Integrating phylogenies into single-cell RNA sequencing analysis allows comparisons across species, genes, and cells Article Swipe
 YOU?
·
· 2024
· Open Access
·
· DOI: https://doi.org/10.1371/journal.pbio.3002633
YOU?
·
· 2024
· Open Access
·
· DOI: https://doi.org/10.1371/journal.pbio.3002633
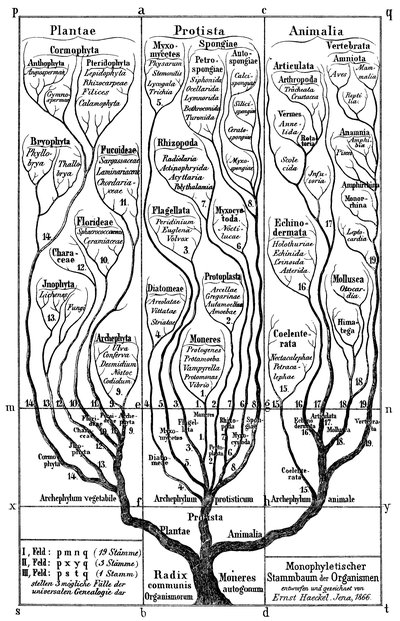
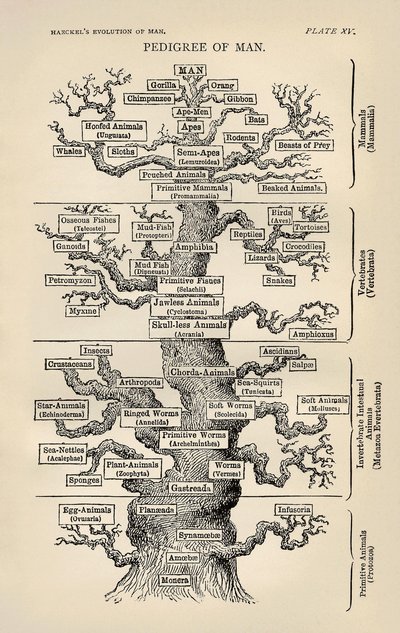
Comparisons of single-cell RNA sequencing (scRNA-seq) data across species can reveal links between cellular gene expression and the evolution of cell functions, features, and phenotypes. These comparisons evoke evolutionary histories, as depicted by phylogenetic trees, that define relationships between species, genes, and cells. This Essay considers each of these in turn, laying out challenges and solutions derived from a phylogenetic comparative approach and relating these solutions to previously proposed methods for the pairwise alignment of cellular dimensional maps. This Essay contends that species trees, gene trees, cell phylogenies, and cell lineages can all be reconciled as descriptions of the same concept—the tree of cellular life. By integrating phylogenetic approaches into scRNA-seq analyses, challenges for building informed comparisons across species can be overcome, and hypotheses about gene and cell evolution can be robustly tested.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.1371/journal.pbio.3002633
- https://journals.plos.org/plosbiology/article/file?id=10.1371/journal.pbio.3002633&type=printable
- OA Status
- gold
- Cited By
- 11
- References
- 55
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4398765394
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4398765394Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1371/journal.pbio.3002633Digital Object Identifier
- Title
-
Integrating phylogenies into single-cell RNA sequencing analysis allows comparisons across species, genes, and cellsWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2024Year of publication
- Publication date
-
2024-05-24Full publication date if available
- Authors
-
Samuel H. Church, Jasmine L. Mah, Casey W. DunnList of authors in order
- Landing page
-
https://doi.org/10.1371/journal.pbio.3002633Publisher landing page
- PDF URL
-
https://journals.plos.org/plosbiology/article/file?id=10.1371/journal.pbio.3002633&type=printableDirect link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
goldOpen access status per OpenAlex
- OA URL
-
https://journals.plos.org/plosbiology/article/file?id=10.1371/journal.pbio.3002633&type=printableDirect OA link when available
- Concepts
-
Biology, Phylogenetic tree, Gene, Phylogenetics, Evolutionary biology, Pairwise comparison, Phylogenetic network, Computational biology, Tree of life (biology), Genetics, Statistics, MathematicsTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
11Total citation count in OpenAlex
- Citations by year (recent)
-
2025: 7, 2024: 4Per-year citation counts (last 5 years)
- References (count)
-
55Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4398765394 |
|---|---|
| doi | https://doi.org/10.1371/journal.pbio.3002633 |
| ids.doi | https://doi.org/10.1371/journal.pbio.3002633 |
| ids.pmid | https://pubmed.ncbi.nlm.nih.gov/38787797 |
| ids.openalex | https://openalex.org/W4398765394 |
| fwci | 5.28271948 |
| mesh[0].qualifier_ui | Q000379 |
| mesh[0].descriptor_ui | D059010 |
| mesh[0].is_major_topic | True |
| mesh[0].qualifier_name | methods |
| mesh[0].descriptor_name | Single-Cell Analysis |
| mesh[1].qualifier_ui | |
| mesh[1].descriptor_ui | D010802 |
| mesh[1].is_major_topic | True |
| mesh[1].qualifier_name | |
| mesh[1].descriptor_name | Phylogeny |
| mesh[2].qualifier_ui | Q000379 |
| mesh[2].descriptor_ui | D017423 |
| mesh[2].is_major_topic | True |
| mesh[2].qualifier_name | methods |
| mesh[2].descriptor_name | Sequence Analysis, RNA |
| mesh[3].qualifier_ui | |
| mesh[3].descriptor_ui | D000818 |
| mesh[3].is_major_topic | False |
| mesh[3].qualifier_name | |
| mesh[3].descriptor_name | Animals |
| mesh[4].qualifier_ui | |
| mesh[4].descriptor_ui | D006801 |
| mesh[4].is_major_topic | False |
| mesh[4].qualifier_name | |
| mesh[4].descriptor_name | Humans |
| mesh[5].qualifier_ui | Q000235 |
| mesh[5].descriptor_ui | D019070 |
| mesh[5].is_major_topic | False |
| mesh[5].qualifier_name | genetics |
| mesh[5].descriptor_name | Cell Lineage |
| mesh[6].qualifier_ui | |
| mesh[6].descriptor_ui | D019143 |
| mesh[6].is_major_topic | False |
| mesh[6].qualifier_name | |
| mesh[6].descriptor_name | Evolution, Molecular |
| mesh[7].qualifier_ui | |
| mesh[7].descriptor_ui | D013045 |
| mesh[7].is_major_topic | False |
| mesh[7].qualifier_name | |
| mesh[7].descriptor_name | Species Specificity |
| mesh[8].qualifier_ui | Q000379 |
| mesh[8].descriptor_ui | D059010 |
| mesh[8].is_major_topic | True |
| mesh[8].qualifier_name | methods |
| mesh[8].descriptor_name | Single-Cell Analysis |
| mesh[9].qualifier_ui | |
| mesh[9].descriptor_ui | D010802 |
| mesh[9].is_major_topic | True |
| mesh[9].qualifier_name | |
| mesh[9].descriptor_name | Phylogeny |
| mesh[10].qualifier_ui | Q000379 |
| mesh[10].descriptor_ui | D017423 |
| mesh[10].is_major_topic | True |
| mesh[10].qualifier_name | methods |
| mesh[10].descriptor_name | Sequence Analysis, RNA |
| mesh[11].qualifier_ui | |
| mesh[11].descriptor_ui | D000818 |
| mesh[11].is_major_topic | False |
| mesh[11].qualifier_name | |
| mesh[11].descriptor_name | Animals |
| mesh[12].qualifier_ui | |
| mesh[12].descriptor_ui | D006801 |
| mesh[12].is_major_topic | False |
| mesh[12].qualifier_name | |
| mesh[12].descriptor_name | Humans |
| mesh[13].qualifier_ui | Q000235 |
| mesh[13].descriptor_ui | D019070 |
| mesh[13].is_major_topic | False |
| mesh[13].qualifier_name | genetics |
| mesh[13].descriptor_name | Cell Lineage |
| mesh[14].qualifier_ui | |
| mesh[14].descriptor_ui | D019143 |
| mesh[14].is_major_topic | False |
| mesh[14].qualifier_name | |
| mesh[14].descriptor_name | Evolution, Molecular |
| mesh[15].qualifier_ui | |
| mesh[15].descriptor_ui | D013045 |
| mesh[15].is_major_topic | False |
| mesh[15].qualifier_name | |
| mesh[15].descriptor_name | Species Specificity |
| mesh[16].qualifier_ui | Q000379 |
| mesh[16].descriptor_ui | D059010 |
| mesh[16].is_major_topic | True |
| mesh[16].qualifier_name | methods |
| mesh[16].descriptor_name | Single-Cell Analysis |
| mesh[17].qualifier_ui | |
| mesh[17].descriptor_ui | D010802 |
| mesh[17].is_major_topic | True |
| mesh[17].qualifier_name | |
| mesh[17].descriptor_name | Phylogeny |
| mesh[18].qualifier_ui | Q000379 |
| mesh[18].descriptor_ui | D017423 |
| mesh[18].is_major_topic | True |
| mesh[18].qualifier_name | methods |
| mesh[18].descriptor_name | Sequence Analysis, RNA |
| mesh[19].qualifier_ui | |
| mesh[19].descriptor_ui | D000818 |
| mesh[19].is_major_topic | False |
| mesh[19].qualifier_name | |
| mesh[19].descriptor_name | Animals |
| mesh[20].qualifier_ui | |
| mesh[20].descriptor_ui | D006801 |
| mesh[20].is_major_topic | False |
| mesh[20].qualifier_name | |
| mesh[20].descriptor_name | Humans |
| mesh[21].qualifier_ui | Q000235 |
| mesh[21].descriptor_ui | D019070 |
| mesh[21].is_major_topic | False |
| mesh[21].qualifier_name | genetics |
| mesh[21].descriptor_name | Cell Lineage |
| mesh[22].qualifier_ui | |
| mesh[22].descriptor_ui | D019143 |
| mesh[22].is_major_topic | False |
| mesh[22].qualifier_name | |
| mesh[22].descriptor_name | Evolution, Molecular |
| mesh[23].qualifier_ui | |
| mesh[23].descriptor_ui | D013045 |
| mesh[23].is_major_topic | False |
| mesh[23].qualifier_name | |
| mesh[23].descriptor_name | Species Specificity |
| type | article |
| title | Integrating phylogenies into single-cell RNA sequencing analysis allows comparisons across species, genes, and cells |
| awards[0].id | https://openalex.org/G2488421719 |
| awards[0].funder_id | https://openalex.org/F4320306076 |
| awards[0].display_name | |
| awards[0].funder_award_id | 2109502 |
| awards[0].funder_display_name | National Science Foundation |
| biblio.issue | 5 |
| biblio.volume | 22 |
| biblio.last_page | e3002633 |
| biblio.first_page | e3002633 |
| grants[0].funder | https://openalex.org/F4320306076 |
| grants[0].award_id | 2109502 |
| grants[0].funder_display_name | National Science Foundation |
| topics[0].id | https://openalex.org/T11289 |
| topics[0].field.id | https://openalex.org/fields/13 |
| topics[0].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[0].score | 0.9998000264167786 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1312 |
| topics[0].subfield.display_name | Molecular Biology |
| topics[0].display_name | Single-cell and spatial transcriptomics |
| topics[1].id | https://openalex.org/T10015 |
| topics[1].field.id | https://openalex.org/fields/13 |
| topics[1].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[1].score | 0.9879999756813049 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1312 |
| topics[1].subfield.display_name | Molecular Biology |
| topics[1].display_name | Genomics and Phylogenetic Studies |
| topics[2].id | https://openalex.org/T12859 |
| topics[2].field.id | https://openalex.org/fields/13 |
| topics[2].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[2].score | 0.9801999926567078 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1304 |
| topics[2].subfield.display_name | Biophysics |
| topics[2].display_name | Cell Image Analysis Techniques |
| funders[0].id | https://openalex.org/F4320306076 |
| funders[0].ror | https://ror.org/021nxhr62 |
| funders[0].display_name | National Science Foundation |
| is_xpac | False |
| apc_list.value | 5300 |
| apc_list.currency | USD |
| apc_list.value_usd | 5300 |
| apc_paid.value | 5300 |
| apc_paid.currency | USD |
| apc_paid.value_usd | 5300 |
| concepts[0].id | https://openalex.org/C86803240 |
| concepts[0].level | 0 |
| concepts[0].score | 0.9260641932487488 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[0].display_name | Biology |
| concepts[1].id | https://openalex.org/C193252679 |
| concepts[1].level | 3 |
| concepts[1].score | 0.8861380219459534 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q242125 |
| concepts[1].display_name | Phylogenetic tree |
| concepts[2].id | https://openalex.org/C104317684 |
| concepts[2].level | 2 |
| concepts[2].score | 0.6676326990127563 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[2].display_name | Gene |
| concepts[3].id | https://openalex.org/C90132467 |
| concepts[3].level | 3 |
| concepts[3].score | 0.63831627368927 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q171184 |
| concepts[3].display_name | Phylogenetics |
| concepts[4].id | https://openalex.org/C78458016 |
| concepts[4].level | 1 |
| concepts[4].score | 0.6304683089256287 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q840400 |
| concepts[4].display_name | Evolutionary biology |
| concepts[5].id | https://openalex.org/C184898388 |
| concepts[5].level | 2 |
| concepts[5].score | 0.6301652193069458 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q1435712 |
| concepts[5].display_name | Pairwise comparison |
| concepts[6].id | https://openalex.org/C26619641 |
| concepts[6].level | 4 |
| concepts[6].score | 0.5748656392097473 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q3142246 |
| concepts[6].display_name | Phylogenetic network |
| concepts[7].id | https://openalex.org/C70721500 |
| concepts[7].level | 1 |
| concepts[7].score | 0.4716973304748535 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q177005 |
| concepts[7].display_name | Computational biology |
| concepts[8].id | https://openalex.org/C124388736 |
| concepts[8].level | 4 |
| concepts[8].score | 0.42581111192703247 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q3247497 |
| concepts[8].display_name | Tree of life (biology) |
| concepts[9].id | https://openalex.org/C54355233 |
| concepts[9].level | 1 |
| concepts[9].score | 0.4147166609764099 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[9].display_name | Genetics |
| concepts[10].id | https://openalex.org/C105795698 |
| concepts[10].level | 1 |
| concepts[10].score | 0.0 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q12483 |
| concepts[10].display_name | Statistics |
| concepts[11].id | https://openalex.org/C33923547 |
| concepts[11].level | 0 |
| concepts[11].score | 0.0 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q395 |
| concepts[11].display_name | Mathematics |
| keywords[0].id | https://openalex.org/keywords/biology |
| keywords[0].score | 0.9260641932487488 |
| keywords[0].display_name | Biology |
| keywords[1].id | https://openalex.org/keywords/phylogenetic-tree |
| keywords[1].score | 0.8861380219459534 |
| keywords[1].display_name | Phylogenetic tree |
| keywords[2].id | https://openalex.org/keywords/gene |
| keywords[2].score | 0.6676326990127563 |
| keywords[2].display_name | Gene |
| keywords[3].id | https://openalex.org/keywords/phylogenetics |
| keywords[3].score | 0.63831627368927 |
| keywords[3].display_name | Phylogenetics |
| keywords[4].id | https://openalex.org/keywords/evolutionary-biology |
| keywords[4].score | 0.6304683089256287 |
| keywords[4].display_name | Evolutionary biology |
| keywords[5].id | https://openalex.org/keywords/pairwise-comparison |
| keywords[5].score | 0.6301652193069458 |
| keywords[5].display_name | Pairwise comparison |
| keywords[6].id | https://openalex.org/keywords/phylogenetic-network |
| keywords[6].score | 0.5748656392097473 |
| keywords[6].display_name | Phylogenetic network |
| keywords[7].id | https://openalex.org/keywords/computational-biology |
| keywords[7].score | 0.4716973304748535 |
| keywords[7].display_name | Computational biology |
| keywords[8].id | https://openalex.org/keywords/tree-of-life |
| keywords[8].score | 0.42581111192703247 |
| keywords[8].display_name | Tree of life (biology) |
| keywords[9].id | https://openalex.org/keywords/genetics |
| keywords[9].score | 0.4147166609764099 |
| keywords[9].display_name | Genetics |
| language | en |
| locations[0].id | doi:10.1371/journal.pbio.3002633 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S154343897 |
| locations[0].source.issn | 1544-9173, 1545-7885 |
| locations[0].source.type | journal |
| locations[0].source.is_oa | True |
| locations[0].source.issn_l | 1544-9173 |
| locations[0].source.is_core | True |
| locations[0].source.is_in_doaj | True |
| locations[0].source.display_name | PLoS Biology |
| locations[0].source.host_organization | https://openalex.org/P4310315706 |
| locations[0].source.host_organization_name | Public Library of Science |
| locations[0].source.host_organization_lineage | https://openalex.org/P4310315706 |
| locations[0].source.host_organization_lineage_names | Public Library of Science |
| locations[0].license | cc-by |
| locations[0].pdf_url | https://journals.plos.org/plosbiology/article/file?id=10.1371/journal.pbio.3002633&type=printable |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | PLOS Biology |
| locations[0].landing_page_url | https://doi.org/10.1371/journal.pbio.3002633 |
| locations[1].id | pmid:38787797 |
| locations[1].is_oa | False |
| locations[1].source.id | https://openalex.org/S4306525036 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | PubMed |
| locations[1].source.host_organization | https://openalex.org/I1299303238 |
| locations[1].source.host_organization_name | National Institutes of Health |
| locations[1].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[1].license | |
| locations[1].pdf_url | |
| locations[1].version | publishedVersion |
| locations[1].raw_type | |
| locations[1].license_id | |
| locations[1].is_accepted | True |
| locations[1].is_published | True |
| locations[1].raw_source_name | PLoS biology |
| locations[1].landing_page_url | https://pubmed.ncbi.nlm.nih.gov/38787797 |
| locations[2].id | pmh:oai:pubmedcentral.nih.gov:11125556 |
| locations[2].is_oa | True |
| locations[2].source.id | https://openalex.org/S2764455111 |
| locations[2].source.issn | |
| locations[2].source.type | repository |
| locations[2].source.is_oa | False |
| locations[2].source.issn_l | |
| locations[2].source.is_core | False |
| locations[2].source.is_in_doaj | False |
| locations[2].source.display_name | PubMed Central |
| locations[2].source.host_organization | https://openalex.org/I1299303238 |
| locations[2].source.host_organization_name | National Institutes of Health |
| locations[2].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[2].license | other-oa |
| locations[2].pdf_url | |
| locations[2].version | submittedVersion |
| locations[2].raw_type | Text |
| locations[2].license_id | https://openalex.org/licenses/other-oa |
| locations[2].is_accepted | False |
| locations[2].is_published | False |
| locations[2].raw_source_name | PLoS Biol |
| locations[2].landing_page_url | https://www.ncbi.nlm.nih.gov/pmc/articles/11125556 |
| indexed_in | crossref, doaj, pubmed |
| authorships[0].author.id | https://openalex.org/A5029311537 |
| authorships[0].author.orcid | https://orcid.org/0000-0002-8451-103X |
| authorships[0].author.display_name | Samuel H. Church |
| authorships[0].countries | US |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I32971472 |
| authorships[0].affiliations[0].raw_affiliation_string | Department of Ecology and Evolutionary Biology, Yale University, New Haven, Connecticut, United States of America |
| authorships[0].institutions[0].id | https://openalex.org/I32971472 |
| authorships[0].institutions[0].ror | https://ror.org/03v76x132 |
| authorships[0].institutions[0].type | education |
| authorships[0].institutions[0].lineage | https://openalex.org/I32971472 |
| authorships[0].institutions[0].country_code | US |
| authorships[0].institutions[0].display_name | Yale University |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Samuel H. Church |
| authorships[0].is_corresponding | True |
| authorships[0].raw_affiliation_strings | Department of Ecology and Evolutionary Biology, Yale University, New Haven, Connecticut, United States of America |
| authorships[1].author.id | https://openalex.org/A5078455912 |
| authorships[1].author.orcid | https://orcid.org/0000-0003-3044-4131 |
| authorships[1].author.display_name | Jasmine L. Mah |
| authorships[1].countries | US |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I32971472 |
| authorships[1].affiliations[0].raw_affiliation_string | Department of Ecology and Evolutionary Biology, Yale University, New Haven, Connecticut, United States of America |
| authorships[1].institutions[0].id | https://openalex.org/I32971472 |
| authorships[1].institutions[0].ror | https://ror.org/03v76x132 |
| authorships[1].institutions[0].type | education |
| authorships[1].institutions[0].lineage | https://openalex.org/I32971472 |
| authorships[1].institutions[0].country_code | US |
| authorships[1].institutions[0].display_name | Yale University |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Jasmine L. Mah |
| authorships[1].is_corresponding | True |
| authorships[1].raw_affiliation_strings | Department of Ecology and Evolutionary Biology, Yale University, New Haven, Connecticut, United States of America |
| authorships[2].author.id | https://openalex.org/A5068691883 |
| authorships[2].author.orcid | https://orcid.org/0000-0003-0628-5150 |
| authorships[2].author.display_name | Casey W. Dunn |
| authorships[2].countries | US |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I32971472 |
| authorships[2].affiliations[0].raw_affiliation_string | Department of Ecology and Evolutionary Biology, Yale University, New Haven, Connecticut, United States of America |
| authorships[2].institutions[0].id | https://openalex.org/I32971472 |
| authorships[2].institutions[0].ror | https://ror.org/03v76x132 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I32971472 |
| authorships[2].institutions[0].country_code | US |
| authorships[2].institutions[0].display_name | Yale University |
| authorships[2].author_position | last |
| authorships[2].raw_author_name | Casey W. Dunn |
| authorships[2].is_corresponding | True |
| authorships[2].raw_affiliation_strings | Department of Ecology and Evolutionary Biology, Yale University, New Haven, Connecticut, United States of America |
| has_content.pdf | True |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://journals.plos.org/plosbiology/article/file?id=10.1371/journal.pbio.3002633&type=printable |
| open_access.oa_status | gold |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Integrating phylogenies into single-cell RNA sequencing analysis allows comparisons across species, genes, and cells |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T11289 |
| primary_topic.field.id | https://openalex.org/fields/13 |
| primary_topic.field.display_name | Biochemistry, Genetics and Molecular Biology |
| primary_topic.score | 0.9998000264167786 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1312 |
| primary_topic.subfield.display_name | Molecular Biology |
| primary_topic.display_name | Single-cell and spatial transcriptomics |
| related_works | https://openalex.org/W4254268221, https://openalex.org/W2038264393, https://openalex.org/W147282674, https://openalex.org/W2059637021, https://openalex.org/W2065083790, https://openalex.org/W4210644918, https://openalex.org/W2604174992, https://openalex.org/W2057191541, https://openalex.org/W2150194765, https://openalex.org/W3025857586 |
| cited_by_count | 11 |
| counts_by_year[0].year | 2025 |
| counts_by_year[0].cited_by_count | 7 |
| counts_by_year[1].year | 2024 |
| counts_by_year[1].cited_by_count | 4 |
| locations_count | 3 |
| best_oa_location.id | doi:10.1371/journal.pbio.3002633 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S154343897 |
| best_oa_location.source.issn | 1544-9173, 1545-7885 |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | True |
| best_oa_location.source.issn_l | 1544-9173 |
| best_oa_location.source.is_core | True |
| best_oa_location.source.is_in_doaj | True |
| best_oa_location.source.display_name | PLoS Biology |
| best_oa_location.source.host_organization | https://openalex.org/P4310315706 |
| best_oa_location.source.host_organization_name | Public Library of Science |
| best_oa_location.source.host_organization_lineage | https://openalex.org/P4310315706 |
| best_oa_location.source.host_organization_lineage_names | Public Library of Science |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | https://journals.plos.org/plosbiology/article/file?id=10.1371/journal.pbio.3002633&type=printable |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | journal-article |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | PLOS Biology |
| best_oa_location.landing_page_url | https://doi.org/10.1371/journal.pbio.3002633 |
| primary_location.id | doi:10.1371/journal.pbio.3002633 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S154343897 |
| primary_location.source.issn | 1544-9173, 1545-7885 |
| primary_location.source.type | journal |
| primary_location.source.is_oa | True |
| primary_location.source.issn_l | 1544-9173 |
| primary_location.source.is_core | True |
| primary_location.source.is_in_doaj | True |
| primary_location.source.display_name | PLoS Biology |
| primary_location.source.host_organization | https://openalex.org/P4310315706 |
| primary_location.source.host_organization_name | Public Library of Science |
| primary_location.source.host_organization_lineage | https://openalex.org/P4310315706 |
| primary_location.source.host_organization_lineage_names | Public Library of Science |
| primary_location.license | cc-by |
| primary_location.pdf_url | https://journals.plos.org/plosbiology/article/file?id=10.1371/journal.pbio.3002633&type=printable |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | PLOS Biology |
| primary_location.landing_page_url | https://doi.org/10.1371/journal.pbio.3002633 |
| publication_date | 2024-05-24 |
| publication_year | 2024 |
| referenced_works | https://openalex.org/W2269040222, https://openalex.org/W3160097893, https://openalex.org/W2809507176, https://openalex.org/W3122547168, https://openalex.org/W3157099244, https://openalex.org/W4372335730, https://openalex.org/W3159175200, https://openalex.org/W3012842442, https://openalex.org/W2001157929, https://openalex.org/W2116624183, https://openalex.org/W2553676377, https://openalex.org/W2072977772, https://openalex.org/W4327575912, https://openalex.org/W2169821963, https://openalex.org/W3020787789, https://openalex.org/W2782262419, https://openalex.org/W2013410948, https://openalex.org/W4317759286, https://openalex.org/W1967814055, https://openalex.org/W2029102865, https://openalex.org/W2526917659, https://openalex.org/W2968069281, https://openalex.org/W2912390237, https://openalex.org/W2138181932, https://openalex.org/W2565054888, https://openalex.org/W4210519385, https://openalex.org/W2506408709, https://openalex.org/W2119338396, https://openalex.org/W3134710066, https://openalex.org/W2075338411, https://openalex.org/W2171939839, https://openalex.org/W4390615391, https://openalex.org/W2103682580, https://openalex.org/W3210441301, https://openalex.org/W2300048866, https://openalex.org/W2948469692, https://openalex.org/W2794480084, https://openalex.org/W2953405766, https://openalex.org/W2611974824, https://openalex.org/W4388569079, https://openalex.org/W2069089843, https://openalex.org/W2949237386, https://openalex.org/W2288153033, https://openalex.org/W2794891507, https://openalex.org/W2998065416, https://openalex.org/W3009609875, https://openalex.org/W2997280954, https://openalex.org/W3111378927, https://openalex.org/W1986367828, https://openalex.org/W2171193618, https://openalex.org/W193391966, https://openalex.org/W3105577581, https://openalex.org/W156146614, https://openalex.org/W4233459387, https://openalex.org/W4252783855 |
| referenced_works_count | 55 |
| abstract_inverted_index.a | 58 |
| abstract_inverted_index.By | 105 |
| abstract_inverted_index.as | 30, 95 |
| abstract_inverted_index.be | 93, 120, 130 |
| abstract_inverted_index.by | 32 |
| abstract_inverted_index.in | 49 |
| abstract_inverted_index.of | 1, 19, 47, 74, 97, 102 |
| abstract_inverted_index.to | 66 |
| abstract_inverted_index.RNA | 3 |
| abstract_inverted_index.all | 92 |
| abstract_inverted_index.and | 16, 23, 41, 54, 62, 88, 122, 126 |
| abstract_inverted_index.can | 9, 91, 119, 129 |
| abstract_inverted_index.for | 70, 113 |
| abstract_inverted_index.out | 52 |
| abstract_inverted_index.the | 17, 71, 98 |
| abstract_inverted_index.This | 43, 78 |
| abstract_inverted_index.cell | 20, 86, 89, 127 |
| abstract_inverted_index.data | 6 |
| abstract_inverted_index.each | 46 |
| abstract_inverted_index.from | 57 |
| abstract_inverted_index.gene | 14, 84, 125 |
| abstract_inverted_index.into | 109 |
| abstract_inverted_index.same | 99 |
| abstract_inverted_index.that | 35, 81 |
| abstract_inverted_index.tree | 101 |
| abstract_inverted_index.Essay | 44, 79 |
| abstract_inverted_index.These | 25 |
| abstract_inverted_index.about | 124 |
| abstract_inverted_index.evoke | 27 |
| abstract_inverted_index.life. | 104 |
| abstract_inverted_index.links | 11 |
| abstract_inverted_index.maps. | 77 |
| abstract_inverted_index.these | 48, 64 |
| abstract_inverted_index.turn, | 50 |
| abstract_inverted_index.across | 7, 117 |
| abstract_inverted_index.cells. | 42 |
| abstract_inverted_index.define | 36 |
| abstract_inverted_index.genes, | 40 |
| abstract_inverted_index.laying | 51 |
| abstract_inverted_index.reveal | 10 |
| abstract_inverted_index.trees, | 34, 83, 85 |
| abstract_inverted_index.between | 12, 38 |
| abstract_inverted_index.derived | 56 |
| abstract_inverted_index.methods | 69 |
| abstract_inverted_index.species | 8, 82, 118 |
| abstract_inverted_index.tested. | 132 |
| abstract_inverted_index.approach | 61 |
| abstract_inverted_index.building | 114 |
| abstract_inverted_index.cellular | 13, 75, 103 |
| abstract_inverted_index.contends | 80 |
| abstract_inverted_index.depicted | 31 |
| abstract_inverted_index.informed | 115 |
| abstract_inverted_index.lineages | 90 |
| abstract_inverted_index.pairwise | 72 |
| abstract_inverted_index.proposed | 68 |
| abstract_inverted_index.relating | 63 |
| abstract_inverted_index.robustly | 131 |
| abstract_inverted_index.species, | 39 |
| abstract_inverted_index.alignment | 73 |
| abstract_inverted_index.analyses, | 111 |
| abstract_inverted_index.considers | 45 |
| abstract_inverted_index.evolution | 18, 128 |
| abstract_inverted_index.features, | 22 |
| abstract_inverted_index.overcome, | 121 |
| abstract_inverted_index.scRNA-seq | 110 |
| abstract_inverted_index.solutions | 55, 65 |
| abstract_inverted_index.approaches | 108 |
| abstract_inverted_index.challenges | 53, 112 |
| abstract_inverted_index.expression | 15 |
| abstract_inverted_index.functions, | 21 |
| abstract_inverted_index.histories, | 29 |
| abstract_inverted_index.hypotheses | 123 |
| abstract_inverted_index.previously | 67 |
| abstract_inverted_index.reconciled | 94 |
| abstract_inverted_index.sequencing | 4 |
| abstract_inverted_index.(scRNA-seq) | 5 |
| abstract_inverted_index.Comparisons | 0 |
| abstract_inverted_index.comparative | 60 |
| abstract_inverted_index.comparisons | 26, 116 |
| abstract_inverted_index.dimensional | 76 |
| abstract_inverted_index.integrating | 106 |
| abstract_inverted_index.phenotypes. | 24 |
| abstract_inverted_index.single-cell | 2 |
| abstract_inverted_index.descriptions | 96 |
| abstract_inverted_index.evolutionary | 28 |
| abstract_inverted_index.phylogenetic | 33, 59, 107 |
| abstract_inverted_index.phylogenies, | 87 |
| abstract_inverted_index.concept—the | 100 |
| abstract_inverted_index.relationships | 37 |
| cited_by_percentile_year.max | 99 |
| cited_by_percentile_year.min | 97 |
| corresponding_author_ids | https://openalex.org/A5029311537, https://openalex.org/A5068691883, https://openalex.org/A5078455912 |
| countries_distinct_count | 1 |
| institutions_distinct_count | 3 |
| corresponding_institution_ids | https://openalex.org/I32971472 |
| sustainable_development_goals[0].id | https://metadata.un.org/sdg/15 |
| sustainable_development_goals[0].score | 0.5299999713897705 |
| sustainable_development_goals[0].display_name | Life in Land |
| citation_normalized_percentile.value | 0.93179619 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | True |