other-oa |
| locations[4].pdf_url | |
| locations[4].version | submittedVersion |
| locations[4].raw_type | Text |
| locations[4].license_id | https://openalex.org/licenses/other-oa |
| locations[4].is_accepted | False |
| locations[4].is_published | False |
| locations[4].raw_source_name | PLoS Pathog |
| locations[4].landing_page_url | https://www.ncbi.nlm.nih.gov/pmc/articles/8496794 |
| indexed_in | crossref, doaj, pubmed |
| authorships[0].author.id | https://openalex.org/A5085851600 |
| authorships[0].author.orcid | https://orcid.org/0000-0001-7785-0976 |
| authorships[0].author.display_name | Heather M. Froggatt |
| authorships[0].countries | US |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I170897317 |
| authorships[0].affiliations[0].raw_affiliation_string | Department of Molecular Genetics and Microbiology Duke University School of Medicine Durham, North Carolina, United States of America |
| authorships[0].institutions[0].id | https://openalex.org/I170897317 |
| authorships[0].institutions[0].ror | https://ror.org/00py81415 |
| authorships[0].institutions[0].type | education |
| authorships[0].institutions[0].lineage | https://openalex.org/I170897317 |
| authorships[0].institutions[0].country_code | US |
| authorships[0].institutions[0].display_name | Duke University |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Heather M. Froggatt |
| authorships[0].is_corresponding | True |
| authorships[0].raw_affiliation_strings | Department of Molecular Genetics and Microbiology Duke University School of Medicine Durham, North Carolina, United States of America |
| authorships[1].author.id | https://openalex.org/A5039935202 |
| authorships[1].author.orcid | https://orcid.org/0000-0002-4868-4494 |
| authorships[1].author.display_name | Kaitlyn N. Burke |
| authorships[1].countries | US |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I170897317 |
| authorships[1].affiliations[0].raw_affiliation_string | Department of Molecular Genetics and Microbiology Duke University School of Medicine Durham, North Carolina, United States of America |
| authorships[1].institutions[0].id | https://openalex.org/I170897317 |
| authorships[1].institutions[0].ror | https://ror.org/00py81415 |
| authorships[1].institutions[0].type | education |
| authorships[1].institutions[0].lineage | https://openalex.org/I170897317 |
| authorships[1].institutions[0].country_code | US |
| authorships[1].institutions[0].display_name | Duke University |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Kaitlyn N. Burke |
| authorships[1].is_corresponding | True |
| authorships[1].raw_affiliation_strings | Department of Molecular Genetics and Microbiology Duke University School of Medicine Durham, North Carolina, United States of America |
| authorships[2].author.id | https://openalex.org/A5029193160 |
| authorships[2].author.orcid | https://orcid.org/0000-0003-3838-1868 |
| authorships[2].author.display_name | Ryan R. Chaparian |
| authorships[2].countries | US |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I170897317 |
| authorships[2].affiliations[0].raw_affiliation_string | Department of Molecular Genetics and Microbiology Duke University School of Medicine Durham, North Carolina, United States of America |
| authorships[2].institutions[0].id | https://openalex.org/I170897317 |
| authorships[2].institutions[0].ror | https://ror.org/00py81415 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I170897317 |
| authorships[2].institutions[0].country_code | US |
| authorships[2].institutions[0].display_name | Duke University |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Ryan R. Chaparian |
| authorships[2].is_corresponding | True |
| authorships[2].raw_affiliation_strings | Department of Molecular Genetics and Microbiology Duke University School of Medicine Durham, North Carolina, United States of America |
| authorships[3].author.id | https://openalex.org/A5029379023 |
| authorships[3].author.orcid | https://orcid.org/0000-0003-4072-6634 |
| authorships[3].author.display_name | Hector A. Miranda |
| authorships[3].countries | US |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I170897317 |
| authorships[3].affiliations[0].raw_affiliation_string | Department of Molecular Genetics and Microbiology Duke University School of Medicine Durham, North Carolina, United States of America |
| authorships[3].institutions[0].id | https://openalex.org/I170897317 |
| authorships[3].institutions[0].ror | https://ror.org/00py81415 |
| authorships[3].institutions[0].type | education |
| authorships[3].institutions[0].lineage | https://openalex.org/I170897317 |
| authorships[3].institutions[0].country_code | US |
| authorships[3].institutions[0].display_name | Duke University |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Hector A. Miranda |
| authorships[3].is_corresponding | True |
| authorships[3].raw_affiliation_strings | Department of Molecular Genetics and Microbiology Duke University School of Medicine Durham, North Carolina, United States of America |
| authorships[4].author.id | https://openalex.org/A5101670196 |
| authorships[4].author.orcid | https://orcid.org/0000-0002-1805-7203 |
| authorships[4].author.display_name | Xinyu Zhu |
| authorships[4].countries | US |
| authorships[4].affiliations[0].institution_ids | https://openalex.org/I170897317 |
| authorships[4].affiliations[0].raw_affiliation_string | Department of Molecular Genetics and Microbiology Duke University School of Medicine Durham, North Carolina, United States of America |
| authorships[4].institutions[0].id | https://openalex.org/I170897317 |
| authorships[4].institutions[0].ror | https://ror.org/00py81415 |
| authorships[4].institutions[0].type | education |
| authorships[4].institutions[0].lineage | https://openalex.org/I170897317 |
| authorships[4].institutions[0].country_code | US |
| authorships[4].institutions[0].display_name | Duke University |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Xinyu Zhu |
| authorships[4].is_corresponding | True |
| authorships[4].raw_affiliation_strings | Department of Molecular Genetics and Microbiology Duke University School of Medicine Durham, North Carolina, United States of America |
| authorships[5].author.id | https://openalex.org/A5027001919 |
| authorships[5].author.orcid | https://orcid.org/0000-0002-8095-5012 |
| authorships[5].author.display_name | Benjamin S. Chambers |
| authorships[5].countries | US |
| authorships[5].affiliations[0].institution_ids | https://openalex.org/I170897317 |
| authorships[5].affiliations[0].raw_affiliation_string | Department of Molecular Genetics and Microbiology Duke University School of Medicine Durham, North Carolina, United States of America |
| authorships[5].institutions[0].id | https://openalex.org/I170897317 |
| authorships[5].institutions[0].ror | https://ror.org/00py81415 |
| authorships[5].institutions[0].type | education |
| authorships[5].institutions[0].lineage | https://openalex.org/I170897317 |
| authorships[5].institutions[0].country_code | US |
| authorships[5].institutions[0].display_name | Duke University |
| authorships[5].author_position | middle |
| authorships[5].raw_author_name | Benjamin S. Chambers |
| authorships[5].is_corresponding | True |
| authorships[5].raw_affiliation_strings | Department of Molecular Genetics and Microbiology Duke University School of Medicine Durham, North Carolina, United States of America |
| authorships[6].author.id | https://openalex.org/A5064828779 |
| authorships[6].author.orcid | https://orcid.org/0000-0002-5307-3428 |
| authorships[6].author.display_name | Nicholas S. Heaton |
| authorships[6].countries | US |
| authorships[6].affiliations[0].institution_ids | https://openalex.org/I170897317 |
| authorships[6].affiliations[0].raw_affiliation_string | Department of Molecular Genetics and Microbiology Duke University School of Medicine Durham, North Carolina, United States of America |
| authorships[6].affiliations[1].institution_ids | https://openalex.org/I170897317 |
| authorships[6].affiliations[1].raw_affiliation_string | Duke Human Vaccine Institute Duke University School of Medicine Durham, North Carolina, United States of America |
| authorships[6].institutions[0].id | https://openalex.org/I170897317 |
| authorships[6].institutions[0].ror | https://ror.org/00py81415 |
| authorships[6].institutions[0].type | education |
| authorships[6].institutions[0].lineage | https://openalex.org/I170897317 |
| authorships[6].institutions[0].country_code | US |
| authorships[6].institutions[0].display_name | Duke University |
| authorships[6].author_position | last |
| authorships[6].raw_author_name | Nicholas S. Heaton |
| authorships[6].is_corresponding | True |
