ISCO: Intelligent Framework for Accurate Segmentation and Comparative Analysis of Organoids Article Swipe
 YOU?
·
· 2024
· Open Access
·
· DOI: https://doi.org/10.1101/2024.12.24.630244
YOU?
·
· 2024
· Open Access
·
· DOI: https://doi.org/10.1101/2024.12.24.630244
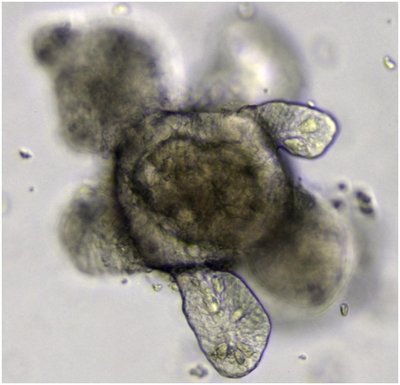
Organoids are self-organizing 3D cell clusters that closely mimic the structure and function of in vivo tissues and organs. Quantifying organoid morphology is critical for advancing our understanding of organ development, drug discovery, and toxicity assessment. Recent advances in microscopy have provided powerful tools to capture detailed morphological features of organoids, yet manual image analysis remains labor-intensive and time-consuming. In response, we present a comprehensive microscopy-based analysis pipeline that utilizes SegmentAnything 2.1 to accurately segment individual organoids. Additionally, we introduce a suite of morphological features—including perimeter, area, radius, non-smoothness, and non-circularity—that enable researchers to quantitatively and automatically analyze organoid structures. To further standardize organoid analysis across the field, Intelligent segmentation and comparison of organoids(ISCO), an intelligent AI-driven open-source algorithm, is developed with the aim of establishing a comprehensive toolset for organoid characterization.
Related Topics
- Type
- preprint
- Language
- en
- Landing Page
- https://doi.org/10.1101/2024.12.24.630244
- OA Status
- green
- References
- 5
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4405740483
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4405740483Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1101/2024.12.24.630244Digital Object Identifier
- Title
-
ISCO: Intelligent Framework for Accurate Segmentation and Comparative Analysis of OrganoidsWork title
- Type
-
preprintOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2024Year of publication
- Publication date
-
2024-12-24Full publication date if available
- Authors
-
Jian Zhou, Zihao Fu, Xiaohua Ni, Qi Luo, Gen YangList of authors in order
- Landing page
-
https://doi.org/10.1101/2024.12.24.630244Publisher landing page
- Open access
-
YesWhether a free full text is available
- OA status
-
greenOpen access status per OpenAlex
- OA URL
-
https://doi.org/10.1101/2024.12.24.630244Direct OA link when available
- Concepts
-
Organoid, Segmentation, Computer science, Computational biology, Artificial intelligence, Biology, Cell biologyTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
0Total citation count in OpenAlex
- References (count)
-
5Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4405740483 |
|---|---|
| doi | https://doi.org/10.1101/2024.12.24.630244 |
| ids.doi | https://doi.org/10.1101/2024.12.24.630244 |
| ids.openalex | https://openalex.org/W4405740483 |
| fwci | 0.0 |
| type | preprint |
| title | ISCO: Intelligent Framework for Accurate Segmentation and Comparative Analysis of Organoids |
| biblio.issue | |
| biblio.volume | |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T10640 |
| topics[0].field.id | https://openalex.org/fields/16 |
| topics[0].field.display_name | Chemistry |
| topics[0].score | 0.7379000186920166 |
| topics[0].domain.id | https://openalex.org/domains/3 |
| topics[0].domain.display_name | Physical Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1602 |
| topics[0].subfield.display_name | Analytical Chemistry |
| topics[0].display_name | Spectroscopy and Chemometric Analyses |
| topics[1].id | https://openalex.org/T13937 |
| topics[1].field.id | https://openalex.org/fields/13 |
| topics[1].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[1].score | 0.7279000282287598 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1312 |
| topics[1].subfield.display_name | Molecular Biology |
| topics[1].display_name | Genetics, Bioinformatics, and Biomedical Research |
| topics[2].id | https://openalex.org/T12282 |
| topics[2].field.id | https://openalex.org/fields/22 |
| topics[2].field.display_name | Engineering |
| topics[2].score | 0.6897000074386597 |
| topics[2].domain.id | https://openalex.org/domains/3 |
| topics[2].domain.display_name | Physical Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/2210 |
| topics[2].subfield.display_name | Mechanical Engineering |
| topics[2].display_name | Mineral Processing and Grinding |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C31695470 |
| concepts[0].level | 2 |
| concepts[0].score | 0.8339677453041077 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q11293125 |
| concepts[0].display_name | Organoid |
| concepts[1].id | https://openalex.org/C89600930 |
| concepts[1].level | 2 |
| concepts[1].score | 0.6898863315582275 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q1423946 |
| concepts[1].display_name | Segmentation |
| concepts[2].id | https://openalex.org/C41008148 |
| concepts[2].level | 0 |
| concepts[2].score | 0.5871887803077698 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q21198 |
| concepts[2].display_name | Computer science |
| concepts[3].id | https://openalex.org/C70721500 |
| concepts[3].level | 1 |
| concepts[3].score | 0.460651159286499 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q177005 |
| concepts[3].display_name | Computational biology |
| concepts[4].id | https://openalex.org/C154945302 |
| concepts[4].level | 1 |
| concepts[4].score | 0.36294496059417725 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q11660 |
| concepts[4].display_name | Artificial intelligence |
| concepts[5].id | https://openalex.org/C86803240 |
| concepts[5].level | 0 |
| concepts[5].score | 0.1805589199066162 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[5].display_name | Biology |
| concepts[6].id | https://openalex.org/C95444343 |
| concepts[6].level | 1 |
| concepts[6].score | 0.1136401891708374 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q7141 |
| concepts[6].display_name | Cell biology |
| keywords[0].id | https://openalex.org/keywords/organoid |
| keywords[0].score | 0.8339677453041077 |
| keywords[0].display_name | Organoid |
| keywords[1].id | https://openalex.org/keywords/segmentation |
| keywords[1].score | 0.6898863315582275 |
| keywords[1].display_name | Segmentation |
| keywords[2].id | https://openalex.org/keywords/computer-science |
| keywords[2].score | 0.5871887803077698 |
| keywords[2].display_name | Computer science |
| keywords[3].id | https://openalex.org/keywords/computational-biology |
| keywords[3].score | 0.460651159286499 |
| keywords[3].display_name | Computational biology |
| keywords[4].id | https://openalex.org/keywords/artificial-intelligence |
| keywords[4].score | 0.36294496059417725 |
| keywords[4].display_name | Artificial intelligence |
| keywords[5].id | https://openalex.org/keywords/biology |
| keywords[5].score | 0.1805589199066162 |
| keywords[5].display_name | Biology |
| keywords[6].id | https://openalex.org/keywords/cell-biology |
| keywords[6].score | 0.1136401891708374 |
| keywords[6].display_name | Cell biology |
| language | en |
| locations[0].id | doi:10.1101/2024.12.24.630244 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4306402567 |
| locations[0].source.issn | |
| locations[0].source.type | repository |
| locations[0].source.is_oa | False |
| locations[0].source.issn_l | |
| locations[0].source.is_core | False |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| locations[0].source.host_organization | https://openalex.org/I2750212522 |
| locations[0].source.host_organization_name | Cold Spring Harbor Laboratory |
| locations[0].source.host_organization_lineage | https://openalex.org/I2750212522 |
| locations[0].license | cc-by-nc-nd |
| locations[0].pdf_url | |
| locations[0].version | acceptedVersion |
| locations[0].raw_type | posted-content |
| locations[0].license_id | https://openalex.org/licenses/cc-by-nc-nd |
| locations[0].is_accepted | True |
| locations[0].is_published | False |
| locations[0].raw_source_name | |
| locations[0].landing_page_url | https://doi.org/10.1101/2024.12.24.630244 |
| indexed_in | crossref |
| authorships[0].author.id | https://openalex.org/A5027790964 |
| authorships[0].author.orcid | https://orcid.org/0000-0002-1534-2279 |
| authorships[0].author.display_name | Jian Zhou |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | JIAN ZHOU |
| authorships[0].is_corresponding | False |
| authorships[1].author.id | https://openalex.org/A5101393043 |
| authorships[1].author.orcid | |
| authorships[1].author.display_name | Zihao Fu |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Zihao Fu |
| authorships[1].is_corresponding | False |
| authorships[2].author.id | https://openalex.org/A5103060163 |
| authorships[2].author.orcid | https://orcid.org/0000-0002-9527-6426 |
| authorships[2].author.display_name | Xiaohua Ni |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Xiaoying Ni |
| authorships[2].is_corresponding | False |
| authorships[3].author.id | https://openalex.org/A5101625460 |
| authorships[3].author.orcid | https://orcid.org/0000-0002-0393-6352 |
| authorships[3].author.display_name | Qi Luo |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Qi Luo |
| authorships[3].is_corresponding | False |
| authorships[4].author.id | https://openalex.org/A5040160528 |
| authorships[4].author.orcid | |
| authorships[4].author.display_name | Gen Yang |
| authorships[4].author_position | last |
| authorships[4].raw_author_name | Gen Yang |
| authorships[4].is_corresponding | False |
| has_content.pdf | False |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://doi.org/10.1101/2024.12.24.630244 |
| open_access.oa_status | green |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | ISCO: Intelligent Framework for Accurate Segmentation and Comparative Analysis of Organoids |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T10640 |
| primary_topic.field.id | https://openalex.org/fields/16 |
| primary_topic.field.display_name | Chemistry |
| primary_topic.score | 0.7379000186920166 |
| primary_topic.domain.id | https://openalex.org/domains/3 |
| primary_topic.domain.display_name | Physical Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1602 |
| primary_topic.subfield.display_name | Analytical Chemistry |
| primary_topic.display_name | Spectroscopy and Chemometric Analyses |
| related_works | https://openalex.org/W4391375266, https://openalex.org/W2899084033, https://openalex.org/W2748952813, https://openalex.org/W4386439204, https://openalex.org/W4401794395, https://openalex.org/W3133804457, https://openalex.org/W3033121461, https://openalex.org/W3094248195, https://openalex.org/W2811397904, https://openalex.org/W3047600752 |
| cited_by_count | 0 |
| locations_count | 1 |
| best_oa_location.id | doi:10.1101/2024.12.24.630244 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4306402567 |
| best_oa_location.source.issn | |
| best_oa_location.source.type | repository |
| best_oa_location.source.is_oa | False |
| best_oa_location.source.issn_l | |
| best_oa_location.source.is_core | False |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| best_oa_location.source.host_organization | https://openalex.org/I2750212522 |
| best_oa_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| best_oa_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| best_oa_location.license | cc-by-nc-nd |
| best_oa_location.pdf_url | |
| best_oa_location.version | acceptedVersion |
| best_oa_location.raw_type | posted-content |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by-nc-nd |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | False |
| best_oa_location.raw_source_name | |
| best_oa_location.landing_page_url | https://doi.org/10.1101/2024.12.24.630244 |
| primary_location.id | doi:10.1101/2024.12.24.630244 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4306402567 |
| primary_location.source.issn | |
| primary_location.source.type | repository |
| primary_location.source.is_oa | False |
| primary_location.source.issn_l | |
| primary_location.source.is_core | False |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| primary_location.source.host_organization | https://openalex.org/I2750212522 |
| primary_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| primary_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| primary_location.license | cc-by-nc-nd |
| primary_location.pdf_url | |
| primary_location.version | acceptedVersion |
| primary_location.raw_type | posted-content |
| primary_location.license_id | https://openalex.org/licenses/cc-by-nc-nd |
| primary_location.is_accepted | True |
| primary_location.is_published | False |
| primary_location.raw_source_name | |
| primary_location.landing_page_url | https://doi.org/10.1101/2024.12.24.630244 |
| publication_date | 2024-12-24 |
| publication_year | 2024 |
| referenced_works | https://openalex.org/W3197097711, https://openalex.org/W4391446966, https://openalex.org/W4308624890, https://openalex.org/W3115787212, https://openalex.org/W3011139338 |
| referenced_works_count | 5 |
| abstract_inverted_index.a | 64, 81, 127 |
| abstract_inverted_index.3D | 4 |
| abstract_inverted_index.In | 60 |
| abstract_inverted_index.To | 101 |
| abstract_inverted_index.an | 115 |
| abstract_inverted_index.in | 15, 39 |
| abstract_inverted_index.is | 23, 120 |
| abstract_inverted_index.of | 14, 29, 50, 83, 113, 125 |
| abstract_inverted_index.to | 45, 73, 94 |
| abstract_inverted_index.we | 62, 79 |
| abstract_inverted_index.2.1 | 72 |
| abstract_inverted_index.aim | 124 |
| abstract_inverted_index.and | 12, 18, 34, 58, 90, 96, 111 |
| abstract_inverted_index.are | 2 |
| abstract_inverted_index.for | 25, 130 |
| abstract_inverted_index.our | 27 |
| abstract_inverted_index.the | 10, 107, 123 |
| abstract_inverted_index.yet | 52 |
| abstract_inverted_index.cell | 5 |
| abstract_inverted_index.drug | 32 |
| abstract_inverted_index.have | 41 |
| abstract_inverted_index.that | 7, 69 |
| abstract_inverted_index.vivo | 16 |
| abstract_inverted_index.with | 122 |
| abstract_inverted_index.area, | 87 |
| abstract_inverted_index.image | 54 |
| abstract_inverted_index.mimic | 9 |
| abstract_inverted_index.organ | 30 |
| abstract_inverted_index.suite | 82 |
| abstract_inverted_index.tools | 44 |
| abstract_inverted_index.Recent | 37 |
| abstract_inverted_index.across | 106 |
| abstract_inverted_index.enable | 92 |
| abstract_inverted_index.field, | 108 |
| abstract_inverted_index.manual | 53 |
| abstract_inverted_index.analyze | 98 |
| abstract_inverted_index.capture | 46 |
| abstract_inverted_index.closely | 8 |
| abstract_inverted_index.further | 102 |
| abstract_inverted_index.organs. | 19 |
| abstract_inverted_index.present | 63 |
| abstract_inverted_index.radius, | 88 |
| abstract_inverted_index.remains | 56 |
| abstract_inverted_index.segment | 75 |
| abstract_inverted_index.tissues | 17 |
| abstract_inverted_index.toolset | 129 |
| abstract_inverted_index.Abstract | 0 |
| abstract_inverted_index.advances | 38 |
| abstract_inverted_index.analysis | 55, 67, 105 |
| abstract_inverted_index.clusters | 6 |
| abstract_inverted_index.critical | 24 |
| abstract_inverted_index.detailed | 47 |
| abstract_inverted_index.features | 49 |
| abstract_inverted_index.function | 13 |
| abstract_inverted_index.organoid | 21, 99, 104, 131 |
| abstract_inverted_index.pipeline | 68 |
| abstract_inverted_index.powerful | 43 |
| abstract_inverted_index.provided | 42 |
| abstract_inverted_index.toxicity | 35 |
| abstract_inverted_index.utilizes | 70 |
| abstract_inverted_index.AI-driven | 117 |
| abstract_inverted_index.Organoids | 1 |
| abstract_inverted_index.advancing | 26 |
| abstract_inverted_index.developed | 121 |
| abstract_inverted_index.introduce | 80 |
| abstract_inverted_index.response, | 61 |
| abstract_inverted_index.structure | 11 |
| abstract_inverted_index.accurately | 74 |
| abstract_inverted_index.algorithm, | 119 |
| abstract_inverted_index.comparison | 112 |
| abstract_inverted_index.discovery, | 33 |
| abstract_inverted_index.individual | 76 |
| abstract_inverted_index.microscopy | 40 |
| abstract_inverted_index.morphology | 22 |
| abstract_inverted_index.organoids, | 51 |
| abstract_inverted_index.organoids. | 77 |
| abstract_inverted_index.perimeter, | 86 |
| abstract_inverted_index.Intelligent | 109 |
| abstract_inverted_index.Quantifying | 20 |
| abstract_inverted_index.assessment. | 36 |
| abstract_inverted_index.intelligent | 116 |
| abstract_inverted_index.open-source | 118 |
| abstract_inverted_index.researchers | 93 |
| abstract_inverted_index.standardize | 103 |
| abstract_inverted_index.structures. | 100 |
| abstract_inverted_index.development, | 31 |
| abstract_inverted_index.establishing | 126 |
| abstract_inverted_index.segmentation | 110 |
| abstract_inverted_index.Additionally, | 78 |
| abstract_inverted_index.automatically | 97 |
| abstract_inverted_index.comprehensive | 65, 128 |
| abstract_inverted_index.morphological | 48, 84 |
| abstract_inverted_index.understanding | 28 |
| abstract_inverted_index.quantitatively | 95 |
| abstract_inverted_index.SegmentAnything | 71 |
| abstract_inverted_index.labor-intensive | 57 |
| abstract_inverted_index.non-smoothness, | 89 |
| abstract_inverted_index.self-organizing | 3 |
| abstract_inverted_index.time-consuming. | 59 |
| abstract_inverted_index.microscopy-based | 66 |
| abstract_inverted_index.organoids(ISCO), | 114 |
| abstract_inverted_index.characterization. | 132 |
| abstract_inverted_index.features—including | 85 |
| abstract_inverted_index.non-circularity—that | 91 |
| cited_by_percentile_year | |
| countries_distinct_count | 0 |
| institutions_distinct_count | 5 |
| sustainable_development_goals[0].id | https://metadata.un.org/sdg/8 |
| sustainable_development_goals[0].score | 0.46000000834465027 |
| sustainable_development_goals[0].display_name | Decent work and economic growth |
| citation_normalized_percentile.value | 0.30585106 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |