Loss of charge at solvent exposed Lys residues does not induce the aggregation of superoxide dismutase 1 Article Swipe
 YOU?
·
· 2018
· Open Access
·
· DOI: https://doi.org/10.1101/301309
YOU?
·
· 2018
· Open Access
·
· DOI: https://doi.org/10.1101/301309
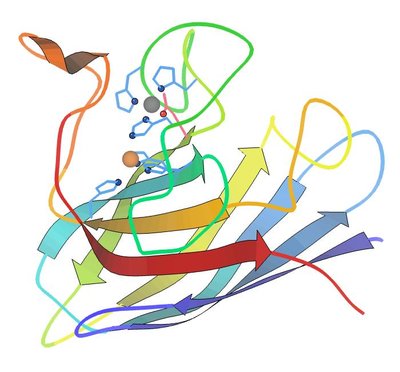
Mutations in superoxide dismutase 1 (SOD1) associated with familial amyotrophic lateral sclerosis (fALS) induce the protein to misfold and aggregate. To date, missense mutations at more than 80 different amino acid positions have been associated with disease. How these mutations perturb native structure to heighten the propensity to misfold and aggregate is unclear. One potential mechanism that has been suggested is that when mutations occur at positions occupied by charged amino acids, then repulsive forces that would inhibit aberrant protein:protein interactions would be reduced. Mutations at twenty-one charged residues in SOD1 have been associated with fALS. Here, we examined whether loss of positively charged surface Lys residues would induce the misfolding and aggregation of SOD1. We randomly mutated four different Lys residues (K30, K36, K75, K91) in SOD1 and expressed these variants as fusion proteins with yellow fluorescent protein (YFP). We also assessed whether these mutations induced binding to a conformation-restricted SOD1 antibody, designated C4F6, which recognizes non-natively folded protein. Our findings indicate that SOD1 generally tolerates mutations at surface exposed lysine residues, and that loss of positive charge is insufficient to induce aggregation. Our findings may explain why mutations at these Lys residues have not been identified in ALS patients.
Related Topics
- Type
- preprint
- Language
- en
- Landing Page
- https://doi.org/10.1101/301309
- https://www.biorxiv.org/content/biorxiv/early/2018/04/14/301309.full.pdf
- OA Status
- green
- References
- 70
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W2797955520
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W2797955520Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1101/301309Digital Object Identifier
- Title
-
Loss of charge at solvent exposed Lys residues does not induce the aggregation of superoxide dismutase 1Work title
- Type
-
preprintOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2018Year of publication
- Publication date
-
2018-04-14Full publication date if available
- Authors
-
Keith Crosby, Anthony M. Crown, Brittany L. Roberts, Hilda Brown, Jacob I. Ayers, David BorcheltList of authors in order
- Landing page
-
https://doi.org/10.1101/301309Publisher landing page
- PDF URL
-
https://www.biorxiv.org/content/biorxiv/early/2018/04/14/301309.full.pdfDirect link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
greenOpen access status per OpenAlex
- OA URL
-
https://www.biorxiv.org/content/biorxiv/early/2018/04/14/301309.full.pdfDirect OA link when available
- Concepts
-
SOD1, Missense mutation, Superoxide dismutase, Protein aggregation, Mutation, Amino acid, Chemistry, Biochemistry, Molecular biology, Genetics, Biology, Gene, Oxidative stressTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
0Total citation count in OpenAlex
- References (count)
-
70Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W2797955520 |
|---|---|
| doi | https://doi.org/10.1101/301309 |
| ids.doi | https://doi.org/10.1101/301309 |
| ids.mag | 2797955520 |
| ids.openalex | https://openalex.org/W2797955520 |
| fwci | |
| type | preprint |
| title | Loss of charge at solvent exposed Lys residues does not induce the aggregation of superoxide dismutase 1 |
| biblio.issue | |
| biblio.volume | |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T10855 |
| topics[0].field.id | https://openalex.org/fields/27 |
| topics[0].field.display_name | Medicine |
| topics[0].score | 1.0 |
| topics[0].domain.id | https://openalex.org/domains/4 |
| topics[0].domain.display_name | Health Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/2728 |
| topics[0].subfield.display_name | Neurology |
| topics[0].display_name | Amyotrophic Lateral Sclerosis Research |
| topics[1].id | https://openalex.org/T12400 |
| topics[1].field.id | https://openalex.org/fields/27 |
| topics[1].field.display_name | Medicine |
| topics[1].score | 0.972000002861023 |
| topics[1].domain.id | https://openalex.org/domains/4 |
| topics[1].domain.display_name | Health Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/2716 |
| topics[1].subfield.display_name | Genetics |
| topics[1].display_name | Neurogenetic and Muscular Disorders Research |
| topics[2].id | https://openalex.org/T10085 |
| topics[2].field.id | https://openalex.org/fields/27 |
| topics[2].field.display_name | Medicine |
| topics[2].score | 0.9657999873161316 |
| topics[2].domain.id | https://openalex.org/domains/4 |
| topics[2].domain.display_name | Health Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/2728 |
| topics[2].subfield.display_name | Neurology |
| topics[2].display_name | Parkinson's Disease Mechanisms and Treatments |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C2777615887 |
| concepts[0].level | 4 |
| concepts[0].score | 0.8688570261001587 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q15328137 |
| concepts[0].display_name | SOD1 |
| concepts[1].id | https://openalex.org/C75563809 |
| concepts[1].level | 4 |
| concepts[1].score | 0.657257080078125 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q2656896 |
| concepts[1].display_name | Missense mutation |
| concepts[2].id | https://openalex.org/C2775838275 |
| concepts[2].level | 3 |
| concepts[2].score | 0.650001585483551 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q410776 |
| concepts[2].display_name | Superoxide dismutase |
| concepts[3].id | https://openalex.org/C136238340 |
| concepts[3].level | 2 |
| concepts[3].score | 0.577212393283844 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q7251455 |
| concepts[3].display_name | Protein aggregation |
| concepts[4].id | https://openalex.org/C501734568 |
| concepts[4].level | 3 |
| concepts[4].score | 0.5747652053833008 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q42918 |
| concepts[4].display_name | Mutation |
| concepts[5].id | https://openalex.org/C515207424 |
| concepts[5].level | 2 |
| concepts[5].score | 0.5699641704559326 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q8066 |
| concepts[5].display_name | Amino acid |
| concepts[6].id | https://openalex.org/C185592680 |
| concepts[6].level | 0 |
| concepts[6].score | 0.524770200252533 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q2329 |
| concepts[6].display_name | Chemistry |
| concepts[7].id | https://openalex.org/C55493867 |
| concepts[7].level | 1 |
| concepts[7].score | 0.4593120217323303 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q7094 |
| concepts[7].display_name | Biochemistry |
| concepts[8].id | https://openalex.org/C153911025 |
| concepts[8].level | 1 |
| concepts[8].score | 0.36535030603408813 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q7202 |
| concepts[8].display_name | Molecular biology |
| concepts[9].id | https://openalex.org/C54355233 |
| concepts[9].level | 1 |
| concepts[9].score | 0.35546523332595825 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[9].display_name | Genetics |
| concepts[10].id | https://openalex.org/C86803240 |
| concepts[10].level | 0 |
| concepts[10].score | 0.352750301361084 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[10].display_name | Biology |
| concepts[11].id | https://openalex.org/C104317684 |
| concepts[11].level | 2 |
| concepts[11].score | 0.27062100172042847 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[11].display_name | Gene |
| concepts[12].id | https://openalex.org/C2776151105 |
| concepts[12].level | 2 |
| concepts[12].score | 0.16884064674377441 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q898814 |
| concepts[12].display_name | Oxidative stress |
| keywords[0].id | https://openalex.org/keywords/sod1 |
| keywords[0].score | 0.8688570261001587 |
| keywords[0].display_name | SOD1 |
| keywords[1].id | https://openalex.org/keywords/missense-mutation |
| keywords[1].score | 0.657257080078125 |