| authorships[6].raw_affiliation_strings | Department of Molecular Genetics and Microbiology Duke University School of Medicine Durham, North Carolina, United States of America, Duke Human Vaccine Institute Duke University School of Medicine Durham, North Carolina, United States of America |
| has_content.pdf | True |
| has_content.grobid_xml | True |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://journals.plos.org/plospathogens/article/file?id=10.1371/journal.ppat.1009951&type=printable |
| open_access.oa_status | gold |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Influenza A virus segments five and six can harbor artificial introns allowing expanded coding capacity |
| has_fulltext | True |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T10167 |
| primary_topic.field.id | https://openalex.org/fields/27 |
| primary_topic.field.display_name | Medicine |
| primary_topic.score | 0.9998000264167786 |
| primary_topic.domain.id | https://openalex.org/domains/4 |
| primary_topic.domain.display_name | Health Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/2713 |
| primary_topic.subfield.display_name | Epidemiology |
| primary_topic.display_name | Influenza Virus Research Studies |
| related_works | https://openalex.org/W3090446747, https://openalex.org/W2083872838, https://openalex.org/W2045666137, https://openalex.org/W2056412942, https://openalex.org/W2087463521, https://openalex.org/W2093797995, https://openalex.org/W4211196276, https://openalex.org/W2171581216, https://openalex.org/W2036321075, https://openalex.org/W4240704896 |
| cited_by_count | 7 |
| counts_by_year[0].year | 2025 |
| counts_by_year[0].cited_by_count | 2 |
| counts_by_year[1].year | 2023 |
| counts_by_year[1].cited_by_count | 3 |
| counts_by_year[2].year | 2022 |
| counts_by_year[2].cited_by_count | 2 |
| locations_count | 5 |
| best_oa_location.id | doi:10.1371/journal.ppat.1009951 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S2004986 |
| best_oa_location.source.issn | 1553-7366, 1553-7374 |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | True |
| best_oa_location.source.issn_l | 1553-7366 |
| best_oa_location.source.is_core | True |
| best_oa_location.source.is_in_doaj | True |
| best_oa_location.source.display_name | PLoS Pathogens |
| best_oa_location.source.host_organization | https://openalex.org/P4310315706 |
| best_oa_location.source.host_organization_name | Public Library of Science |
| best_oa_location.source.host_organization_lineage | https://openalex.org/P4310315706 |
| best_oa_location.source.host_organization_lineage_names | Public Library of Science |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | https://journals.plos.org/plospathogens/article/file?id=10.1371/journal.ppat.1009951&type=printable |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | journal-article |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | PLOS Pathogens |
| best_oa_location.landing_page_url | https://doi.org/10.1371/journal.ppat.1009951 |
| primary_location.id | doi:10.1371/journal.ppat.1009951 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S2004986 |
| primary_location.source.issn | 1553-7366, 1553-7374 |
| primary_location.source.type | journal |
| primary_location.source.is_oa | True |
| primary_location.source.issn_l | 1553-7366 |
| primary_location.source.is_core | True |
| primary_location.source.is_in_doaj | True |
| primary_location.source.display_name | PLoS Pathogens |
| primary_location.source.host_organization | https://openalex.org/P4310315706 |
| primary_location.source.host_organization_name | Public Library of Science |
| primary_location.source.host_organization_lineage | https://openalex.org/P4310315706 |
| primary_location.source.host_organization_lineage_names | Public Library of Science |
| primary_location.license | cc-by |
| primary_location.pdf_url | https://journals.plos.org/plospathogens/article/file?id=10.1371/journal.ppat.1009951&type=printable |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | PLOS Pathogens |
| primary_location.landing_page_url | https://doi.org/10.1371/journal.ppat.1009951 |
| publication_date | 2021-09-27 |
| publication_year | 2021 |
| referenced_works | https://openalex.org/W2141838466, https://openalex.org/W2811479613, https://openalex.org/W2160546938, https://openalex.org/W2004586645, https://openalex.org/W2100900365, https://openalex.org/W6746694070, https://openalex.org/W2958139469, https://openalex.org/W2597445526, https://openalex.org/W3036094002, https://openalex.org/W2968073481, https://openalex.org/W2992458104, https://openalex.org/W2029575848, https://openalex.org/W2100560524, https://openalex.org/W68443390, https://openalex.org/W2261974718, https://openalex.org/W2058740609, https://openalex.org/W2344192488, https://openalex.org/W2283604714, https://openalex.org/W2003564963, https://openalex.org/W2080277033, https://openalex.org/W2155713267, https://openalex.org/W2805745501, https://openalex.org/W2325427926, https://openalex.org/W1970877840, https://openalex.org/W1970801348, https://openalex.org/W1166101103, https://openalex.org/W2098526680, https://openalex.org/W2069488928, https://openalex.org/W2090697124, https://openalex.org/W2897317761, https://openalex.org/W1563046203, https://openalex.org/W2800987064, https://openalex.org/W2566587919, https://openalex.org/W3037827718, https://openalex.org/W194294989, https://openalex.org/W2152237596, https://openalex.org/W1992308631, https://openalex.org/W2047664825, https://openalex.org/W2409472203, https://openalex.org/W2017224181, https://openalex.org/W2474039476, https://openalex.org/W2525128349, https://openalex.org/W2105360874, https://openalex.org/W2006737337, https://openalex.org/W2009443782, https://openalex.org/W2069401360, https://openalex.org/W2118150714, https://openalex.org/W1977743637, https://openalex.org/W2772022440, https://openalex.org/W2127096557, https://openalex.org/W2568007245 |
| referenced_works_count | 51 |
| abstract_inverted_index.A | 1 |
| abstract_inverted_index.a | 61, 118 |
| abstract_inverted_index.As | 60 |
| abstract_inverted_index.NA | 87 |
| abstract_inverted_index.NP | 153 |
| abstract_inverted_index.To | 108 |
| abstract_inverted_index.an | 80 |
| abstract_inverted_index.be | 140, 149, 210 |
| abstract_inverted_index.do | 20 |
| abstract_inverted_index.in | 122, 157, 185 |
| abstract_inverted_index.is | 68, 198 |
| abstract_inverted_index.of | 44, 57, 111, 166, 176, 201 |
| abstract_inverted_index.on | 105, 173 |
| abstract_inverted_index.or | 179 |
| abstract_inverted_index.to | 32, 151, 163, 182, 212 |
| abstract_inverted_index.we | 76, 100, 116, 145 |
| abstract_inverted_index.M2. | 40 |
| abstract_inverted_index.NEP | 38 |
| abstract_inverted_index.ORF | 127 |
| abstract_inverted_index.RNA | 10, 45 |
| abstract_inverted_index.The | 12 |
| abstract_inverted_index.and | 39, 78, 156, 206 |
| abstract_inverted_index.are | 29 |
| abstract_inverted_index.can | 148, 209 |
| abstract_inverted_index.for | 219 |
| abstract_inverted_index.led | 162 |
| abstract_inverted_index.not | 21, 69, 134 |
| abstract_inverted_index.six | 13 |
| abstract_inverted_index.the | 25, 34, 49, 55, 84, 112, 125, 131, 136, 152, 174, 180, 194 |
| abstract_inverted_index.two | 26 |
| abstract_inverted_index.was | 91, 133 |
| abstract_inverted_index.why | 66 |
| abstract_inverted_index.This | 89, 142 |
| abstract_inverted_index.also | 146 |
| abstract_inverted_index.and, | 93 |
| abstract_inverted_index.data | 190 |
| abstract_inverted_index.gene | 121 |
| abstract_inverted_index.high | 164 |
| abstract_inverted_index.into | 83 |
| abstract_inverted_index.mRNA | 17 |
| abstract_inverted_index.more | 70, 199 |
| abstract_inverted_index.only | 102 |
| abstract_inverted_index.show | 147, 192 |
| abstract_inverted_index.step | 63 |
| abstract_inverted_index.such | 128 |
| abstract_inverted_index.take | 109 |
| abstract_inverted_index.than | 203 |
| abstract_inverted_index.that | 19, 129, 193 |
| abstract_inverted_index.this | 207 |
| abstract_inverted_index.were | 97 |
| abstract_inverted_index.when | 130 |
| abstract_inverted_index.with | 124, 170, 217 |
| abstract_inverted_index.RNAs, | 115 |
| abstract_inverted_index.These | 189 |
| abstract_inverted_index.Thus, | 41 |
| abstract_inverted_index.cause | 183 |
| abstract_inverted_index.first | 62 |
| abstract_inverted_index.frame | 123 |
| abstract_inverted_index.mRNAs | 96 |
| abstract_inverted_index.minor | 103 |
| abstract_inverted_index.sense | 9 |
| abstract_inverted_index.their | 4 |
| abstract_inverted_index.viral | 36, 42, 50, 95, 106, 114, 126, 159, 177, 196, 214 |
| abstract_inverted_index.which | 144 |
| abstract_inverted_index.would | 139 |
| abstract_inverted_index.across | 6, 73 |
| abstract_inverted_index.coding | 51 |
| abstract_inverted_index.eight, | 7 |
| abstract_inverted_index.encode | 3 |
| abstract_inverted_index.genome | 197 |
| abstract_inverted_index.intron | 82, 132 |
| abstract_inverted_index.levels | 165 |
| abstract_inverted_index.number | 56 |
| abstract_inverted_index.ability | 181 |
| abstract_inverted_index.applied | 150 |
| abstract_inverted_index.broadly | 71 |
| abstract_inverted_index.develop | 213 |
| abstract_inverted_index.disease | 184 |
| abstract_inverted_index.effects | 104, 172 |
| abstract_inverted_index.encoded | 117 |
| abstract_inverted_index.expands | 48 |
| abstract_inverted_index.genetic | 160, 215 |
| abstract_inverted_index.genomes | 5 |
| abstract_inverted_index.genomic | 58, 74 |
| abstract_inverted_index.largest | 14 |
| abstract_inverted_index.minimal | 171 |
| abstract_inverted_index.produce | 16, 33 |
| abstract_inverted_index.protein | 138, 168 |
| abstract_inverted_index.removed | 135 |
| abstract_inverted_index.segment | 155 |
| abstract_inverted_index.splice; | 23 |
| abstract_inverted_index.spliced | 31 |
| abstract_inverted_index.towards | 64 |
| abstract_inverted_index.utility | 218 |
| abstract_inverted_index.viruses | 2 |
| abstract_inverted_index.without | 53 |
| abstract_inverted_index.actively | 30 |
| abstract_inverted_index.although | 94 |
| abstract_inverted_index.animals. | 188 |
| abstract_inverted_index.capacity | 52 |
| abstract_inverted_index.designed | 77 |
| abstract_inverted_index.encoding | 154 |
| abstract_inverted_index.fitness. | 107 |
| abstract_inverted_index.however, | 24 |
| abstract_inverted_index.infected | 187 |
| abstract_inverted_index.inserted | 79 |
| abstract_inverted_index.kinetics | 175 |
| abstract_inverted_index.negative | 8 |
| abstract_inverted_index.normally | 85 |
| abstract_inverted_index.observed | 101 |
| abstract_inverted_index.proteins | 37 |
| abstract_inverted_index.reporter | 119, 137, 167 |
| abstract_inverted_index.segment. | 88 |
| abstract_inverted_index.segments | 15, 28 |
| abstract_inverted_index.smallest | 27 |
| abstract_inverted_index.spliced, | 99 |
| abstract_inverted_index.splicing | 46, 67, 202 |
| abstract_inverted_index.together | 191 |
| abstract_inverted_index.tolerant | 200 |
| abstract_inverted_index.utilized | 72 |
| abstract_inverted_index.Influenza | 0 |
| abstract_inverted_index.advantage | 110 |
| abstract_inverted_index.approach, | 143 |
| abstract_inverted_index.different | 158 |
| abstract_inverted_index.essential | 35 |
| abstract_inverted_index.generally | 22 |
| abstract_inverted_index.influenza | 195 |
| abstract_inverted_index.insertion | 90 |
| abstract_inverted_index.knowledge | 208 |
| abstract_inverted_index.leveraged | 211 |
| abstract_inverted_index.platforms | 216 |
| abstract_inverted_index.produced. | 141 |
| abstract_inverted_index.segments, | 75 |
| abstract_inverted_index.segments. | 11, 59 |
| abstract_inverted_index.tolerated | 92 |
| abstract_inverted_index.unspliced | 113 |
| abstract_inverted_index.artificial | 81 |
| abstract_inverted_index.expression | 169 |
| abstract_inverted_index.increasing | 54 |
| abstract_inverted_index.luciferase | 120 |
| abstract_inverted_index.previously | 204 |
| abstract_inverted_index.appreciated | 205 |
| abstract_inverted_index.effectively | 47 |
| abstract_inverted_index.nonsplicing | 86 |
| abstract_inverted_index.replication | 178 |
| abstract_inverted_index.transcripts | 18 |
| abstract_inverted_index.utilization | 43 |
| abstract_inverted_index.backgrounds, | 161 |
| abstract_inverted_index.incompletely | 98 |
| abstract_inverted_index.applications. | 221 |
| abstract_inverted_index.biotechnology | 220 |
| abstract_inverted_index.understanding | 65 |
| abstract_inverted_index.experimentally | 186 |
| cited_by_percentile_year.max | 97 |
| cited_by_percentile_year.min | 94 |
| corresponding_author_ids | https://openalex.org/A5029193160, https://openalex.org/A5101670196, https://openalex.org/A5085851600, https://openalex.org/A5039935202, https://openalex.org/A5064828779, https://openalex.org/A5029379023, https://openalex.org/A5027001919 |
| countries_distinct_count | 1 |
| institutions_distinct_count | 7 |
| corresponding_institution_ids | https://openalex.org/I170897317 |
| citation_normalized_percentile.value | 0.72924306 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |