Ligand Binding Prediction Using Protein Structure Graphs and Residual Graph Attention Networks Article Swipe
 YOU?
·
· 2022
· Open Access
·
· DOI: https://doi.org/10.3390/molecules27165114
YOU?
·
· 2022
· Open Access
·
· DOI: https://doi.org/10.3390/molecules27165114
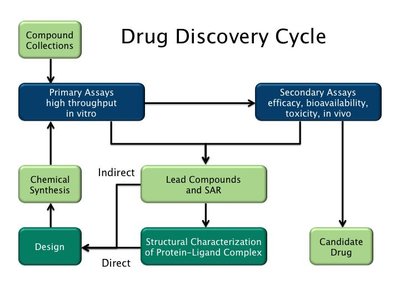
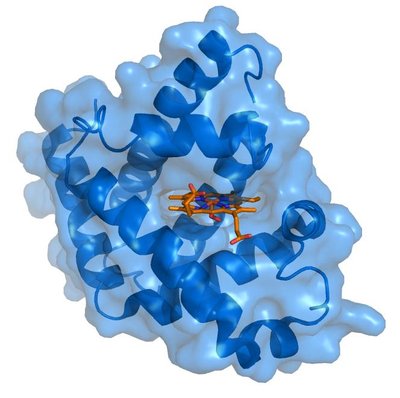
Computational prediction of ligand–target interactions is a crucial part of modern drug discovery as it helps to bypass high costs and labor demands of in vitro and in vivo screening. As the wealth of bioactivity data accumulates, it provides opportunities for the development of deep learning (DL) models with increasing predictive powers. Conventionally, such models were either limited to the use of very simplified representations of proteins or ineffective voxelization of their 3D structures. Herein, we present the development of the PSG-BAR (Protein Structure Graph-Binding Affinity Regression) approach that utilizes 3D structural information of the proteins along with 2D graph representations of ligands. The method also introduces attention scores to selectively weight protein regions that are most important for ligand binding. Results: The developed approach demonstrates the state-of-the-art performance on several binding affinity benchmarking datasets. The attention-based pooling of protein graphs enables identification of surface residues as critical residues for protein–ligand binding. Finally, we validate our model predictions against an experimental assay on a viral main protease (Mpro)—the hallmark target of SARS-CoV-2 coronavirus.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.3390/molecules27165114
- https://www.mdpi.com/1420-3049/27/16/5114/pdf?version=1660214839
- OA Status
- gold
- Cited By
- 19
- References
- 63
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4290998851
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4290998851Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.3390/molecules27165114Digital Object Identifier
- Title
-
Ligand Binding Prediction Using Protein Structure Graphs and Residual Graph Attention NetworksWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2022Year of publication
- Publication date
-
2022-08-11Full publication date if available
- Authors
-
Mohit Pandey, Mariia Radaeva, Hazem Mslati, Olivia Garland, Michael Fernández, Martin Ester, Artem CherkasovList of authors in order
- Landing page
-
https://doi.org/10.3390/molecules27165114Publisher landing page
- PDF URL
-
https://www.mdpi.com/1420-3049/27/16/5114/pdf?version=1660214839Direct link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
goldOpen access status per OpenAlex
- OA URL
-
https://www.mdpi.com/1420-3049/27/16/5114/pdf?version=1660214839Direct OA link when available
- Concepts
-
Computational biology, Drug discovery, Pooling, Ligand (biochemistry), Graph, Computer science, Protein ligand, Target protein, Chemistry, Artificial intelligence, Machine learning, Biology, Biochemistry, Theoretical computer science, Receptor, GeneTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
19Total citation count in OpenAlex
- Citations by year (recent)
-
2025: 4, 2024: 10, 2023: 4, 2022: 1Per-year citation counts (last 5 years)
- References (count)
-
63Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4290998851 |
|---|---|
| doi | https://doi.org/10.3390/molecules27165114 |
| ids.doi | https://doi.org/10.3390/molecules27165114 |
| ids.pmid | https://pubmed.ncbi.nlm.nih.gov/36014351 |
| ids.openalex | https://openalex.org/W4290998851 |
| fwci | 5.00360751 |
| mesh[0].qualifier_ui | |
| mesh[0].descriptor_ui | D000086382 |
| mesh[0].is_major_topic | True |
| mesh[0].qualifier_name | |
| mesh[0].descriptor_name | COVID-19 |
| mesh[1].qualifier_ui | |
| mesh[1].descriptor_ui | D006801 |
| mesh[1].is_major_topic | False |
| mesh[1].qualifier_name | |
| mesh[1].descriptor_name | Humans |
| mesh[2].qualifier_ui | |
| mesh[2].descriptor_ui | D008024 |
| mesh[2].is_major_topic | False |
| mesh[2].qualifier_name | |
| mesh[2].descriptor_name | Ligands |
| mesh[3].qualifier_ui | |
| mesh[3].descriptor_ui | D011485 |
| mesh[3].is_major_topic | False |
| mesh[3].qualifier_name | |
| mesh[3].descriptor_name | Protein Binding |
| mesh[4].qualifier_ui | Q000737 |
| mesh[4].descriptor_ui | D011506 |
| mesh[4].is_major_topic | False |
| mesh[4].qualifier_name | chemistry |
| mesh[4].descriptor_name | Proteins |
| mesh[5].qualifier_ui | |
| mesh[5].descriptor_ui | D000086402 |
| mesh[5].is_major_topic | True |
| mesh[5].qualifier_name | |
| mesh[5].descriptor_name | SARS-CoV-2 |
| mesh[6].qualifier_ui | |
| mesh[6].descriptor_ui | D000086382 |
| mesh[6].is_major_topic | True |
| mesh[6].qualifier_name | |
| mesh[6].descriptor_name | COVID-19 |
| mesh[7].qualifier_ui | |
| mesh[7].descriptor_ui | D006801 |
| mesh[7].is_major_topic | False |
| mesh[7].qualifier_name | |
| mesh[7].descriptor_name | Humans |
| mesh[8].qualifier_ui | |
| mesh[8].descriptor_ui | D008024 |
| mesh[8].is_major_topic | False |
| mesh[8].qualifier_name | |
| mesh[8].descriptor_name | Ligands |
| mesh[9].qualifier_ui | |
| mesh[9].descriptor_ui | D011485 |
| mesh[9].is_major_topic | False |
| mesh[9].qualifier_name | |
| mesh[9].descriptor_name | Protein Binding |
| mesh[10].qualifier_ui | Q000737 |
| mesh[10].descriptor_ui | D011506 |
| mesh[10].is_major_topic | False |
| mesh[10].qualifier_name | chemistry |
| mesh[10].descriptor_name | Proteins |
| mesh[11].qualifier_ui | |
| mesh[11].descriptor_ui | D000086402 |
| mesh[11].is_major_topic | True |
| mesh[11].qualifier_name | |
| mesh[11].descriptor_name | SARS-CoV-2 |
| type | article |
| title | Ligand Binding Prediction Using Protein Structure Graphs and Residual Graph Attention Networks |
| biblio.issue | 16 |
| biblio.volume | 27 |
| biblio.last_page | 5114 |
| biblio.first_page | 5114 |
| topics[0].id | https://openalex.org/T10211 |
| topics[0].field.id | https://openalex.org/fields/17 |
| topics[0].field.display_name | Computer Science |
| topics[0].score | 0.9998999834060669 |
| topics[0].domain.id | https://openalex.org/domains/3 |
| topics[0].domain.display_name | Physical Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1703 |
| topics[0].subfield.display_name | Computational Theory and Mathematics |
| topics[0].display_name | Computational Drug Discovery Methods |
| topics[1].id | https://openalex.org/T10044 |
| topics[1].field.id | https://openalex.org/fields/13 |
| topics[1].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[1].score | 0.9993000030517578 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1312 |
| topics[1].subfield.display_name | Molecular Biology |
| topics[1].display_name | Protein Structure and Dynamics |
| topics[2].id | https://openalex.org/T10887 |
| topics[2].field.id | https://openalex.org/fields/13 |
| topics[2].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[2].score | 0.9898999929428101 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1312 |
| topics[2].subfield.display_name | Molecular Biology |
| topics[2].display_name | Bioinformatics and Genomic Networks |
| is_xpac | False |
| apc_list.value | 2300 |
| apc_list.currency | CHF |
| apc_list.value_usd | 2490 |
| apc_paid.value | 2300 |
| apc_paid.currency | CHF |
| apc_paid.value_usd | 2490 |
| concepts[0].id | https://openalex.org/C70721500 |
| concepts[0].level | 1 |
| concepts[0].score | 0.576322615146637 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q177005 |
| concepts[0].display_name | Computational biology |
| concepts[1].id | https://openalex.org/C74187038 |
| concepts[1].level | 2 |
| concepts[1].score | 0.5706411600112915 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q1418791 |
| concepts[1].display_name | Drug discovery |
| concepts[2].id | https://openalex.org/C70437156 |
| concepts[2].level | 2 |
| concepts[2].score | 0.5643045902252197 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q7228652 |
| concepts[2].display_name | Pooling |
| concepts[3].id | https://openalex.org/C116569031 |
| concepts[3].level | 3 |
| concepts[3].score | 0.5622912645339966 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q899107 |
| concepts[3].display_name | Ligand (biochemistry) |
| concepts[4].id | https://openalex.org/C132525143 |
| concepts[4].level | 2 |
| concepts[4].score | 0.5296837091445923 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q141488 |
| concepts[4].display_name | Graph |
| concepts[5].id | https://openalex.org/C41008148 |
| concepts[5].level | 0 |
| concepts[5].score | 0.5178933143615723 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q21198 |
| concepts[5].display_name | Computer science |
| concepts[6].id | https://openalex.org/C109095088 |
| concepts[6].level | 2 |
| concepts[6].score | 0.4835802912712097 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q899107 |
| concepts[6].display_name | Protein ligand |
| concepts[7].id | https://openalex.org/C2780430158 |
| concepts[7].level | 3 |
| concepts[7].score | 0.4160418212413788 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q7685902 |
| concepts[7].display_name | Target protein |
| concepts[8].id | https://openalex.org/C185592680 |
| concepts[8].level | 0 |
| concepts[8].score | 0.38809090852737427 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q2329 |
| concepts[8].display_name | Chemistry |
| concepts[9].id | https://openalex.org/C154945302 |
| concepts[9].level | 1 |
| concepts[9].score | 0.3748767375946045 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q11660 |
| concepts[9].display_name | Artificial intelligence |
| concepts[10].id | https://openalex.org/C119857082 |
| concepts[10].level | 1 |
| concepts[10].score | 0.3723667860031128 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q2539 |
| concepts[10].display_name | Machine learning |
| concepts[11].id | https://openalex.org/C86803240 |
| concepts[11].level | 0 |
| concepts[11].score | 0.26499515771865845 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[11].display_name | Biology |
| concepts[12].id | https://openalex.org/C55493867 |
| concepts[12].level | 1 |
| concepts[12].score | 0.21235612034797668 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q7094 |
| concepts[12].display_name | Biochemistry |
| concepts[13].id | https://openalex.org/C80444323 |
| concepts[13].level | 1 |
| concepts[13].score | 0.19042861461639404 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q2878974 |
| concepts[13].display_name | Theoretical computer science |
| concepts[14].id | https://openalex.org/C170493617 |
| concepts[14].level | 2 |
| concepts[14].score | 0.1096111536026001 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q208467 |
| concepts[14].display_name | Receptor |
| concepts[15].id | https://openalex.org/C104317684 |
| concepts[15].level | 2 |
| concepts[15].score | 0.0 |
| concepts[15].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[15].display_name | Gene |
| keywords[0].id | https://openalex.org/keywords/computational-biology |
| keywords[0].score | 0.576322615146637 |
| keywords[0].display_name | Computational biology |
| keywords[1].id | https://openalex.org/keywords/drug-discovery |
| keywords[1].score | 0.5706411600112915 |
| keywords[1].display_name | Drug discovery |
| keywords[2].id | https://openalex.org/keywords/pooling |
| keywords[2].score | 0.5643045902252197 |
| keywords[2].display_name | Pooling |
| keywords[3].id | https://openalex.org/keywords/ligand |
| keywords[3].score | 0.5622912645339966 |
| keywords[3].display_name | Ligand (biochemistry) |
| keywords[4].id | https://openalex.org/keywords/graph |
| keywords[4].score | 0.5296837091445923 |
| keywords[4].display_name | Graph |
| keywords[5].id | https://openalex.org/keywords/computer-science |
| keywords[5].score | 0.5178933143615723 |
| keywords[5].display_name | Computer science |
| keywords[6].id | https://openalex.org/keywords/protein-ligand |
| keywords[6].score | 0.4835802912712097 |
| keywords[6].display_name | Protein ligand |
| keywords[7].id | https://openalex.org/keywords/target-protein |
| keywords[7].score | 0.4160418212413788 |
| keywords[7].display_name | Target protein |
| keywords[8].id | https://openalex.org/keywords/chemistry |
| keywords[8].score | 0.38809090852737427 |
| keywords[8].display_name | Chemistry |
| keywords[9].id | https://openalex.org/keywords/artificial-intelligence |
| keywords[9].score | 0.3748767375946045 |
| keywords[9].display_name | Artificial intelligence |
| keywords[10].id | https://openalex.org/keywords/machine-learning |
| keywords[10].score | 0.3723667860031128 |
| keywords[10].display_name | Machine learning |
| keywords[11].id | https://openalex.org/keywords/biology |
| keywords[11].score | 0.26499515771865845 |
| keywords[11].display_name | Biology |
| keywords[12].id | https://openalex.org/keywords/biochemistry |
| keywords[12].score | 0.21235612034797668 |
| keywords[12].display_name | Biochemistry |
| keywords[13].id | https://openalex.org/keywords/theoretical-computer-science |
| keywords[13].score | 0.19042861461639404 |
| keywords[13].display_name | Theoretical computer science |
| keywords[14].id | https://openalex.org/keywords/receptor |
| keywords[14].score | 0.1096111536026001 |
| keywords[14].display_name | Receptor |
| language | en |
| locations[0].id | doi:10.3390/molecules27165114 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S108911230 |
| locations[0].source.issn | 1420-3049, 1433-1373 |
| locations[0].source.type | journal |
| locations[0].source.is_oa | True |
| locations[0].source.issn_l | 1420-3049 |
| locations[0].source.is_core | True |
| locations[0].source.is_in_doaj | True |
| locations[0].source.display_name | Molecules |
| locations[0].source.host_organization | https://openalex.org/P4310310987 |
| locations[0].source.host_organization_name | Multidisciplinary Digital Publishing Institute |
| locations[0].source.host_organization_lineage | https://openalex.org/P4310310987 |
| locations[0].source.host_organization_lineage_names | Multidisciplinary Digital Publishing Institute |
| locations[0].license | cc-by |
| locations[0].pdf_url | https://www.mdpi.com/1420-3049/27/16/5114/pdf?version=1660214839 |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | Molecules |
| locations[0].landing_page_url | https://doi.org/10.3390/molecules27165114 |
| locations[1].id | pmid:36014351 |
| locations[1].is_oa | False |
| locations[1].source.id | https://openalex.org/S4306525036 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | PubMed |
| locations[1].source.host_organization | https://openalex.org/I1299303238 |
| locations[1].source.host_organization_name | National Institutes of Health |
| locations[1].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[1].license | |
| locations[1].pdf_url | |
| locations[1].version | publishedVersion |
| locations[1].raw_type | |
| locations[1].license_id | |
| locations[1].is_accepted | True |
| locations[1].is_published | True |
| locations[1].raw_source_name | Molecules (Basel, Switzerland) |
| locations[1].landing_page_url | https://pubmed.ncbi.nlm.nih.gov/36014351 |
| locations[2].id | pmh:oai:circle.library.ubc.ca:2429/85156 |
| locations[2].is_oa | True |
| locations[2].source.id | https://openalex.org/S4377196297 |
| locations[2].source.issn | |
| locations[2].source.type | repository |
| locations[2].source.is_oa | False |
| locations[2].source.issn_l | |
| locations[2].source.is_core | False |
| locations[2].source.is_in_doaj | False |
| locations[2].source.display_name | cIRcle (University of British Columbia) |
| locations[2].source.host_organization | https://openalex.org/I141945490 |
| locations[2].source.host_organization_name | University of British Columbia |
| locations[2].source.host_organization_lineage | https://openalex.org/I141945490 |
| locations[2].license | cc-by |
| locations[2].pdf_url | |
| locations[2].version | submittedVersion |
| locations[2].raw_type | Text |
| locations[2].license_id | https://openalex.org/licenses/cc-by |
| locations[2].is_accepted | False |
| locations[2].is_published | False |
| locations[2].raw_source_name | |
| locations[2].landing_page_url | http://hdl.handle.net/2429/85156 |
| locations[3].id | pmh:oai:doaj.org/article:f60c71814bfc4a21be54e803f827c010 |
| locations[3].is_oa | True |
| locations[3].source.id | https://openalex.org/S4306401280 |
| locations[3].source.issn | |
| locations[3].source.type | repository |
| locations[3].source.is_oa | False |
| locations[3].source.issn_l | |
| locations[3].source.is_core | False |
| locations[3].source.is_in_doaj | False |
| locations[3].source.display_name | DOAJ (DOAJ: Directory of Open Access Journals) |
| locations[3].source.host_organization | |
| locations[3].source.host_organization_name | |
| locations[3].license | cc-by-sa |
| locations[3].pdf_url | |
| locations[3].version | submittedVersion |
| locations[3].raw_type | article |
| locations[3].license_id | https://openalex.org/licenses/cc-by-sa |
| locations[3].is_accepted | False |
| locations[3].is_published | False |
| locations[3].raw_source_name | Molecules, Vol 27, Iss 16, p 5114 (2022) |
| locations[3].landing_page_url | https://doaj.org/article/f60c71814bfc4a21be54e803f827c010 |
| locations[4].id | pmh:oai:mdpi.com:/1420-3049/27/16/5114/ |
| locations[4].is_oa | True |
| locations[4].source.id | https://openalex.org/S4306400947 |
| locations[4].source.issn | |
| locations[4].source.type | repository |
| locations[4].source.is_oa | True |
| locations[4].source.issn_l | |
| locations[4].source.is_core | False |
| locations[4].source.is_in_doaj | False |
| locations[4].source.display_name | MDPI (MDPI AG) |
| locations[4].source.host_organization | https://openalex.org/I4210097602 |
| locations[4].source.host_organization_name | Multidisciplinary Digital Publishing Institute (Switzerland) |
| locations[4].source.host_organization_lineage | https://openalex.org/I4210097602 |
| locations[4].license | cc-by |
| locations[4].pdf_url | |
| locations[4].version | submittedVersion |
| locations[4].raw_type | Text |
| locations[4].license_id | https://openalex.org/licenses/cc-by |
| locations[4].is_accepted | False |
| locations[4].is_published | False |
| locations[4].raw_source_name | Molecules; Volume 27; Issue 16; Pages: 5114 |
| locations[4].landing_page_url | https://dx.doi.org/10.3390/molecules27165114 |
| locations[5].id | pmh:oai:pubmedcentral.nih.gov:9416537 |
| locations[5].is_oa | True |
| locations[5].source.id | https://openalex.org/S2764455111 |
| locations[5].source.issn | |
| locations[5].source.type | repository |
| locations[5].source.is_oa | False |
| locations[5].source.issn_l | |
| locations[5].source.is_core | False |
| locations[5].source.is_in_doaj | False |
| locations[5].source.display_name | PubMed Central |
| locations[5].source.host_organization | https://openalex.org/I1299303238 |
| locations[5].source.host_organization_name | National Institutes of Health |
| locations[5].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[5].license | other-oa |
| locations[5].pdf_url | |
| locations[5].version | submittedVersion |
| locations[5].raw_type | Text |
| locations[5].license_id | https://openalex.org/licenses/other-oa |
| locations[5].is_accepted | False |
| locations[5].is_published | False |
| locations[5].raw_source_name | Molecules |
| locations[5].landing_page_url | https://www.ncbi.nlm.nih.gov/pmc/articles/9416537 |
| indexed_in | crossref, doaj, pubmed |
| authorships[0].author.id | https://openalex.org/A5014143640 |
| authorships[0].author.orcid | https://orcid.org/0000-0002-2562-7155 |
| authorships[0].author.display_name | Mohit Pandey |
| authorships[0].countries | CA |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I141945490 |
| authorships[0].affiliations[0].raw_affiliation_string | Vancouver Prostate Centre, Department of Urologic Sciences, University of British Columbia, Vancouver, BC V6T 1Z2, Canada |
| authorships[0].institutions[0].id | https://openalex.org/I141945490 |
| authorships[0].institutions[0].ror | https://ror.org/03rmrcq20 |
| authorships[0].institutions[0].type | education |
| authorships[0].institutions[0].lineage | https://openalex.org/I141945490, https://openalex.org/I4210128534, https://openalex.org/I4210135497, https://openalex.org/I4387154919 |
| authorships[0].institutions[0].country_code | CA |
| authorships[0].institutions[0].display_name | University of British Columbia |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Mohit Pandey |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Vancouver Prostate Centre, Department of Urologic Sciences, University of British Columbia, Vancouver, BC V6T 1Z2, Canada |
| authorships[1].author.id | https://openalex.org/A5005139941 |
| authorships[1].author.orcid | https://orcid.org/0000-0002-1707-9304 |
| authorships[1].author.display_name | Mariia Radaeva |
| authorships[1].countries | CA |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I141945490 |
| authorships[1].affiliations[0].raw_affiliation_string | Vancouver Prostate Centre, Department of Urologic Sciences, University of British Columbia, Vancouver, BC V6T 1Z2, Canada |
| authorships[1].institutions[0].id | https://openalex.org/I141945490 |
| authorships[1].institutions[0].ror | https://ror.org/03rmrcq20 |
| authorships[1].institutions[0].type | education |
| authorships[1].institutions[0].lineage | https://openalex.org/I141945490, https://openalex.org/I4210128534, https://openalex.org/I4210135497, https://openalex.org/I4387154919 |
| authorships[1].institutions[0].country_code | CA |
| authorships[1].institutions[0].display_name | University of British Columbia |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Mariia Radaeva |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | Vancouver Prostate Centre, Department of Urologic Sciences, University of British Columbia, Vancouver, BC V6T 1Z2, Canada |
| authorships[2].author.id | https://openalex.org/A5061108056 |
| authorships[2].author.orcid | https://orcid.org/0000-0003-2456-4022 |
| authorships[2].author.display_name | Hazem Mslati |
| authorships[2].countries | CA |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I141945490 |
| authorships[2].affiliations[0].raw_affiliation_string | Vancouver Prostate Centre, Department of Urologic Sciences, University of British Columbia, Vancouver, BC V6T 1Z2, Canada |
| authorships[2].institutions[0].id | https://openalex.org/I141945490 |
| authorships[2].institutions[0].ror | https://ror.org/03rmrcq20 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I141945490, https://openalex.org/I4210128534, https://openalex.org/I4210135497, https://openalex.org/I4387154919 |
| authorships[2].institutions[0].country_code | CA |
| authorships[2].institutions[0].display_name | University of British Columbia |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Hazem Mslati |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | Vancouver Prostate Centre, Department of Urologic Sciences, University of British Columbia, Vancouver, BC V6T 1Z2, Canada |
| authorships[3].author.id | https://openalex.org/A5063044696 |
| authorships[3].author.orcid | https://orcid.org/0000-0003-1903-6276 |
| authorships[3].author.display_name | Olivia Garland |
| authorships[3].countries | CA |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I141945490 |
| authorships[3].affiliations[0].raw_affiliation_string | Vancouver Prostate Centre, Department of Urologic Sciences, University of British Columbia, Vancouver, BC V6T 1Z2, Canada |
| authorships[3].institutions[0].id | https://openalex.org/I141945490 |
| authorships[3].institutions[0].ror | https://ror.org/03rmrcq20 |
| authorships[3].institutions[0].type | education |
| authorships[3].institutions[0].lineage | https://openalex.org/I141945490, https://openalex.org/I4210128534, https://openalex.org/I4210135497, https://openalex.org/I4387154919 |
| authorships[3].institutions[0].country_code | CA |
| authorships[3].institutions[0].display_name | University of British Columbia |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Olivia Garland |
| authorships[3].is_corresponding | False |
| authorships[3].raw_affiliation_strings | Vancouver Prostate Centre, Department of Urologic Sciences, University of British Columbia, Vancouver, BC V6T 1Z2, Canada |
| authorships[4].author.id | https://openalex.org/A5029378374 |
| authorships[4].author.orcid | https://orcid.org/0000-0003-2273-733X |
| authorships[4].author.display_name | Michael Fernández |
| authorships[4].countries | CA |
| authorships[4].affiliations[0].institution_ids | https://openalex.org/I141945490 |
| authorships[4].affiliations[0].raw_affiliation_string | Vancouver Prostate Centre, Department of Urologic Sciences, University of British Columbia, Vancouver, BC V6T 1Z2, Canada |
| authorships[4].institutions[0].id | https://openalex.org/I141945490 |
| authorships[4].institutions[0].ror | https://ror.org/03rmrcq20 |
| authorships[4].institutions[0].type | education |
| authorships[4].institutions[0].lineage | https://openalex.org/I141945490, https://openalex.org/I4210128534, https://openalex.org/I4210135497, https://openalex.org/I4387154919 |
| authorships[4].institutions[0].country_code | CA |
| authorships[4].institutions[0].display_name | University of British Columbia |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Michael Fernandez |
| authorships[4].is_corresponding | False |
| authorships[4].raw_affiliation_strings | Vancouver Prostate Centre, Department of Urologic Sciences, University of British Columbia, Vancouver, BC V6T 1Z2, Canada |
| authorships[5].author.id | https://openalex.org/A5018267399 |
| authorships[5].author.orcid | https://orcid.org/0000-0001-7732-2815 |
| authorships[5].author.display_name | Martin Ester |
| authorships[5].countries | CA |
| authorships[5].affiliations[0].institution_ids | https://openalex.org/I18014758 |
| authorships[5].affiliations[0].raw_affiliation_string | School of Computing Science, Simon Fraser University, Burnaby, BC V5A 1S6, Canada |
| authorships[5].institutions[0].id | https://openalex.org/I18014758 |
| authorships[5].institutions[0].ror | https://ror.org/0213rcc28 |
| authorships[5].institutions[0].type | education |
| authorships[5].institutions[0].lineage | https://openalex.org/I18014758 |
| authorships[5].institutions[0].country_code | CA |
| authorships[5].institutions[0].display_name | Simon Fraser University |
| authorships[5].author_position | middle |
| authorships[5].raw_author_name | Martin Ester |
| authorships[5].is_corresponding | False |
| authorships[5].raw_affiliation_strings | School of Computing Science, Simon Fraser University, Burnaby, BC V5A 1S6, Canada |
| authorships[6].author.id | https://openalex.org/A5070580886 |
| authorships[6].author.orcid | https://orcid.org/0000-0002-1599-1439 |
| authorships[6].author.display_name | Artem Cherkasov |
| authorships[6].countries | CA |
| authorships[6].affiliations[0].institution_ids | https://openalex.org/I141945490 |
| authorships[6].affiliations[0].raw_affiliation_string | Vancouver Prostate Centre, Department of Urologic Sciences, University of British Columbia, Vancouver, BC V6T 1Z2, Canada |
| authorships[6].institutions[0].id | https://openalex.org/I141945490 |
| authorships[6].institutions[0].ror | https://ror.org/03rmrcq20 |
| authorships[6].institutions[0].type | education |
| authorships[6].institutions[0].lineage | https://openalex.org/I141945490, https://openalex.org/I4210128534, https://openalex.org/I4210135497, https://openalex.org/I4387154919 |
| authorships[6].institutions[0].country_code | CA |
| authorships[6].institutions[0].display_name | University of British Columbia |
| authorships[6].author_position | last |
| authorships[6].raw_author_name | Artem Cherkasov |
| authorships[6].is_corresponding | True |
| authorships[6].raw_affiliation_strings | Vancouver Prostate Centre, Department of Urologic Sciences, University of British Columbia, Vancouver, BC V6T 1Z2, Canada |
| has_content.pdf | True |
| has_content.grobid_xml | True |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://www.mdpi.com/1420-3049/27/16/5114/pdf?version=1660214839 |
| open_access.oa_status | gold |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Ligand Binding Prediction Using Protein Structure Graphs and Residual Graph Attention Networks |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-09T23:09:16.995542 |
| primary_topic.id | https://openalex.org/T10211 |
| primary_topic.field.id | https://openalex.org/fields/17 |
| primary_topic.field.display_name | Computer Science |
| primary_topic.score | 0.9998999834060669 |
| primary_topic.domain.id | https://openalex.org/domains/3 |
| primary_topic.domain.display_name | Physical Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1703 |
| primary_topic.subfield.display_name | Computational Theory and Mathematics |
| primary_topic.display_name | Computational Drug Discovery Methods |
| related_works | https://openalex.org/W1485106526, https://openalex.org/W2011432577, https://openalex.org/W3008230527, https://openalex.org/W3131522312, https://openalex.org/W2597496502, https://openalex.org/W2172993862, https://openalex.org/W4388563702, https://openalex.org/W1969950105, https://openalex.org/W1971119335, https://openalex.org/W3033139795 |
| cited_by_count | 19 |
| counts_by_year[0].year | 2025 |
| counts_by_year[0].cited_by_count | 4 |
| counts_by_year[1].year | 2024 |
| counts_by_year[1].cited_by_count | 10 |
| counts_by_year[2].year | 2023 |
| counts_by_year[2].cited_by_count | 4 |
| counts_by_year[3].year | 2022 |
| counts_by_year[3].cited_by_count | 1 |
| locations_count | 6 |
| best_oa_location.id | doi:10.3390/molecules27165114 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S108911230 |
| best_oa_location.source.issn | 1420-3049, 1433-1373 |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | True |
| best_oa_location.source.issn_l | 1420-3049 |
| best_oa_location.source.is_core | True |
| best_oa_location.source.is_in_doaj | True |
| best_oa_location.source.display_name | Molecules |
| best_oa_location.source.host_organization | https://openalex.org/P4310310987 |
| best_oa_location.source.host_organization_name | Multidisciplinary Digital Publishing Institute |
| best_oa_location.source.host_organization_lineage | https://openalex.org/P4310310987 |
| best_oa_location.source.host_organization_lineage_names | Multidisciplinary Digital Publishing Institute |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | https://www.mdpi.com/1420-3049/27/16/5114/pdf?version=1660214839 |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | journal-article |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | Molecules |
| best_oa_location.landing_page_url | https://doi.org/10.3390/molecules27165114 |
| primary_location.id | doi:10.3390/molecules27165114 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S108911230 |
| primary_location.source.issn | 1420-3049, 1433-1373 |
| primary_location.source.type | journal |
| primary_location.source.is_oa | True |
| primary_location.source.issn_l | 1420-3049 |
| primary_location.source.is_core | True |
| primary_location.source.is_in_doaj | True |
| primary_location.source.display_name | Molecules |
| primary_location.source.host_organization | https://openalex.org/P4310310987 |
| primary_location.source.host_organization_name | Multidisciplinary Digital Publishing Institute |
| primary_location.source.host_organization_lineage | https://openalex.org/P4310310987 |
| primary_location.source.host_organization_lineage_names | Multidisciplinary Digital Publishing Institute |
| primary_location.license | cc-by |
| primary_location.pdf_url | https://www.mdpi.com/1420-3049/27/16/5114/pdf?version=1660214839 |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | Molecules |
| primary_location.landing_page_url | https://doi.org/10.3390/molecules27165114 |
| publication_date | 2022-08-11 |
| publication_year | 2022 |
| referenced_works | https://openalex.org/W3004974539, https://openalex.org/W2166550586, https://openalex.org/W3189831819, https://openalex.org/W2175130792, https://openalex.org/W3187882175, https://openalex.org/W3023126697, https://openalex.org/W3027975282, https://openalex.org/W3201539868, https://openalex.org/W2608559058, https://openalex.org/W3094640617, https://openalex.org/W3135935512, https://openalex.org/W4291288469, https://openalex.org/W3201055938, https://openalex.org/W2129434099, https://openalex.org/W3012107310, https://openalex.org/W3088265803, https://openalex.org/W2592742128, https://openalex.org/W3033099853, https://openalex.org/W2785947426, https://openalex.org/W2899788782, https://openalex.org/W2033757486, https://openalex.org/W29860408, https://openalex.org/W2048816080, https://openalex.org/W2051160906, https://openalex.org/W2114162221, https://openalex.org/W3177828909, https://openalex.org/W2784213390, https://openalex.org/W2781821160, https://openalex.org/W2969996838, https://openalex.org/W3175786839, https://openalex.org/W3140330522, https://openalex.org/W3032123378, https://openalex.org/W2035585923, https://openalex.org/W2068495101, https://openalex.org/W2076779488, https://openalex.org/W2086286404, https://openalex.org/W2106612397, https://openalex.org/W2204695023, https://openalex.org/W1993046136, https://openalex.org/W3133246485, https://openalex.org/W2605952223, https://openalex.org/W2166744107, https://openalex.org/W1984379900, https://openalex.org/W2791796577, https://openalex.org/W3156288747, https://openalex.org/W3043507132, https://openalex.org/W2290847742, https://openalex.org/W3146944767, https://openalex.org/W2955251621, https://openalex.org/W2256119113, https://openalex.org/W3000369508, https://openalex.org/W2809216727, https://openalex.org/W3109916301, https://openalex.org/W3167575787, https://openalex.org/W3109493217, https://openalex.org/W3214234430, https://openalex.org/W3035943921, https://openalex.org/W2343614482, https://openalex.org/W6659061787, https://openalex.org/W3094771832, https://openalex.org/W3100157108, https://openalex.org/W2035175173, https://openalex.org/W2161572568 |
| referenced_works_count | 63 |
| abstract_inverted_index.a | 6, 163 |
| abstract_inverted_index.2D | 98 |
| abstract_inverted_index.3D | 72, 90 |
| abstract_inverted_index.As | 30 |
| abstract_inverted_index.an | 159 |
| abstract_inverted_index.as | 13, 146 |
| abstract_inverted_index.in | 24, 27 |
| abstract_inverted_index.is | 5 |
| abstract_inverted_index.it | 14, 37 |
| abstract_inverted_index.of | 2, 9, 23, 33, 43, 61, 65, 70, 79, 93, 101, 138, 143, 170 |
| abstract_inverted_index.on | 129, 162 |
| abstract_inverted_index.or | 67 |
| abstract_inverted_index.to | 16, 58, 109 |
| abstract_inverted_index.we | 75, 153 |
| abstract_inverted_index.The | 103, 122, 135 |
| abstract_inverted_index.and | 20, 26 |
| abstract_inverted_index.are | 115 |
| abstract_inverted_index.for | 40, 118, 149 |
| abstract_inverted_index.our | 155 |
| abstract_inverted_index.the | 31, 41, 59, 77, 80, 94, 126 |
| abstract_inverted_index.use | 60 |
| abstract_inverted_index.(DL) | 46 |
| abstract_inverted_index.also | 105 |
| abstract_inverted_index.data | 35 |
| abstract_inverted_index.deep | 44 |
| abstract_inverted_index.drug | 11 |
| abstract_inverted_index.high | 18 |
| abstract_inverted_index.main | 165 |
| abstract_inverted_index.most | 116 |
| abstract_inverted_index.part | 8 |
| abstract_inverted_index.such | 53 |
| abstract_inverted_index.that | 88, 114 |
| abstract_inverted_index.very | 62 |
| abstract_inverted_index.vivo | 28 |
| abstract_inverted_index.were | 55 |
| abstract_inverted_index.with | 48, 97 |
| abstract_inverted_index.along | 96 |
| abstract_inverted_index.assay | 161 |
| abstract_inverted_index.costs | 19 |
| abstract_inverted_index.graph | 99 |
| abstract_inverted_index.helps | 15 |
| abstract_inverted_index.labor | 21 |
| abstract_inverted_index.model | 156 |
| abstract_inverted_index.their | 71 |
| abstract_inverted_index.viral | 164 |
| abstract_inverted_index.vitro | 25 |
| abstract_inverted_index.bypass | 17 |
| abstract_inverted_index.either | 56 |
| abstract_inverted_index.graphs | 140 |
| abstract_inverted_index.ligand | 119 |
| abstract_inverted_index.method | 104 |
| abstract_inverted_index.models | 47, 54 |
| abstract_inverted_index.modern | 10 |
| abstract_inverted_index.scores | 108 |
| abstract_inverted_index.target | 169 |
| abstract_inverted_index.wealth | 32 |
| abstract_inverted_index.weight | 111 |
| abstract_inverted_index.Herein, | 74 |
| abstract_inverted_index.PSG-BAR | 81 |
| abstract_inverted_index.against | 158 |
| abstract_inverted_index.binding | 131 |
| abstract_inverted_index.crucial | 7 |
| abstract_inverted_index.demands | 22 |
| abstract_inverted_index.enables | 141 |
| abstract_inverted_index.limited | 57 |
| abstract_inverted_index.pooling | 137 |
| abstract_inverted_index.powers. | 51 |
| abstract_inverted_index.present | 76 |
| abstract_inverted_index.protein | 112, 139 |
| abstract_inverted_index.regions | 113 |
| abstract_inverted_index.several | 130 |
| abstract_inverted_index.surface | 144 |
| abstract_inverted_index.(Protein | 82 |
| abstract_inverted_index.Affinity | 85 |
| abstract_inverted_index.Finally, | 152 |
| abstract_inverted_index.Results: | 121 |
| abstract_inverted_index.affinity | 132 |
| abstract_inverted_index.approach | 87, 124 |
| abstract_inverted_index.binding. | 120, 151 |
| abstract_inverted_index.critical | 147 |
| abstract_inverted_index.hallmark | 168 |
| abstract_inverted_index.learning | 45 |
| abstract_inverted_index.ligands. | 102 |
| abstract_inverted_index.protease | 166 |
| abstract_inverted_index.proteins | 66, 95 |
| abstract_inverted_index.provides | 38 |
| abstract_inverted_index.residues | 145, 148 |
| abstract_inverted_index.utilizes | 89 |
| abstract_inverted_index.validate | 154 |
| abstract_inverted_index.Structure | 83 |
| abstract_inverted_index.attention | 107 |
| abstract_inverted_index.datasets. | 134 |
| abstract_inverted_index.developed | 123 |
| abstract_inverted_index.discovery | 12 |
| abstract_inverted_index.important | 117 |
| abstract_inverted_index.SARS-CoV-2 | 171 |
| abstract_inverted_index.increasing | 49 |
| abstract_inverted_index.introduces | 106 |
| abstract_inverted_index.prediction | 1 |
| abstract_inverted_index.predictive | 50 |
| abstract_inverted_index.screening. | 29 |
| abstract_inverted_index.simplified | 63 |
| abstract_inverted_index.structural | 91 |
| abstract_inverted_index.Regression) | 86 |
| abstract_inverted_index.bioactivity | 34 |
| abstract_inverted_index.development | 42, 78 |
| abstract_inverted_index.ineffective | 68 |
| abstract_inverted_index.information | 92 |
| abstract_inverted_index.performance | 128 |
| abstract_inverted_index.predictions | 157 |
| abstract_inverted_index.selectively | 110 |
| abstract_inverted_index.structures. | 73 |
| abstract_inverted_index.(Mpro)—the | 167 |
| abstract_inverted_index.accumulates, | 36 |
| abstract_inverted_index.benchmarking | 133 |
| abstract_inverted_index.coronavirus. | 172 |
| abstract_inverted_index.demonstrates | 125 |
| abstract_inverted_index.experimental | 160 |
| abstract_inverted_index.interactions | 4 |
| abstract_inverted_index.voxelization | 69 |
| abstract_inverted_index.Computational | 0 |
| abstract_inverted_index.Graph-Binding | 84 |
| abstract_inverted_index.opportunities | 39 |
| abstract_inverted_index.identification | 142 |
| abstract_inverted_index.Conventionally, | 52 |
| abstract_inverted_index.attention-based | 136 |
| abstract_inverted_index.ligand–target | 3 |
| abstract_inverted_index.representations | 64, 100 |
| abstract_inverted_index.protein–ligand | 150 |
| abstract_inverted_index.state-of-the-art | 127 |
| cited_by_percentile_year.max | 99 |
| cited_by_percentile_year.min | 89 |
| corresponding_author_ids | https://openalex.org/A5070580886 |
| countries_distinct_count | 1 |
| institutions_distinct_count | 7 |
| corresponding_institution_ids | https://openalex.org/I141945490 |
| sustainable_development_goals[0].id | https://metadata.un.org/sdg/8 |
| sustainable_development_goals[0].score | 0.5899999737739563 |
| sustainable_development_goals[0].display_name | Decent work and economic growth |
| citation_normalized_percentile.value | 0.92464194 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | True |