| keywords[1].display_name | Missense mutation |
| keywords[2].id | https://openalex.org/keywords/superoxide-dismutase |
| keywords[2].score | 0.650001585483551 |
| keywords[2].display_name | Superoxide dismutase |
| keywords[3].id | https://openalex.org/keywords/protein-aggregation |
| keywords[3].score | 0.577212393283844 |
| keywords[3].display_name | Protein aggregation |
| keywords[4].id | https://openalex.org/keywords/mutation |
| keywords[4].score | 0.5747652053833008 |
| keywords[4].display_name | Mutation |
| keywords[5].id | https://openalex.org/keywords/amino-acid |
| keywords[5].score | 0.5699641704559326 |
| keywords[5].display_name | Amino acid |
| keywords[6].id | https://openalex.org/keywords/chemistry |
| keywords[6].score | 0.524770200252533 |
| keywords[6].display_name | Chemistry |
| keywords[7].id | https://openalex.org/keywords/biochemistry |
| keywords[7].score | 0.4593120217323303 |
| keywords[7].display_name | Biochemistry |
| keywords[8].id | https://openalex.org/keywords/molecular-biology |
| keywords[8].score | 0.36535030603408813 |
| keywords[8].display_name | Molecular biology |
| keywords[9].id | https://openalex.org/keywords/genetics |
| keywords[9].score | 0.35546523332595825 |
| keywords[9].display_name | Genetics |
| keywords[10].id | https://openalex.org/keywords/biology |
| keywords[10].score | 0.352750301361084 |
| keywords[10].display_name | Biology |
| keywords[11].id | https://openalex.org/keywords/gene |
| keywords[11].score | 0.27062100172042847 |
| keywords[11].display_name | Gene |
| keywords[12].id | https://openalex.org/keywords/oxidative-stress |
| keywords[12].score | 0.16884064674377441 |
| keywords[12].display_name | Oxidative stress |
| language | en |
| locations[0].id | doi:10.1101/301309 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4306402567 |
| locations[0].source.issn | |
| locations[0].source.type | repository |
| locations[0].source.is_oa | False |
| locations[0].source.issn_l | |
| locations[0].source.is_core | False |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| locations[0].source.host_organization | https://openalex.org/I2750212522 |
| locations[0].source.host_organization_name | Cold Spring Harbor Laboratory |
| locations[0].source.host_organization_lineage | https://openalex.org/I2750212522 |
| locations[0].license | |
| locations[0].pdf_url | https://www.biorxiv.org/content/biorxiv/early/2018/04/14/301309.full.pdf |
| locations[0].version | acceptedVersion |
| locations[0].raw_type | posted-content |
| locations[0].license_id | |
| locations[0].is_accepted | True |
| locations[0].is_published | False |
| locations[0].raw_source_name | |
| locations[0].landing_page_url | https://doi.org/10.1101/301309 |
| indexed_in | crossref |
| authorships[0].author.id | https://openalex.org/A5047632041 |
| authorships[0].author.orcid | |
| authorships[0].author.display_name | Keith Crosby |
| authorships[0].countries | US |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I33213144 |
| authorships[0].affiliations[0].raw_affiliation_string | Department of Neuroscience, Center for Translational Research in Neurodegenerative Disease, University of Florida, Gainesville, FL 3260. |
| authorships[0].institutions[0].id | https://openalex.org/I33213144 |
| authorships[0].institutions[0].ror | https://ror.org/02y3ad647 |
| authorships[0].institutions[0].type | education |
| authorships[0].institutions[0].lineage | https://openalex.org/I33213144 |
| authorships[0].institutions[0].country_code | US |
| authorships[0].institutions[0].display_name | University of Florida |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Keith Crosby |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Department of Neuroscience, Center for Translational Research in Neurodegenerative Disease, University of Florida, Gainesville, FL 3260. |
| authorships[1].author.id | https://openalex.org/A5041560909 |
| authorships[1].author.orcid | https://orcid.org/0000-0003-3884-6223 |
| authorships[1].author.display_name | Anthony M. Crown |
| authorships[1].countries | US |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I103163165, https://openalex.org/I33213144 |
| authorships[1].affiliations[0].raw_affiliation_string | College of Arts and Sciences, University of Florida, Gainesville, FL 3610. |
| authorships[1].institutions[0].id | https://openalex.org/I103163165 |
| authorships[1].institutions[0].ror | https://ror.org/05g3dte14 |
| authorships[1].institutions[0].type | education |
| authorships[1].institutions[0].lineage | https://openalex.org/I103163165 |
| authorships[1].institutions[0].country_code | US |
| authorships[1].institutions[0].display_name | Florida State University |
| authorships[1].institutions[1].id | https://openalex.org/I33213144 |
| authorships[1].institutions[1].ror | https://ror.org/02y3ad647 |
| authorships[1].institutions[1].type | education |
| authorships[1].institutions[1].lineage | https://openalex.org/I33213144 |
| authorships[1].institutions[1].country_code | US |
| authorships[1].institutions[1].display_name | University of Florida |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Anthony M. Crown |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | College of Arts and Sciences, University of Florida, Gainesville, FL 3610. |
| authorships[2].author.id | https://openalex.org/A5008806082 |
| authorships[2].author.orcid | |
| authorships[2].author.display_name | Brittany L. Roberts |
| authorships[2].countries | US |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I103163165, https://openalex.org/I33213144 |
| authorships[2].affiliations[0].raw_affiliation_string | College of Arts and Sciences, University of Florida, Gainesville, FL 3610. |
| authorships[2].institutions[0].id | https://openalex.org/I103163165 |
| authorships[2].institutions[0].ror | https://ror.org/05g3dte14 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I103163165 |
| authorships[2].institutions[0].country_code | US |
| authorships[2].institutions[0].display_name | Florida State University |
| authorships[2].institutions[1].id | https://openalex.org/I33213144 |
| authorships[2].institutions[1].ror | https://ror.org/02y3ad647 |
| authorships[2].institutions[1].type | education |
| authorships[2].institutions[1].lineage | https://openalex.org/I33213144 |
| authorships[2].institutions[1].country_code | US |
| authorships[2].institutions[1].display_name | University of Florida |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Brittany L. Roberts |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | College of Arts and Sciences, University of Florida, Gainesville, FL 3610. |
| authorships[3].author.id | https://openalex.org/A5001791933 |
| authorships[3].author.orcid | |
| authorships[3].author.display_name | Hilda Brown |
| authorships[3].countries | US |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I2800717037 |
| authorships[3].affiliations[0].raw_affiliation_string | SantaFe HealthCare Alzheimer's Disease Research Center, McKnight Brain Institute, University of Florida, Gainesville, FL 2610. |
| authorships[3].affiliations[1].institution_ids | https://openalex.org/I33213144 |
| authorships[3].affiliations[1].raw_affiliation_string | Department of Neuroscience, Center for Translational Research in Neurodegenerative Disease, University of Florida, Gainesville, FL 3260. |
| authorships[3].institutions[0].id | https://openalex.org/I33213144 |
| authorships[3].institutions[0].ror | https://ror.org/02y3ad647 |
| authorships[3].institutions[0].type | education |
| authorships[3].institutions[0].lineage | https://openalex.org/I33213144 |
| authorships[3].institutions[0].country_code | US |
| authorships[3].institutions[0].display_name | University of Florida |
| authorships[3].institutions[1].id | https://openalex.org/I2800717037 |
| authorships[3].institutions[1].ror | https://ror.org/04tk2gy88 |
| authorships[3].institutions[1].type | education |
| authorships[3].institutions[1].lineage | https://openalex.org/I2800717037 |
| authorships[3].institutions[1].country_code | US |
| authorships[3].institutions[1].display_name | University of Florida Health |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Hilda Brown |
| authorships[3].is_corresponding | False |
| authorships[3].raw_affiliation_strings | Department of Neuroscience, Center for Translational Research in Neurodegenerative Disease, University of Florida, Gainesville, FL 3260., SantaFe HealthCare Alzheimer's Disease Research Center, McKnight Brain Institute, University of Florida, Gainesville, FL 2610. |
| authorships[4].author.id | https://openalex.org/A5034424187 |
| authorships[4].author.orcid | https://orcid.org/0000-0003-4544-2366 |
| authorships[4].author.display_name | Jacob I. Ayers |
| authorships[4].countries | US |
| authorships[4].affiliations[0].institution_ids | https://openalex.org/I33213144 |
| authorships[4].affiliations[0].raw_affiliation_string | Department of Neuroscience, Center for Translational Research in Neurodegenerative Disease, University of Florida, Gainesville, FL 3260. |
| authorships[4].institutions[0].id | https://openalex.org/I33213144 |
| authorships[4].institutions[0].ror | https://ror.org/02y3ad647 |
| authorships[4].institutions[0].type | education |
| authorships[4].institutions[0].lineage | https://openalex.org/I33213144 |
| authorships[4].institutions[0].country_code | US |
| authorships[4].institutions[0].display_name | University of Florida |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Jacob I. Ayers |
| authorships[4].is_corresponding | False |
| authorships[4].raw_affiliation_strings | Department of Neuroscience, Center for Translational Research in Neurodegenerative Disease, University of Florida, Gainesville, FL 3260. |
| authorships[5].author.id | https://openalex.org/A5038629622 |
| authorships[5].author.orcid | https://orcid.org/0000-0002-0813-6979 |
| authorships[5].author.display_name | David Borchelt |
| authorships[5].countries | US |
| authorships[5].affiliations[0].institution_ids | https://openalex.org/I2800717037 |
| authorships[5].affiliations[0].raw_affiliation_string | SantaFe HealthCare Alzheimer's Disease Research Center, McKnight Brain Institute, University of Florida, Gainesville, FL 2610. |
| authorships[5].affiliations[1].institution_ids | https://openalex.org/I33213144 |
| authorships[5].affiliations[1].raw_affiliation_string | Department of Neuroscience, Center for Translational Research in Neurodegenerative Disease, University of Florida, Gainesville, FL 3260. |
| authorships[5].affiliations[2].institution_ids | https://openalex.org/I103163165, https://openalex.org/I33213144 |
| authorships[5].affiliations[2].raw_affiliation_string | College of Arts and Sciences, University of Florida, Gainesville, FL 3610. |
| authorships[5].institutions[0].id | https://openalex.org/I103163165 |
| authorships[5].institutions[0].ror | https://ror.org/05g3dte14 |
| authorships[5].institutions[0].type | education |
| authorships[5].institutions[0].lineage | https://openalex.org/I103163165 |
| authorships[5].institutions[0].country_code | US |
| authorships[5].institutions[0].display_name | Florida State University |
| authorships[5].institutions[1].id | https://openalex.org/I33213144 |
| authorships[5].institutions[1].ror | https://ror.org/02y3ad647 |
| authorships[5].institutions[1].type | education |
| authorships[5].institutions[1].lineage | https://openalex.org/I33213144 |
| authorships[5].institutions[1].country_code | US |
| authorships[5].institutions[1].display_name | University of Florida |
| authorships[5].institutions[2].id | https://openalex.org/I2800717037 |
| authorships[5].institutions[2].ror | https://ror.org/04tk2gy88 |
| authorships[5].institutions[2].type | education |
| authorships[5].institutions[2].lineage | https://openalex.org/I2800717037 |
| authorships[5].institutions[2].country_code | US |
| authorships[5].institutions[2].display_name | University of Florida Health |
| authorships[5].author_position | last |
| authorships[5].raw_author_name | David R. Borchelt |
| authorships[5].is_corresponding | True |
| authorships[5].raw_affiliation_strings | College of Arts and Sciences, University of Florida, Gainesville, FL 3610., Department of Neuroscience, Center for Translational Research in Neurodegenerative Disease, University of Florida, Gainesville, FL 3260., SantaFe HealthCare Alzheimer's Disease Research Center, McKnight Brain Institute, University of Florida, Gainesville, FL 2610. |
| has_content.pdf | True |
| has_content.grobid_xml | True |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://www.biorxiv.org/content/biorxiv/early/2018/04/14/301309.full.pdf |
| open_access.oa_status | green |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Loss of charge at solvent exposed Lys residues does not induce the aggregation of superoxide dismutase 1 |
| has_fulltext | True |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T10855 |
| primary_topic.field.id | https://openalex.org/fields/27 |
| primary_topic.field.display_name | Medicine |
| primary_topic.score | 1.0 |
| primary_topic.domain.id | https://openalex.org/domains/4 |
| primary_topic.domain.display_name | Health Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/2728 |
| primary_topic.subfield.display_name | Neurology |
| primary_topic.display_name | Amyotrophic Lateral Sclerosis Research |
| related_works | https://openalex.org/W2073260254, https://openalex.org/W2160523206, https://openalex.org/W1978123724, https://openalex.org/W2070740797, https://openalex.org/W2091731575, https://openalex.org/W2779809725, https://openalex.org/W3042514551, https://openalex.org/W2079503671, https://openalex.org/W2048401333, https://openalex.org/W2167113152 |
| cited_by_count | 0 |
| locations_count | 1 |
| best_oa_location.id | doi:10.1101/301309 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4306402567 |
| best_oa_location.source.issn | |
| best_oa_location.source.type | repository |
| best_oa_location.source.is_oa | False |
| best_oa_location.source.issn_l | |
| best_oa_location.source.is_core | False |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| best_oa_location.source.host_organization | https://openalex.org/I2750212522 |
| best_oa_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| best_oa_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| best_oa_location.license | |
| best_oa_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2018/04/14/301309.full.pdf |
| best_oa_location.version | acceptedVersion |
| best_oa_location.raw_type | posted-content |
| best_oa_location.license_id | |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | False |
| best_oa_location.raw_source_name | |
| best_oa_location.landing_page_url | https://doi.org/10.1101/301309 |
| primary_location.id | doi:10.1101/301309 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4306402567 |
| primary_location.source.issn | |
| primary_location.source.type | repository |
| primary_location.source.is_oa | False |
| primary_location.source.issn_l | |
| primary_location.source.is_core | False |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| primary_location.source.host_organization | https://openalex.org/I2750212522 |
| primary_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| primary_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| primary_location.license | |
| primary_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2018/04/14/301309.full.pdf |
| primary_location.version | acceptedVersion |
| primary_location.raw_type | posted-content |
| primary_location.license_id | |
| primary_location.is_accepted | True |
| primary_location.is_published | False |
| primary_location.raw_source_name | |
| primary_location.landing_page_url | https://doi.org/10.1101/301309 |
| publication_date | 2018-04-14 |
| publication_year | 2018 |
| referenced_works | https://openalex.org/W2096386031, https://openalex.org/W4211129887, https://openalex.org/W1968735793, https://openalex.org/W2052274307, https://openalex.org/W2135275443, https://openalex.org/W2133714529, https://openalex.org/W2032107666, https://openalex.org/W2059014219, https://openalex.org/W2133408527, https://openalex.org/W2053712580, https://openalex.org/W2106392233, https://openalex.org/W2150430904, https://openalex.org/W2096335797, https://openalex.org/W2107768418, https://openalex.org/W2022367303, https://openalex.org/W2067847915, https://openalex.org/W2110580010, https://openalex.org/W2085880812, https://openalex.org/W2099040799, https://openalex.org/W2122495582, https://openalex.org/W2024059905, https://openalex.org/W1504486296, https://openalex.org/W2086985844, https://openalex.org/W1599352650, https://openalex.org/W2040228307, https://openalex.org/W2049897515, https://openalex.org/W2092082741, https://openalex.org/W1965265324, https://openalex.org/W1985396041, https://openalex.org/W2040621074, https://openalex.org/W2151969338, https://openalex.org/W2007813328, https://openalex.org/W1975906144, https://openalex.org/W2030793256, https://openalex.org/W2107034810, https://openalex.org/W1975578769, https://openalex.org/W2157076525, https://openalex.org/W1966202673, https://openalex.org/W2149428117, https://openalex.org/W2164056914, https://openalex.org/W2158728223, https://openalex.org/W2592243741, https://openalex.org/W2162589295, https://openalex.org/W2143366726, https://openalex.org/W2130435783, https://openalex.org/W2056447311, https://openalex.org/W2022933182, https://openalex.org/W1973324345, https://openalex.org/W1987050708, https://openalex.org/W2167113152, https://openalex.org/W1999913981, https://openalex.org/W2082970207, https://openalex.org/W1989227252, https://openalex.org/W1996162600, https://openalex.org/W2107672209, https://openalex.org/W2022721388, https://openalex.org/W1968280815, https://openalex.org/W2107904341, https://openalex.org/W2004175178, https://openalex.org/W2067288722, https://openalex.org/W2115981836, https://openalex.org/W1999638129, https://openalex.org/W2057838250, https://openalex.org/W1964377967, https://openalex.org/W2012851706, https://openalex.org/W1940436875, https://openalex.org/W2586253893, https://openalex.org/W2128740394, https://openalex.org/W2045479131, https://openalex.org/W2464376366 |
| referenced_works_count | 70 |
| abstract_inverted_index.1 | 5 |
| abstract_inverted_index.a | 150 |
| abstract_inverted_index.80 | 28 |
| abstract_inverted_index.To | 21 |
| abstract_inverted_index.We | 116, 141 |
| abstract_inverted_index.as | 133 |
| abstract_inverted_index.at | 25, 66, 86, 169, 191 |
| abstract_inverted_index.be | 83 |
| abstract_inverted_index.by | 69 |
| abstract_inverted_index.in | 2, 90, 127, 199 |
| abstract_inverted_index.is | 52, 61, 180 |
| abstract_inverted_index.of | 102, 114, 177 |
| abstract_inverted_index.to | 17, 44, 48, 149, 182 |
| abstract_inverted_index.we | 98 |
| abstract_inverted_index.ALS | 200 |
| abstract_inverted_index.How | 38 |
| abstract_inverted_index.Lys | 106, 121, 193 |
| abstract_inverted_index.One | 54 |
| abstract_inverted_index.Our | 161, 185 |
| abstract_inverted_index.and | 19, 50, 112, 129, 174 |
| abstract_inverted_index.has | 58 |
| abstract_inverted_index.may | 187 |
| abstract_inverted_index.not | 196 |
| abstract_inverted_index.the | 15, 46, 110 |
| abstract_inverted_index.why | 189 |
| abstract_inverted_index.K36, | 124 |
| abstract_inverted_index.K75, | 125 |
| abstract_inverted_index.K91) | 126 |
| abstract_inverted_index.SOD1 | 91, 128, 152, 165 |
| abstract_inverted_index.acid | 31 |
| abstract_inverted_index.also | 142 |
| abstract_inverted_index.been | 34, 59, 93, 197 |
| abstract_inverted_index.four | 119 |
| abstract_inverted_index.have | 33, 92, 195 |
| abstract_inverted_index.loss | 101, 176 |
| abstract_inverted_index.more | 26 |
| abstract_inverted_index.than | 27 |
| abstract_inverted_index.that | 57, 62, 76, 164, 175 |
| abstract_inverted_index.then | 73 |
| abstract_inverted_index.when | 63 |
| abstract_inverted_index.with | 8, 36, 95, 136 |
| abstract_inverted_index.(K30, | 123 |
| abstract_inverted_index.C4F6, | 155 |
| abstract_inverted_index.Here, | 97 |
| abstract_inverted_index.SOD1. | 115 |
| abstract_inverted_index.amino | 30, 71 |
| abstract_inverted_index.date, | 22 |
| abstract_inverted_index.fALS. | 96 |
| abstract_inverted_index.occur | 65 |
| abstract_inverted_index.these | 39, 131, 145, 192 |
| abstract_inverted_index.which | 156 |
| abstract_inverted_index.would | 77, 82, 108 |
| abstract_inverted_index.(SOD1) | 6 |
| abstract_inverted_index.(YFP). | 140 |
| abstract_inverted_index.(fALS) | 13 |
| abstract_inverted_index.acids, | 72 |
| abstract_inverted_index.charge | 179 |
| abstract_inverted_index.folded | 159 |
| abstract_inverted_index.forces | 75 |
| abstract_inverted_index.fusion | 134 |
| abstract_inverted_index.induce | 14, 109, 183 |
| abstract_inverted_index.lysine | 172 |
| abstract_inverted_index.native | 42 |
| abstract_inverted_index.yellow | 137 |
| abstract_inverted_index.binding | 148 |
| abstract_inverted_index.charged | 70, 88, 104 |
| abstract_inverted_index.explain | 188 |
| abstract_inverted_index.exposed | 171 |
| abstract_inverted_index.induced | 147 |
| abstract_inverted_index.inhibit | 78 |
| abstract_inverted_index.lateral | 11 |
| abstract_inverted_index.misfold | 18, 49 |
| abstract_inverted_index.mutated | 118 |
| abstract_inverted_index.perturb | 41 |
| abstract_inverted_index.protein | 16, 139 |
| abstract_inverted_index.surface | 105, 170 |
| abstract_inverted_index.whether | 100, 144 |
| abstract_inverted_index.ABSTRACT | 0 |
| abstract_inverted_index.aberrant | 79 |
| abstract_inverted_index.assessed | 143 |
| abstract_inverted_index.disease. | 37 |
| abstract_inverted_index.examined | 99 |
| abstract_inverted_index.familial | 9 |
| abstract_inverted_index.findings | 162, 186 |
| abstract_inverted_index.heighten | 45 |
| abstract_inverted_index.indicate | 163 |
| abstract_inverted_index.missense | 23 |
| abstract_inverted_index.occupied | 68 |
| abstract_inverted_index.positive | 178 |
| abstract_inverted_index.protein. | 160 |
| abstract_inverted_index.proteins | 135 |
| abstract_inverted_index.randomly | 117 |
| abstract_inverted_index.reduced. | 84 |
| abstract_inverted_index.residues | 89, 107, 122, 194 |
| abstract_inverted_index.unclear. | 53 |
| abstract_inverted_index.variants | 132 |
| abstract_inverted_index.Mutations | 1, 85 |
| abstract_inverted_index.aggregate | 51 |
| abstract_inverted_index.antibody, | 153 |
| abstract_inverted_index.different | 29, 120 |
| abstract_inverted_index.dismutase | 4 |
| abstract_inverted_index.expressed | 130 |
| abstract_inverted_index.generally | 166 |
| abstract_inverted_index.mechanism | 56 |
| abstract_inverted_index.mutations | 24, 40, 64, 146, 168, 190 |
| abstract_inverted_index.patients. | 201 |
| abstract_inverted_index.positions | 32, 67 |
| abstract_inverted_index.potential | 55 |
| abstract_inverted_index.repulsive | 74 |
| abstract_inverted_index.residues, | 173 |
| abstract_inverted_index.sclerosis | 12 |
| abstract_inverted_index.structure | 43 |
| abstract_inverted_index.suggested | 60 |
| abstract_inverted_index.tolerates | 167 |
| abstract_inverted_index.aggregate. | 20 |
| abstract_inverted_index.associated | 7, 35, 94 |
| abstract_inverted_index.designated | 154 |
| abstract_inverted_index.identified | 198 |
| abstract_inverted_index.misfolding | 111 |
| abstract_inverted_index.positively | 103 |
| abstract_inverted_index.propensity | 47 |
| abstract_inverted_index.recognizes | 157 |
| abstract_inverted_index.superoxide | 3 |
| abstract_inverted_index.twenty-one | 87 |
| abstract_inverted_index.aggregation | 113 |
| abstract_inverted_index.amyotrophic | 10 |
| abstract_inverted_index.fluorescent | 138 |
| abstract_inverted_index.aggregation. | 184 |
| abstract_inverted_index.insufficient | 181 |
| abstract_inverted_index.interactions | 81 |
| abstract_inverted_index.non-natively | 158 |
| abstract_inverted_index.protein:protein | 80 |
| abstract_inverted_index.conformation-restricted | 151 |
| cited_by_percentile_year | |
| corresponding_author_ids | https://openalex.org/A5038629622 |
| countries_distinct_count | 1 |
| institutions_distinct_count | 6 |
| corresponding_institution_ids | https://openalex.org/I103163165, https://openalex.org/I2800717037, https://openalex.org/I33213144 |
| citation_normalized_percentile.value | 0.04670454 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |