Machine learning algorithm to characterize antimicrobial resistance associated with the International Space Station surface microbiome Article Swipe
 YOU?
·
· 2022
· Open Access
·
· DOI: https://doi.org/10.1101/2022.02.07.479455
YOU?
·
· 2022
· Open Access
·
· DOI: https://doi.org/10.1101/2022.02.07.479455
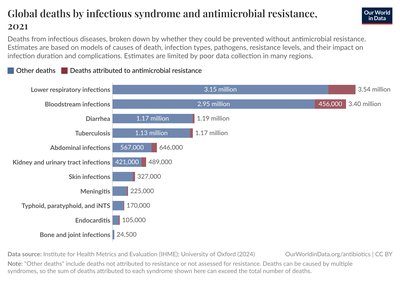
Background Antimicrobial Resistance (AMR) has a detrimental impact on human health on Earth and it is equally concerning in other environments such as space due to microgravity, radiation and confinement, especially for long-distance space travel. The International Space Station (ISS) is ideal for investigating microbial diversity and virulence. The shotgun metagenomics data of the ISS generated during the Microbial Tracking – 1 (MT-1) project and resulting metagenome-assembled genomes (MAGs) across three flights in eight different locations during 12 months were used in this study. The objective of this study was to identify the AMR genes associated with whole genomes of 227 cultivable strains, 21 shotgun metagenome sequences, and 24 MAGs retrieved from the ISS environmental samples that were treated with propidium monoazide (PMA; viable microbes). Results We have analyzed the data using a deep learning model, allowing us to go beyond traditional cut-offs based only on high DNA sequence similarity and extending the catalog of AMR genes. Our results in PMA treated samples revealed AMR dominance in the last flight for Kalamiella piersonii , a bacteria related to urinary tract infection in humans. The analysis of 227 pure strains isolated from the MT-1 project revealed hundreds of antibiotic resistance genes from many isolates, including two top-ranking species that corresponded to strains of Enterobacter bugandensis and Bacillus cereus . Computational predictions were experimentally validated by antibiotic resistance profiles in these two species, showing a high degree of concordance. Specifically, disc assay data confirmed the high resistance of these two pathogens to various beta-lactam antibiotics. Conclusion Overall, our computational predictions and validation analyses demonstrate the advantages of machine learning to uncover concealed AMR determinants in metagenomics datasets, expanding the understanding of the ISS environmental microbiomes and their pathogenic potential in humans.
Related Topics
- Type
- preprint
- Language
- en
- Landing Page
- https://doi.org/10.1101/2022.02.07.479455
- https://www.biorxiv.org/content/biorxiv/early/2022/02/10/2022.02.07.479455.full.pdf
- OA Status
- green
- Cited By
- 3
- References
- 56
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4211237552
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4211237552Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1101/2022.02.07.479455Digital Object Identifier
- Title
-
Machine learning algorithm to characterize antimicrobial resistance associated with the International Space Station surface microbiomeWork title
- Type
-
preprintOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2022Year of publication
- Publication date
-
2022-02-10Full publication date if available
- Authors
-
Pedro Madrigal, Nitin K. Singh, Jason M. Wood, Elena Gaudioso, Félix Hernández‐del‐Olmo, Christopher E. Mason, Kasthuri Venkateswaran, Afshin BeheshtiList of authors in order
- Landing page
-
https://doi.org/10.1101/2022.02.07.479455Publisher landing page
- PDF URL
-
https://www.biorxiv.org/content/biorxiv/early/2022/02/10/2022.02.07.479455.full.pdfDirect link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
greenOpen access status per OpenAlex
- OA URL
-
https://www.biorxiv.org/content/biorxiv/early/2022/02/10/2022.02.07.479455.full.pdfDirect OA link when available
- Concepts
-
Metagenomics, Antibiotic resistance, Biology, Microbiome, Bacillus cereus, Computational biology, Multilocus sequence typing, Shotgun sequencing, Microbiology, Genome, Genetics, Gene, Bacteria, GenotypeTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
3Total citation count in OpenAlex
- Citations by year (recent)
-
2025: 1, 2024: 1, 2022: 1Per-year citation counts (last 5 years)
- References (count)
-
56Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4211237552 |
|---|---|
| doi | https://doi.org/10.1101/2022.02.07.479455 |
| ids.doi | https://doi.org/10.17863/cam.87843 |
| ids.openalex | https://openalex.org/W4211237552 |
| fwci | 0.53848911 |
| type | preprint |
| title | Machine learning algorithm to characterize antimicrobial resistance associated with the International Space Station surface microbiome |
| biblio.issue | |
| biblio.volume | |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T12005 |
| topics[0].field.id | https://openalex.org/fields/27 |
| topics[0].field.display_name | Medicine |
| topics[0].score | 0.9986000061035156 |
| topics[0].domain.id | https://openalex.org/domains/4 |
| topics[0].domain.display_name | Health Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/2737 |
| topics[0].subfield.display_name | Physiology |
| topics[0].display_name | Spaceflight effects on biology |
| topics[1].id | https://openalex.org/T10066 |
| topics[1].field.id | https://openalex.org/fields/13 |
| topics[1].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[1].score | 0.9534000158309937 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1312 |
| topics[1].subfield.display_name | Molecular Biology |
| topics[1].display_name | Gut microbiota and health |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C15151743 |
| concepts[0].level | 3 |
| concepts[0].score | 0.8138917684555054 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q903778 |
| concepts[0].display_name | Metagenomics |
| concepts[1].id | https://openalex.org/C94665300 |
| concepts[1].level | 3 |
| concepts[1].score | 0.7128213047981262 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q63391344 |
| concepts[1].display_name | Antibiotic resistance |
| concepts[2].id | https://openalex.org/C86803240 |
| concepts[2].level | 0 |
| concepts[2].score | 0.6482937335968018 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[2].display_name | Biology |
| concepts[3].id | https://openalex.org/C143121216 |
| concepts[3].level | 2 |
| concepts[3].score | 0.5779781341552734 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q1330402 |
| concepts[3].display_name | Microbiome |
| concepts[4].id | https://openalex.org/C2777118263 |
| concepts[4].level | 3 |
| concepts[4].score | 0.4770221710205078 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q131307 |
| concepts[4].display_name | Bacillus cereus |
| concepts[5].id | https://openalex.org/C70721500 |
| concepts[5].level | 1 |
| concepts[5].score | 0.46897444128990173 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q177005 |
| concepts[5].display_name | Computational biology |
| concepts[6].id | https://openalex.org/C77434933 |
| concepts[6].level | 4 |
| concepts[6].score | 0.44863834977149963 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q4043440 |
| concepts[6].display_name | Multilocus sequence typing |
| concepts[7].id | https://openalex.org/C101985253 |
| concepts[7].level | 4 |
| concepts[7].score | 0.4469178318977356 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q1073526 |
| concepts[7].display_name | Shotgun sequencing |
| concepts[8].id | https://openalex.org/C89423630 |
| concepts[8].level | 1 |
| concepts[8].score | 0.4157930016517639 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q7193 |
| concepts[8].display_name | Microbiology |
| concepts[9].id | https://openalex.org/C141231307 |
| concepts[9].level | 3 |
| concepts[9].score | 0.39289674162864685 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q7020 |
| concepts[9].display_name | Genome |
| concepts[10].id | https://openalex.org/C54355233 |
| concepts[10].level | 1 |
| concepts[10].score | 0.33105555176734924 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[10].display_name | Genetics |
| concepts[11].id | https://openalex.org/C104317684 |
| concepts[11].level | 2 |
| concepts[11].score | 0.3264762759208679 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[11].display_name | Gene |
| concepts[12].id | https://openalex.org/C523546767 |
| concepts[12].level | 2 |
| concepts[12].score | 0.3066912889480591 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q10876 |
| concepts[12].display_name | Bacteria |
| concepts[13].id | https://openalex.org/C135763542 |
| concepts[13].level | 3 |
| concepts[13].score | 0.11746716499328613 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q106016 |
| concepts[13].display_name | Genotype |
| keywords[0].id | https://openalex.org/keywords/metagenomics |
| keywords[0].score | 0.8138917684555054 |
| keywords[0].display_name | Metagenomics |
| keywords[1].id | https://openalex.org/keywords/antibiotic-resistance |
| keywords[1].score | 0.7128213047981262 |
| keywords[1].display_name | Antibiotic resistance |
| keywords[2].id | https://openalex.org/keywords/biology |
| keywords[2].score | 0.6482937335968018 |
| keywords[2].display_name | Biology |
| keywords[3].id | https://openalex.org/keywords/microbiome |
| keywords[3].score | 0.5779781341552734 |
| keywords[3].display_name | Microbiome |
| keywords[4].id | https://openalex.org/keywords/bacillus-cereus |
| keywords[4].score | 0.4770221710205078 |
| keywords[4].display_name | Bacillus cereus |
| keywords[5].id | https://openalex.org/keywords/computational-biology |
| keywords[5].score | 0.46897444128990173 |
| keywords[5].display_name | Computational biology |
| keywords[6].id | https://openalex.org/keywords/multilocus-sequence-typing |
| keywords[6].score | 0.44863834977149963 |
| keywords[6].display_name | Multilocus sequence typing |
| keywords[7].id | https://openalex.org/keywords/shotgun-sequencing |
| keywords[7].score | 0.4469178318977356 |
| keywords[7].display_name | Shotgun sequencing |
| keywords[8].id | https://openalex.org/keywords/microbiology |
| keywords[8].score | 0.4157930016517639 |
| keywords[8].display_name | Microbiology |
| keywords[9].id | https://openalex.org/keywords/genome |
| keywords[9].score | 0.39289674162864685 |
| keywords[9].display_name | Genome |
| keywords[10].id | https://openalex.org/keywords/genetics |
| keywords[10].score | 0.33105555176734924 |
| keywords[10].display_name | Genetics |
| keywords[11].id | https://openalex.org/keywords/gene |
| keywords[11].score | 0.3264762759208679 |
| keywords[11].display_name | Gene |
| keywords[12].id | https://openalex.org/keywords/bacteria |
| keywords[12].score | 0.3066912889480591 |
| keywords[12].display_name | Bacteria |
| keywords[13].id | https://openalex.org/keywords/genotype |
| keywords[13].score | 0.11746716499328613 |
| keywords[13].display_name | Genotype |
| language | en |
| locations[0].id | doi:10.1101/2022.02.07.479455 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4306402567 |
| locations[0].source.issn | |
| locations[0].source.type | repository |
| locations[0].source.is_oa | False |
| locations[0].source.issn_l | |
| locations[0].source.is_core | False |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| locations[0].source.host_organization | https://openalex.org/I2750212522 |
| locations[0].source.host_organization_name | Cold Spring Harbor Laboratory |
| locations[0].source.host_organization_lineage | https://openalex.org/I2750212522 |
| locations[0].license | |
| locations[0].pdf_url | https://www.biorxiv.org/content/biorxiv/early/2022/02/10/2022.02.07.479455.full.pdf |
| locations[0].version | acceptedVersion |
| locations[0].raw_type | posted-content |
| locations[0].license_id | |
| locations[0].is_accepted | True |
| locations[0].is_published | False |
| locations[0].raw_source_name | |
| locations[0].landing_page_url | https://doi.org/10.1101/2022.02.07.479455 |
| locations[1].id | pmh:oai:www.repository.cam.ac.uk:1810/340406 |
| locations[1].is_oa | True |
| locations[1].source.id | https://openalex.org/S4306401776 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | Apollo (University of Cambridge) |
| locations[1].source.host_organization | https://openalex.org/I241749 |
| locations[1].source.host_organization_name | University of Cambridge |
| locations[1].source.host_organization_lineage | https://openalex.org/I241749 |
| locations[1].license | cc-by |
| locations[1].pdf_url | |
| locations[1].version | submittedVersion |
| locations[1].raw_type | Article |
| locations[1].license_id | https://openalex.org/licenses/cc-by |
| locations[1].is_accepted | False |
| locations[1].is_published | False |
| locations[1].raw_source_name | |
| locations[1].landing_page_url | https://www.repository.cam.ac.uk/handle/1810/340406 |
| locations[2].id | pmh:oai:www.repository.cam.ac.uk:1810/340790 |
| locations[2].is_oa | True |
| locations[2].source.id | https://openalex.org/S4306401776 |
| locations[2].source.issn | |
| locations[2].source.type | repository |
| locations[2].source.is_oa | False |
| locations[2].source.issn_l | |
| locations[2].source.is_core | False |
| locations[2].source.is_in_doaj | False |
| locations[2].source.display_name | Apollo (University of Cambridge) |
| locations[2].source.host_organization | https://openalex.org/I241749 |
| locations[2].source.host_organization_name | University of Cambridge |
| locations[2].source.host_organization_lineage | https://openalex.org/I241749 |
| locations[2].license | cc-by |
| locations[2].pdf_url | |
| locations[2].version | submittedVersion |
| locations[2].raw_type | Article |
| locations[2].license_id | https://openalex.org/licenses/cc-by |
| locations[2].is_accepted | False |
| locations[2].is_published | False |
| locations[2].raw_source_name | |
| locations[2].landing_page_url | https://www.repository.cam.ac.uk/handle/1810/340790 |
| locations[3].id | pmh:oai:www.repository.cam.ac.uk:1810/341325 |
| locations[3].is_oa | True |
| locations[3].source.id | https://openalex.org/S4306401776 |
| locations[3].source.issn | |
| locations[3].source.type | repository |
| locations[3].source.is_oa | False |
| locations[3].source.issn_l | |
| locations[3].source.is_core | False |
| locations[3].source.is_in_doaj | False |
| locations[3].source.display_name | Apollo (University of Cambridge) |
| locations[3].source.host_organization | https://openalex.org/I241749 |
| locations[3].source.host_organization_name | University of Cambridge |
| locations[3].source.host_organization_lineage | https://openalex.org/I241749 |
| locations[3].license | cc-by |
| locations[3].pdf_url | |
| locations[3].version | submittedVersion |
| locations[3].raw_type | Article |
| locations[3].license_id | https://openalex.org/licenses/cc-by |
| locations[3].is_accepted | False |
| locations[3].is_published | False |
| locations[3].raw_source_name | essn: 2049-2618 |
| locations[3].landing_page_url | https://www.repository.cam.ac.uk/handle/1810/341325 |
| locations[4].id | doi:10.17863/cam.87843 |
| locations[4].is_oa | True |
| locations[4].source.id | https://openalex.org/S7407050737 |
| locations[4].source.type | repository |
| locations[4].source.is_oa | False |
| locations[4].source.issn_l | |
| locations[4].source.is_core | False |
| locations[4].source.is_in_doaj | False |
| locations[4].source.display_name | Apollo |
| locations[4].source.host_organization | |
| locations[4].source.host_organization_name | |
| locations[4].license | |
| locations[4].pdf_url | |
| locations[4].version | |
| locations[4].raw_type | article-journal |
| locations[4].license_id | |
| locations[4].is_accepted | False |
| locations[4].is_published | |
| locations[4].raw_source_name | |
| locations[4].landing_page_url | https://doi.org/10.17863/cam.87843 |
| locations[5].id | doi:10.17863/cam.88226 |
| locations[5].is_oa | True |
| locations[5].source.id | https://openalex.org/S7407050737 |
| locations[5].source.type | repository |
| locations[5].source.is_oa | False |
| locations[5].source.issn_l | |
| locations[5].source.is_core | False |
| locations[5].source.is_in_doaj | False |
| locations[5].source.display_name | Apollo |
| locations[5].source.host_organization | |
| locations[5].source.host_organization_name | |
| locations[5].license | |
| locations[5].pdf_url | |
| locations[5].version | |
| locations[5].raw_type | article-journal |
| locations[5].license_id | |
| locations[5].is_accepted | False |
| locations[5].is_published | |
| locations[5].raw_source_name | |
| locations[5].landing_page_url | https://doi.org/10.17863/cam.88226 |
| locations[6].id | doi:10.17863/cam.88751 |
| locations[6].is_oa | True |
| locations[6].source.id | https://openalex.org/S7407050737 |
| locations[6].source.type | repository |
| locations[6].source.is_oa | False |
| locations[6].source.issn_l | |
| locations[6].source.is_core | False |
| locations[6].source.is_in_doaj | False |
| locations[6].source.display_name | Apollo |
| locations[6].source.host_organization | |
| locations[6].source.host_organization_name | |
| locations[6].license | cc-by |
| locations[6].pdf_url | |
| locations[6].version | |
| locations[6].raw_type | article-journal |
| locations[6].license_id | https://openalex.org/licenses/cc-by |
| locations[6].is_accepted | False |
| locations[6].is_published | |
| locations[6].raw_source_name | |
| locations[6].landing_page_url | https://doi.org/10.17863/cam.88751 |
| indexed_in | crossref, datacite |
| authorships[0].author.id | https://openalex.org/A5017572459 |
| authorships[0].author.orcid | https://orcid.org/0000-0003-1959-8199 |
| authorships[0].author.display_name | Pedro Madrigal |
| authorships[0].countries | GB |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I21196054, https://openalex.org/I241749 |
| authorships[0].affiliations[0].raw_affiliation_string | Jeffrey Cheah Biomedical Centre, Wellcome - MRC Cambridge Stem Cell Institute, University of Cambridge, Cambridge Biomedical Campus, Puddicombe Way, Cambridge CB2 0AW, UK |
| authorships[0].institutions[0].id | https://openalex.org/I241749 |
| authorships[0].institutions[0].ror | https://ror.org/013meh722 |
| authorships[0].institutions[0].type | education |
| authorships[0].institutions[0].lineage | https://openalex.org/I241749 |
| authorships[0].institutions[0].country_code | GB |
| authorships[0].institutions[0].display_name | University of Cambridge |
| authorships[0].institutions[1].id | https://openalex.org/I21196054 |
| authorships[0].institutions[1].ror | https://ror.org/05nz0zp31 |
| authorships[0].institutions[1].type | facility |
| authorships[0].institutions[1].lineage | https://openalex.org/I21196054, https://openalex.org/I241749, https://openalex.org/I4210087105, https://openalex.org/I87048295, https://openalex.org/I90344618 |
| authorships[0].institutions[1].country_code | GB |
| authorships[0].institutions[1].display_name | Wellcome/MRC Cambridge Stem Cell Institute |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Pedro Madrigal |
| authorships[0].is_corresponding | True |
| authorships[0].raw_affiliation_strings | Jeffrey Cheah Biomedical Centre, Wellcome - MRC Cambridge Stem Cell Institute, University of Cambridge, Cambridge Biomedical Campus, Puddicombe Way, Cambridge CB2 0AW, UK |
| authorships[1].author.id | https://openalex.org/A5004469409 |
| authorships[1].author.orcid | https://orcid.org/0000-0001-5344-1190 |
| authorships[1].author.display_name | Nitin K. Singh |
| authorships[1].countries | US |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I1334627681 |
| authorships[1].affiliations[0].raw_affiliation_string | Biotechnology and Planetary Protection Group, Jet Propulsion Laboratory, California Institute of Technology, Pasadena, CA 91109, USA |
| authorships[1].institutions[0].id | https://openalex.org/I1334627681 |
| authorships[1].institutions[0].ror | https://ror.org/027k65916 |
| authorships[1].institutions[0].type | facility |
| authorships[1].institutions[0].lineage | https://openalex.org/I122411786, https://openalex.org/I1334627681, https://openalex.org/I4210124779 |
| authorships[1].institutions[0].country_code | US |
| authorships[1].institutions[0].display_name | Jet Propulsion Laboratory |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Nitin K. Singh |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | Biotechnology and Planetary Protection Group, Jet Propulsion Laboratory, California Institute of Technology, Pasadena, CA 91109, USA |
| authorships[2].author.id | https://openalex.org/A5082890592 |
| authorships[2].author.orcid | https://orcid.org/0000-0002-0847-4695 |
| authorships[2].author.display_name | Jason M. Wood |
| authorships[2].countries | US |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I1334627681 |
| authorships[2].affiliations[0].raw_affiliation_string | Biotechnology and Planetary Protection Group, Jet Propulsion Laboratory, California Institute of Technology, Pasadena, CA 91109, USA |
| authorships[2].institutions[0].id | https://openalex.org/I1334627681 |
| authorships[2].institutions[0].ror | https://ror.org/027k65916 |
| authorships[2].institutions[0].type | facility |
| authorships[2].institutions[0].lineage | https://openalex.org/I122411786, https://openalex.org/I1334627681, https://openalex.org/I4210124779 |
| authorships[2].institutions[0].country_code | US |
| authorships[2].institutions[0].display_name | Jet Propulsion Laboratory |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Jason M. Wood |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | Biotechnology and Planetary Protection Group, Jet Propulsion Laboratory, California Institute of Technology, Pasadena, CA 91109, USA |
| authorships[3].author.id | https://openalex.org/A5073237157 |
| authorships[3].author.orcid | https://orcid.org/0000-0003-2258-6623 |
| authorships[3].author.display_name | Elena Gaudioso |
| authorships[3].countries | ES |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I178450904 |
| authorships[3].affiliations[0].raw_affiliation_string | Department of Artificial Intelligence, Computer Science School, Universidad Nacional de Educación a Distancia (UNED), 28040 Madrid, Spain |
| authorships[3].institutions[0].id | https://openalex.org/I178450904 |
| authorships[3].institutions[0].ror | https://ror.org/02msb5n36 |
| authorships[3].institutions[0].type | education |
| authorships[3].institutions[0].lineage | https://openalex.org/I178450904 |
| authorships[3].institutions[0].country_code | ES |
| authorships[3].institutions[0].display_name | Universidad Nacional de Educación a Distancia |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Elena Gaudioso |
| authorships[3].is_corresponding | False |
| authorships[3].raw_affiliation_strings | Department of Artificial Intelligence, Computer Science School, Universidad Nacional de Educación a Distancia (UNED), 28040 Madrid, Spain |
| authorships[4].author.id | https://openalex.org/A5039024023 |
| authorships[4].author.orcid | https://orcid.org/0000-0002-0567-9572 |
| authorships[4].author.display_name | Félix Hernández‐del‐Olmo |
| authorships[4].countries | ES |
| authorships[4].affiliations[0].institution_ids | https://openalex.org/I178450904 |
| authorships[4].affiliations[0].raw_affiliation_string | Department of Artificial Intelligence, Computer Science School, Universidad Nacional de Educación a Distancia (UNED), 28040 Madrid, Spain |
| authorships[4].institutions[0].id | https://openalex.org/I178450904 |
| authorships[4].institutions[0].ror | https://ror.org/02msb5n36 |
| authorships[4].institutions[0].type | education |
| authorships[4].institutions[0].lineage | https://openalex.org/I178450904 |
| authorships[4].institutions[0].country_code | ES |
| authorships[4].institutions[0].display_name | Universidad Nacional de Educación a Distancia |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Félix Hernández-del-Olmo |
| authorships[4].is_corresponding | False |
| authorships[4].raw_affiliation_strings | Department of Artificial Intelligence, Computer Science School, Universidad Nacional de Educación a Distancia (UNED), 28040 Madrid, Spain |
| authorships[5].author.id | https://openalex.org/A5085435765 |
| authorships[5].author.orcid | https://orcid.org/0000-0002-1850-1642 |
| authorships[5].author.display_name | Christopher E. Mason |
| authorships[5].countries | US |
| authorships[5].affiliations[0].institution_ids | https://openalex.org/I4210151462 |
| authorships[5].affiliations[0].raw_affiliation_string | New York Genome Center, NY, 10013, USA |
| authorships[5].affiliations[1].institution_ids | https://openalex.org/I205783295 |
| authorships[5].affiliations[1].raw_affiliation_string | Department of Physiology and Biophysics, Weill Cornell Medical College, New York, NY 10065, USA |
| authorships[5].affiliations[2].institution_ids | https://openalex.org/I205783295, https://openalex.org/I4210097825, https://openalex.org/I4387153466 |
| authorships[5].affiliations[2].raw_affiliation_string | The Feil Family Brain and Mind Research Institute, Weill Cornell Medicine, NY, 10065, USA |
| authorships[5].affiliations[3].institution_ids | https://openalex.org/I205783295, https://openalex.org/I4387153466 |
| authorships[5].affiliations[3].raw_affiliation_string | The HRH Prince Alwaleed Bin Talal Bin Abdulaziz Alsaud Institute for Computational Biomedicine, Weill Cornell Medicine, NY, 1006, USA |
| authorships[5].institutions[0].id | https://openalex.org/I205783295 |
| authorships[5].institutions[0].ror | https://ror.org/05bnh6r87 |
| authorships[5].institutions[0].type | education |
| authorships[5].institutions[0].lineage | https://openalex.org/I205783295 |
| authorships[5].institutions[0].country_code | US |
| authorships[5].institutions[0].display_name | Cornell University |
| authorships[5].institutions[1].id | https://openalex.org/I4210097825 |
| authorships[5].institutions[1].ror | https://ror.org/00we1gw23 |
| authorships[5].institutions[1].type | nonprofit |
| authorships[5].institutions[1].lineage | https://openalex.org/I4210097825 |
| authorships[5].institutions[1].country_code | US |
| authorships[5].institutions[1].display_name | MIND Research Institute |
| authorships[5].institutions[2].id | https://openalex.org/I4210151462 |
| authorships[5].institutions[2].ror | https://ror.org/05wf2ga96 |
| authorships[5].institutions[2].type | facility |
| authorships[5].institutions[2].lineage | https://openalex.org/I4210151462 |
| authorships[5].institutions[2].country_code | US |
| authorships[5].institutions[2].display_name | New York Genome Center |
| authorships[5].institutions[3].id | https://openalex.org/I4387153466 |
| authorships[5].institutions[3].ror | https://ror.org/02r109517 |
| authorships[5].institutions[3].type | education |
| authorships[5].institutions[3].lineage | https://openalex.org/I205783295, https://openalex.org/I4387153466 |
| authorships[5].institutions[3].country_code | US |
| authorships[5].institutions[3].display_name | Weill Cornell Medicine |
| authorships[5].author_position | middle |
| authorships[5].raw_author_name | Christopher E. Mason |
| authorships[5].is_corresponding | False |
| authorships[5].raw_affiliation_strings | Department of Physiology and Biophysics, Weill Cornell Medical College, New York, NY 10065, USA, New York Genome Center, NY, 10013, USA, The Feil Family Brain and Mind Research Institute, Weill Cornell Medicine, NY, 10065, USA, The HRH Prince Alwaleed Bin Talal Bin Abdulaziz Alsaud Institute for Computational Biomedicine, Weill Cornell Medicine, NY, 1006, USA |
| authorships[6].author.id | https://openalex.org/A5047719221 |
| authorships[6].author.orcid | https://orcid.org/0000-0002-6742-0873 |
| authorships[6].author.display_name | Kasthuri Venkateswaran |
| authorships[6].countries | US |
| authorships[6].affiliations[0].institution_ids | https://openalex.org/I1334627681 |
| authorships[6].affiliations[0].raw_affiliation_string | Biotechnology and Planetary Protection Group, Jet Propulsion Laboratory, California Institute of Technology, Pasadena, CA 91109, USA |
| authorships[6].institutions[0].id | https://openalex.org/I1334627681 |
| authorships[6].institutions[0].ror | https://ror.org/027k65916 |
| authorships[6].institutions[0].type | facility |
| authorships[6].institutions[0].lineage | https://openalex.org/I122411786, https://openalex.org/I1334627681, https://openalex.org/I4210124779 |
| authorships[6].institutions[0].country_code | US |
| authorships[6].institutions[0].display_name | Jet Propulsion Laboratory |
| authorships[6].author_position | middle |
| authorships[6].raw_author_name | Kasthuri Venkateswaran |
| authorships[6].is_corresponding | False |
| authorships[6].raw_affiliation_strings | Biotechnology and Planetary Protection Group, Jet Propulsion Laboratory, California Institute of Technology, Pasadena, CA 91109, USA |
| authorships[7].author.id | https://openalex.org/A5077595545 |
| authorships[7].author.orcid | https://orcid.org/0000-0003-4643-531X |
| authorships[7].author.display_name | Afshin Beheshti |
| authorships[7].countries | US |
| authorships[7].affiliations[0].institution_ids | https://openalex.org/I1280536761 |
| authorships[7].affiliations[0].raw_affiliation_string | KBR, Space Biosciences Division, NASA Ames Research Center, Moffett Field, CA, 94035, USA |
| authorships[7].affiliations[1].institution_ids | https://openalex.org/I107606265 |
| authorships[7].affiliations[1].raw_affiliation_string | Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, MA, 02142, USA |
| authorships[7].institutions[0].id | https://openalex.org/I1280536761 |
| authorships[7].institutions[0].ror | https://ror.org/02acart68 |
| authorships[7].institutions[0].type | facility |
| authorships[7].institutions[0].lineage | https://openalex.org/I1280536761, https://openalex.org/I4210124779 |
| authorships[7].institutions[0].country_code | US |
| authorships[7].institutions[0].display_name | Ames Research Center |
| authorships[7].institutions[1].id | https://openalex.org/I107606265 |
| authorships[7].institutions[1].ror | https://ror.org/05a0ya142 |
| authorships[7].institutions[1].type | nonprofit |
| authorships[7].institutions[1].lineage | https://openalex.org/I107606265 |
| authorships[7].institutions[1].country_code | US |
| authorships[7].institutions[1].display_name | Broad Institute |
| authorships[7].author_position | last |
| authorships[7].raw_author_name | Afshin Beheshti |
| authorships[7].is_corresponding | False |
| authorships[7].raw_affiliation_strings | KBR, Space Biosciences Division, NASA Ames Research Center, Moffett Field, CA, 94035, USA, Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, MA, 02142, USA |
| has_content.pdf | True |
| has_content.grobid_xml | True |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://www.biorxiv.org/content/biorxiv/early/2022/02/10/2022.02.07.479455.full.pdf |
| open_access.oa_status | green |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Machine learning algorithm to characterize antimicrobial resistance associated with the International Space Station surface microbiome |
| has_fulltext | True |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T12005 |
| primary_topic.field.id | https://openalex.org/fields/27 |
| primary_topic.field.display_name | Medicine |
| primary_topic.score | 0.9986000061035156 |
| primary_topic.domain.id | https://openalex.org/domains/4 |
| primary_topic.domain.display_name | Health Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/2737 |
| primary_topic.subfield.display_name | Physiology |
| primary_topic.display_name | Spaceflight effects on biology |
| related_works | https://openalex.org/W3043668926, https://openalex.org/W3036436206, https://openalex.org/W4389452281, https://openalex.org/W4238948313, https://openalex.org/W3111832393, https://openalex.org/W3000916974, https://openalex.org/W3134948850, https://openalex.org/W3183189600, https://openalex.org/W4394325869, https://openalex.org/W4394082811 |
| cited_by_count | 3 |
| counts_by_year[0].year | 2025 |
| counts_by_year[0].cited_by_count | 1 |
| counts_by_year[1].year | 2024 |
| counts_by_year[1].cited_by_count | 1 |
| counts_by_year[2].year | 2022 |
| counts_by_year[2].cited_by_count | 1 |
| locations_count | 7 |
| best_oa_location.id | doi:10.1101/2022.02.07.479455 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4306402567 |
| best_oa_location.source.issn | |
| best_oa_location.source.type | repository |
| best_oa_location.source.is_oa | False |
| best_oa_location.source.issn_l | |
| best_oa_location.source.is_core | False |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| best_oa_location.source.host_organization | https://openalex.org/I2750212522 |
| best_oa_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| best_oa_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| best_oa_location.license | |
| best_oa_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2022/02/10/2022.02.07.479455.full.pdf |
| best_oa_location.version | acceptedVersion |
| best_oa_location.raw_type | posted-content |
| best_oa_location.license_id | |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | False |
| best_oa_location.raw_source_name | |
| best_oa_location.landing_page_url | https://doi.org/10.1101/2022.02.07.479455 |
| primary_location.id | doi:10.1101/2022.02.07.479455 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4306402567 |
| primary_location.source.issn | |
| primary_location.source.type | repository |
| primary_location.source.is_oa | False |
| primary_location.source.issn_l | |
| primary_location.source.is_core | False |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| primary_location.source.host_organization | https://openalex.org/I2750212522 |
| primary_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| primary_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| primary_location.license | |
| primary_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2022/02/10/2022.02.07.479455.full.pdf |
| primary_location.version | acceptedVersion |
| primary_location.raw_type | posted-content |
| primary_location.license_id | |
| primary_location.is_accepted | True |
| primary_location.is_published | False |
| primary_location.raw_source_name | |
| primary_location.landing_page_url | https://doi.org/10.1101/2022.02.07.479455 |
| publication_date | 2022-02-10 |
| publication_year | 2022 |
| referenced_works | https://openalex.org/W3107984511, https://openalex.org/W2951274106, https://openalex.org/W2790564955, https://openalex.org/W3017503194, https://openalex.org/W2004548026, https://openalex.org/W2120902911, https://openalex.org/W2734859723, https://openalex.org/W2941799199, https://openalex.org/W3036535414, https://openalex.org/W3037569543, https://openalex.org/W2131271579, https://openalex.org/W2921872415, https://openalex.org/W2940104035, https://openalex.org/W2951912016, https://openalex.org/W2979939041, https://openalex.org/W2035840565, https://openalex.org/W3163970933, https://openalex.org/W3100944007, https://openalex.org/W2105199251, https://openalex.org/W2126070805, https://openalex.org/W2113214455, https://openalex.org/W2077399200, https://openalex.org/W2939018266, https://openalex.org/W2107772251, https://openalex.org/W2923997757, https://openalex.org/W2078021131, https://openalex.org/W2968322804, https://openalex.org/W2273099552, https://openalex.org/W1584790813, https://openalex.org/W1965936382, https://openalex.org/W2965998894, https://openalex.org/W2318383157, https://openalex.org/W3168226731, https://openalex.org/W2146286718, https://openalex.org/W2599417231, https://openalex.org/W2112364185, https://openalex.org/W3081559394, https://openalex.org/W2900984733, https://openalex.org/W2901971963, https://openalex.org/W2899573152, https://openalex.org/W2940870259, https://openalex.org/W2131738221, https://openalex.org/W2899068429, https://openalex.org/W2107766352, https://openalex.org/W263733316, https://openalex.org/W2782741879, https://openalex.org/W3016316470, https://openalex.org/W2005557196, https://openalex.org/W2727689426, https://openalex.org/W2961363174, https://openalex.org/W2168927628, https://openalex.org/W2588775853, https://openalex.org/W2134852597, https://openalex.org/W2748882831, https://openalex.org/W2908881088, https://openalex.org/W3107384450 |
| referenced_works_count | 56 |
| abstract_inverted_index., | 174 |
| abstract_inverted_index.. | 218 |
| abstract_inverted_index.1 | 62 |
| abstract_inverted_index.a | 6, 133, 175, 233 |
| abstract_inverted_index.12 | 78 |
| abstract_inverted_index.21 | 104 |
| abstract_inverted_index.24 | 109 |
| abstract_inverted_index.We | 127 |
| abstract_inverted_index.as | 23 |
| abstract_inverted_index.by | 224 |
| abstract_inverted_index.go | 140 |
| abstract_inverted_index.in | 19, 73, 82, 160, 167, 182, 228, 273, 288 |
| abstract_inverted_index.is | 16, 41 |
| abstract_inverted_index.it | 15 |
| abstract_inverted_index.of | 53, 87, 100, 155, 186, 197, 212, 236, 246, 265, 279 |
| abstract_inverted_index.on | 9, 12, 146 |
| abstract_inverted_index.to | 26, 91, 139, 178, 210, 250, 268 |
| abstract_inverted_index.us | 138 |
| abstract_inverted_index.227 | 101, 187 |
| abstract_inverted_index.AMR | 94, 156, 165, 271 |
| abstract_inverted_index.DNA | 148 |
| abstract_inverted_index.ISS | 55, 114, 281 |
| abstract_inverted_index.Our | 158 |
| abstract_inverted_index.PMA | 161 |
| abstract_inverted_index.The | 36, 49, 85, 184 |
| abstract_inverted_index.and | 14, 29, 47, 65, 108, 151, 215, 259, 284 |
| abstract_inverted_index.due | 25 |
| abstract_inverted_index.for | 32, 43, 171 |
| abstract_inverted_index.has | 5 |
| abstract_inverted_index.our | 256 |
| abstract_inverted_index.the | 54, 58, 93, 113, 130, 153, 168, 192, 243, 263, 277, 280 |
| abstract_inverted_index.two | 205, 230, 248 |
| abstract_inverted_index.was | 90 |
| abstract_inverted_index.– | 61 |
| abstract_inverted_index.MAGs | 110 |
| abstract_inverted_index.MT-1 | 193 |
| abstract_inverted_index.data | 52, 131, 241 |
| abstract_inverted_index.deep | 134 |
| abstract_inverted_index.disc | 239 |
| abstract_inverted_index.from | 112, 191, 201 |
| abstract_inverted_index.have | 128 |
| abstract_inverted_index.high | 147, 234, 244 |
| abstract_inverted_index.last | 169 |
| abstract_inverted_index.many | 202 |
| abstract_inverted_index.only | 145 |
| abstract_inverted_index.pure | 188 |
| abstract_inverted_index.such | 22 |
| abstract_inverted_index.that | 117, 208 |
| abstract_inverted_index.this | 83, 88 |
| abstract_inverted_index.used | 81 |
| abstract_inverted_index.were | 80, 118, 221 |
| abstract_inverted_index.with | 97, 120 |
| abstract_inverted_index.(AMR) | 4 |
| abstract_inverted_index.(ISS) | 40 |
| abstract_inverted_index.(PMA; | 123 |
| abstract_inverted_index.Earth | 13 |
| abstract_inverted_index.Space | 38 |
| abstract_inverted_index.assay | 240 |
| abstract_inverted_index.based | 144 |
| abstract_inverted_index.eight | 74 |
| abstract_inverted_index.genes | 95, 200 |
| abstract_inverted_index.human | 10 |
| abstract_inverted_index.ideal | 42 |
| abstract_inverted_index.other | 20 |
| abstract_inverted_index.space | 24, 34 |
| abstract_inverted_index.study | 89 |
| abstract_inverted_index.their | 285 |
| abstract_inverted_index.these | 229, 247 |
| abstract_inverted_index.three | 71 |
| abstract_inverted_index.tract | 180 |
| abstract_inverted_index.using | 132 |
| abstract_inverted_index.whole | 98 |
| abstract_inverted_index.(MAGs) | 69 |
| abstract_inverted_index.(MT-1) | 63 |
| abstract_inverted_index.across | 70 |
| abstract_inverted_index.beyond | 141 |
| abstract_inverted_index.cereus | 217 |
| abstract_inverted_index.degree | 235 |
| abstract_inverted_index.during | 57, 77 |
| abstract_inverted_index.flight | 170 |
| abstract_inverted_index.genes. | 157 |
| abstract_inverted_index.health | 11 |
| abstract_inverted_index.impact | 8 |
| abstract_inverted_index.model, | 136 |
| abstract_inverted_index.months | 79 |
| abstract_inverted_index.study. | 84 |
| abstract_inverted_index.viable | 124 |
| abstract_inverted_index.Results | 126 |
| abstract_inverted_index.Station | 39 |
| abstract_inverted_index.catalog | 154 |
| abstract_inverted_index.equally | 17 |
| abstract_inverted_index.flights | 72 |
| abstract_inverted_index.genomes | 68, 99 |
| abstract_inverted_index.humans. | 183, 289 |
| abstract_inverted_index.machine | 266 |
| abstract_inverted_index.project | 64, 194 |
| abstract_inverted_index.related | 177 |
| abstract_inverted_index.results | 159 |
| abstract_inverted_index.samples | 116, 163 |
| abstract_inverted_index.shotgun | 50, 105 |
| abstract_inverted_index.showing | 232 |
| abstract_inverted_index.species | 207 |
| abstract_inverted_index.strains | 189, 211 |
| abstract_inverted_index.travel. | 35 |
| abstract_inverted_index.treated | 119, 162 |
| abstract_inverted_index.uncover | 269 |
| abstract_inverted_index.urinary | 179 |
| abstract_inverted_index.various | 251 |
| abstract_inverted_index.ABSTRACT | 0 |
| abstract_inverted_index.Bacillus | 216 |
| abstract_inverted_index.Overall, | 255 |
| abstract_inverted_index.Tracking | 60 |
| abstract_inverted_index.allowing | 137 |
| abstract_inverted_index.analyses | 261 |
| abstract_inverted_index.analysis | 185 |
| abstract_inverted_index.analyzed | 129 |
| abstract_inverted_index.bacteria | 176 |
| abstract_inverted_index.cut-offs | 143 |
| abstract_inverted_index.hundreds | 196 |
| abstract_inverted_index.identify | 92 |
| abstract_inverted_index.isolated | 190 |
| abstract_inverted_index.learning | 135, 267 |
| abstract_inverted_index.profiles | 227 |
| abstract_inverted_index.revealed | 164, 195 |
| abstract_inverted_index.sequence | 149 |
| abstract_inverted_index.species, | 231 |
| abstract_inverted_index.strains, | 103 |
| abstract_inverted_index.Microbial | 59 |
| abstract_inverted_index.concealed | 270 |
| abstract_inverted_index.confirmed | 242 |
| abstract_inverted_index.datasets, | 275 |
| abstract_inverted_index.different | 75 |
| abstract_inverted_index.diversity | 46 |
| abstract_inverted_index.dominance | 166 |
| abstract_inverted_index.expanding | 276 |
| abstract_inverted_index.extending | 152 |
| abstract_inverted_index.generated | 56 |
| abstract_inverted_index.including | 204 |
| abstract_inverted_index.infection | 181 |
| abstract_inverted_index.isolates, | 203 |
| abstract_inverted_index.locations | 76 |
| abstract_inverted_index.microbial | 45 |
| abstract_inverted_index.monoazide | 122 |
| abstract_inverted_index.objective | 86 |
| abstract_inverted_index.pathogens | 249 |
| abstract_inverted_index.piersonii | 173 |
| abstract_inverted_index.potential | 287 |
| abstract_inverted_index.propidium | 121 |
| abstract_inverted_index.radiation | 28 |
| abstract_inverted_index.resulting | 66 |
| abstract_inverted_index.retrieved | 111 |
| abstract_inverted_index.validated | 223 |
| abstract_inverted_index.Background | 1 |
| abstract_inverted_index.Conclusion | 254 |
| abstract_inverted_index.Kalamiella | 172 |
| abstract_inverted_index.Resistance | 3 |
| abstract_inverted_index.advantages | 264 |
| abstract_inverted_index.antibiotic | 198, 225 |
| abstract_inverted_index.associated | 96 |
| abstract_inverted_index.concerning | 18 |
| abstract_inverted_index.cultivable | 102 |
| abstract_inverted_index.especially | 31 |
| abstract_inverted_index.metagenome | 106 |
| abstract_inverted_index.microbes). | 125 |
| abstract_inverted_index.pathogenic | 286 |
| abstract_inverted_index.resistance | 199, 226, 245 |
| abstract_inverted_index.sequences, | 107 |
| abstract_inverted_index.similarity | 150 |
| abstract_inverted_index.validation | 260 |
| abstract_inverted_index.virulence. | 48 |
| abstract_inverted_index.beta-lactam | 252 |
| abstract_inverted_index.bugandensis | 214 |
| abstract_inverted_index.demonstrate | 262 |
| abstract_inverted_index.detrimental | 7 |
| abstract_inverted_index.microbiomes | 283 |
| abstract_inverted_index.predictions | 220, 258 |
| abstract_inverted_index.top-ranking | 206 |
| abstract_inverted_index.traditional | 142 |
| abstract_inverted_index.Enterobacter | 213 |
| abstract_inverted_index.antibiotics. | 253 |
| abstract_inverted_index.concordance. | 237 |
| abstract_inverted_index.confinement, | 30 |
| abstract_inverted_index.corresponded | 209 |
| abstract_inverted_index.determinants | 272 |
| abstract_inverted_index.environments | 21 |
| abstract_inverted_index.metagenomics | 51, 274 |
| abstract_inverted_index.Antimicrobial | 2 |
| abstract_inverted_index.Computational | 219 |
| abstract_inverted_index.International | 37 |
| abstract_inverted_index.Specifically, | 238 |
| abstract_inverted_index.computational | 257 |
| abstract_inverted_index.environmental | 115, 282 |
| abstract_inverted_index.investigating | 44 |
| abstract_inverted_index.long-distance | 33 |
| abstract_inverted_index.microgravity, | 27 |
| abstract_inverted_index.understanding | 278 |
| abstract_inverted_index.experimentally | 222 |
| abstract_inverted_index.metagenome-assembled | 67 |
| cited_by_percentile_year.max | 95 |
| cited_by_percentile_year.min | 89 |
| corresponding_author_ids | https://openalex.org/A5017572459 |
| countries_distinct_count | 3 |
| institutions_distinct_count | 8 |
| corresponding_institution_ids | https://openalex.org/I21196054, https://openalex.org/I241749 |
| citation_normalized_percentile.value | 0.57475169 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |