mBAT-combo: A more powerful test to detect gene-trait associations from GWAS data Article Swipe
 YOU?
·
· 2024
· Open Access
·
· DOI: https://doi.org/10.17615/d56b-ce53
YOU?
·
· 2024
· Open Access
·
· DOI: https://doi.org/10.17615/d56b-ce53
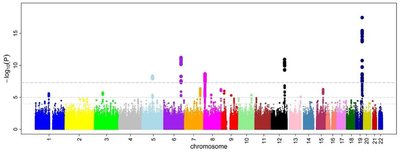
Gene-based association tests aggregate multiple SNP-trait associations into sets defined by gene boundaries and are widely used in post-GWAS analysis. A common approach for gene-based tests is to combine SNPs associations by computing the sum of χ2 statistics. However, this strategy ignores the directions of SNP effects, which could result in a loss of power for SNPs with masking effects, e.g., when the product of two SNP effects and the linkage disequilibrium (LD) correlation is negative. Here, we introduce “mBAT-combo,” a set-based test that is better powered than other methods to detect multi-SNP associations in the context of masking effects. We validate the method through simulations and applications to real data. We find that of 35 blood and urine biomarker traits in the UK Biobank, 34 traits show evidence for masking effects in a total of 4,273 gene-trait pairs, indicating that masking effects is common in complex traits. We further validate the improved power of our method in height, body mass index, and schizophrenia with different GWAS sample sizes and show that on average 95.7% of the genes detected only by mBAT-combo with smaller sample sizes can be identified by the single-SNP approach with a 1.7-fold increase in sample sizes. Eleven genes significant only in mBAT-combo for schizophrenia are confirmed by functionally informed fine-mapping or Mendelian randomization integrating gene expression data. The framework of mBAT-combo can be applied to any set of SNPs to refine trait-association signals hidden in genomic regions with complex LD structures.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.17615/d56b-ce53
- OA Status
- green
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4400034801
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4400034801Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.17615/d56b-ce53Digital Object Identifier
- Title
-
mBAT-combo: A more powerful test to detect gene-trait associations from GWAS dataWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2024Year of publication
- Publication date
-
2024-06-01Full publication date if available
- Authors
-
Li A, Liu S, Amulyasai Bakshi, Longda Jiang, Weihong Chen, Z. Zheng, Sullivan P.F, V. Padma Srivastava M., Wray N.R, Jie Yang, Jianshu ZengList of authors in order
- Landing page
-
https://doi.org/10.17615/d56b-ce53Publisher landing page
- Open access
-
YesWhether a free full text is available
- OA status
-
greenOpen access status per OpenAlex
- OA URL
-
https://doi.org/10.17615/d56b-ce53Direct OA link when available
- Concepts
-
Genome-wide association study, Trait, Computational biology, Genetics, Biology, Gene, Computer science, Single-nucleotide polymorphism, Genotype, Programming languageTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
0Total citation count in OpenAlex
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4400034801 |
|---|---|
| doi | https://doi.org/10.17615/d56b-ce53 |
| ids.doi | https://doi.org/10.17615/d56b-ce53 |
| ids.openalex | https://openalex.org/W4400034801 |
| fwci | 0.0 |
| type | article |
| title | mBAT-combo: A more powerful test to detect gene-trait associations from GWAS data |
| biblio.issue | |
| biblio.volume | |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T10261 |
| topics[0].field.id | https://openalex.org/fields/13 |
| topics[0].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[0].score | 0.9064000248908997 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1311 |
| topics[0].subfield.display_name | Genetics |
| topics[0].display_name | Genetic Associations and Epidemiology |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C106208931 |
| concepts[0].level | 5 |
| concepts[0].score | 0.7584880590438843 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q1098876 |
| concepts[0].display_name | Genome-wide association study |
| concepts[1].id | https://openalex.org/C106934330 |
| concepts[1].level | 2 |
| concepts[1].score | 0.6446062326431274 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q1971873 |
| concepts[1].display_name | Trait |
| concepts[2].id | https://openalex.org/C70721500 |
| concepts[2].level | 1 |
| concepts[2].score | 0.4683273136615753 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q177005 |
| concepts[2].display_name | Computational biology |
| concepts[3].id | https://openalex.org/C54355233 |
| concepts[3].level | 1 |
| concepts[3].score | 0.4631713032722473 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[3].display_name | Genetics |
| concepts[4].id | https://openalex.org/C86803240 |
| concepts[4].level | 0 |
| concepts[4].score | 0.44077929854393005 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[4].display_name | Biology |
| concepts[5].id | https://openalex.org/C104317684 |
| concepts[5].level | 2 |
| concepts[5].score | 0.37028923630714417 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[5].display_name | Gene |
| concepts[6].id | https://openalex.org/C41008148 |
| concepts[6].level | 0 |
| concepts[6].score | 0.21732115745544434 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q21198 |
| concepts[6].display_name | Computer science |
| concepts[7].id | https://openalex.org/C153209595 |
| concepts[7].level | 4 |
| concepts[7].score | 0.1596526801586151 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q501128 |
| concepts[7].display_name | Single-nucleotide polymorphism |
| concepts[8].id | https://openalex.org/C135763542 |
| concepts[8].level | 3 |
| concepts[8].score | 0.06431254744529724 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q106016 |
| concepts[8].display_name | Genotype |
| concepts[9].id | https://openalex.org/C199360897 |
| concepts[9].level | 1 |
| concepts[9].score | 0.0 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q9143 |
| concepts[9].display_name | Programming language |
| keywords[0].id | https://openalex.org/keywords/genome-wide-association-study |
| keywords[0].score | 0.7584880590438843 |
| keywords[0].display_name | Genome-wide association study |
| keywords[1].id | https://openalex.org/keywords/trait |
| keywords[1].score | 0.6446062326431274 |
| keywords[1].display_name | Trait |
| keywords[2].id | https://openalex.org/keywords/computational-biology |
| keywords[2].score | 0.4683273136615753 |
| keywords[2].display_name | Computational biology |
| keywords[3].id | https://openalex.org/keywords/genetics |
| keywords[3].score | 0.4631713032722473 |
| keywords[3].display_name | Genetics |
| keywords[4].id | https://openalex.org/keywords/biology |
| keywords[4].score | 0.44077929854393005 |
| keywords[4].display_name | Biology |
| keywords[5].id | https://openalex.org/keywords/gene |
| keywords[5].score | 0.37028923630714417 |
| keywords[5].display_name | Gene |
| keywords[6].id | https://openalex.org/keywords/computer-science |
| keywords[6].score | 0.21732115745544434 |
| keywords[6].display_name | Computer science |
| keywords[7].id | https://openalex.org/keywords/single-nucleotide-polymorphism |
| keywords[7].score | 0.1596526801586151 |
| keywords[7].display_name | Single-nucleotide polymorphism |
| keywords[8].id | https://openalex.org/keywords/genotype |
| keywords[8].score | 0.06431254744529724 |
| keywords[8].display_name | Genotype |
| language | en |
| locations[0].id | doi:10.17615/d56b-ce53 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S7407051488 |
| locations[0].source.type | repository |
| locations[0].source.is_oa | False |
| locations[0].source.issn_l | |
| locations[0].source.is_core | False |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | UNC Libraries |
| locations[0].source.host_organization | |
| locations[0].source.host_organization_name | |
| locations[0].license | |
| locations[0].pdf_url | |
| locations[0].version | |
| locations[0].raw_type | article-journal |
| locations[0].license_id | |
| locations[0].is_accepted | False |
| locations[0].is_published | |
| locations[0].raw_source_name | |
| locations[0].landing_page_url | https://doi.org/10.17615/d56b-ce53 |
| indexed_in | datacite |
| authorships[0].author.id | https://openalex.org/A5109298218 |
| authorships[0].author.orcid | |
| authorships[0].author.display_name | Li A |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Li A |
| authorships[0].is_corresponding | False |
| authorships[1].author.id | https://openalex.org/A5104136001 |
| authorships[1].author.orcid | |
| authorships[1].author.display_name | Liu S |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Liu S |
| authorships[1].is_corresponding | False |
| authorships[2].author.id | https://openalex.org/A5108972332 |
| authorships[2].author.orcid | |
| authorships[2].author.display_name | Amulyasai Bakshi |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Bakshi A |
| authorships[2].is_corresponding | False |
| authorships[3].author.id | https://openalex.org/A5065532777 |
| authorships[3].author.orcid | https://orcid.org/0000-0003-4964-6497 |
| authorships[3].author.display_name | Longda Jiang |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Jiang L |
| authorships[3].is_corresponding | False |
| authorships[4].author.id | https://openalex.org/A5100679542 |
| authorships[4].author.orcid | https://orcid.org/0000-0001-8689-7311 |
| authorships[4].author.display_name | Weihong Chen |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Chen W |
| authorships[4].is_corresponding | False |
| authorships[5].author.id | https://openalex.org/A5111340769 |
| authorships[5].author.orcid | |
| authorships[5].author.display_name | Z. Zheng |
| authorships[5].author_position | middle |
| authorships[5].raw_author_name | Zheng Z |
| authorships[5].is_corresponding | False |
| authorships[6].author.id | https://openalex.org/A5104335584 |
| authorships[6].author.orcid | |
| authorships[6].author.display_name | Sullivan P.F |
| authorships[6].author_position | middle |
| authorships[6].raw_author_name | Sullivan P.F |
| authorships[6].is_corresponding | False |
| authorships[7].author.id | https://openalex.org/A5100544523 |
| authorships[7].author.orcid | |
| authorships[7].author.display_name | V. Padma Srivastava M. |
| authorships[7].author_position | middle |
| authorships[7].raw_author_name | Visscher P.M |
| authorships[7].is_corresponding | False |
| authorships[8].author.id | https://openalex.org/A5113265325 |
| authorships[8].author.orcid | |
| authorships[8].author.display_name | Wray N.R |
| authorships[8].author_position | middle |
| authorships[8].raw_author_name | Wray N.R |
| authorships[8].is_corresponding | False |
| authorships[9].author.id | https://openalex.org/A5029432491 |
| authorships[9].author.orcid | https://orcid.org/0000-0003-2241-5066 |
| authorships[9].author.display_name | Jie Yang |
| authorships[9].author_position | middle |
| authorships[9].raw_author_name | Yang J |
| authorships[9].is_corresponding | False |
| authorships[10].author.id | https://openalex.org/A5100589768 |
| authorships[10].author.orcid | https://orcid.org/0009-0005-5326-4442 |
| authorships[10].author.display_name | Jianshu Zeng |
| authorships[10].author_position | last |
| authorships[10].raw_author_name | Zeng J |
| authorships[10].is_corresponding | False |
| has_content.pdf | False |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://doi.org/10.17615/d56b-ce53 |
| open_access.oa_status | green |
| open_access.any_repository_has_fulltext | False |
| created_date | 2024-06-27T00:00:00 |
| display_name | mBAT-combo: A more powerful test to detect gene-trait associations from GWAS data |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-06T06:51:31.235846 |
| primary_topic.id | https://openalex.org/T10261 |
| primary_topic.field.id | https://openalex.org/fields/13 |
| primary_topic.field.display_name | Biochemistry, Genetics and Molecular Biology |
| primary_topic.score | 0.9064000248908997 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1311 |
| primary_topic.subfield.display_name | Genetics |
| primary_topic.display_name | Genetic Associations and Epidemiology |
| related_works | https://openalex.org/W4391375266, https://openalex.org/W1539935214, https://openalex.org/W4390666975, https://openalex.org/W1864400744, https://openalex.org/W2736520836, https://openalex.org/W3093265873, https://openalex.org/W3209211130, https://openalex.org/W1966248182, https://openalex.org/W2971313479, https://openalex.org/W2116256947 |
| cited_by_count | 0 |
| locations_count | 1 |
| best_oa_location.id | doi:10.17615/d56b-ce53 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S7407051488 |
| best_oa_location.source.type | repository |
| best_oa_location.source.is_oa | False |
| best_oa_location.source.issn_l | |
| best_oa_location.source.is_core | False |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | UNC Libraries |
| best_oa_location.source.host_organization | |
| best_oa_location.source.host_organization_name | |
| best_oa_location.license | |
| best_oa_location.pdf_url | |
| best_oa_location.version | |
| best_oa_location.raw_type | article-journal |
| best_oa_location.license_id | |
| best_oa_location.is_accepted | False |
| best_oa_location.is_published | False |
| best_oa_location.raw_source_name | |
| best_oa_location.landing_page_url | https://doi.org/10.17615/d56b-ce53 |
| primary_location.id | doi:10.17615/d56b-ce53 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S7407051488 |
| primary_location.source.type | repository |
| primary_location.source.is_oa | False |
| primary_location.source.issn_l | |
| primary_location.source.is_core | False |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | UNC Libraries |
| primary_location.source.host_organization | |
| primary_location.source.host_organization_name | |
| primary_location.license | |
| primary_location.pdf_url | |
| primary_location.version | |
| primary_location.raw_type | article-journal |
| primary_location.license_id | |
| primary_location.is_accepted | False |
| primary_location.is_published | False |
| primary_location.raw_source_name | |
| primary_location.landing_page_url | https://doi.org/10.17615/d56b-ce53 |
| publication_date | 2024-06-01 |
| publication_year | 2024 |
| referenced_works_count | 0 |
| abstract_inverted_index.A | 20 |
| abstract_inverted_index.a | 51, 80, 133, 194 |
| abstract_inverted_index.34 | 125 |
| abstract_inverted_index.35 | 115 |
| abstract_inverted_index.LD | 243 |
| abstract_inverted_index.UK | 123 |
| abstract_inverted_index.We | 100, 111, 148 |
| abstract_inverted_index.be | 187, 226 |
| abstract_inverted_index.by | 10, 31, 180, 189, 210 |
| abstract_inverted_index.in | 17, 50, 94, 121, 132, 145, 157, 197, 204, 238 |
| abstract_inverted_index.is | 26, 74, 84, 143 |
| abstract_inverted_index.of | 35, 44, 53, 64, 97, 114, 135, 154, 175, 223, 231 |
| abstract_inverted_index.on | 172 |
| abstract_inverted_index.or | 214 |
| abstract_inverted_index.to | 27, 90, 108, 228, 233 |
| abstract_inverted_index.we | 77 |
| abstract_inverted_index.SNP | 45, 66 |
| abstract_inverted_index.The | 221 |
| abstract_inverted_index.and | 13, 68, 106, 117, 162, 169 |
| abstract_inverted_index.any | 229 |
| abstract_inverted_index.are | 14, 208 |
| abstract_inverted_index.can | 186, 225 |
| abstract_inverted_index.for | 23, 55, 129, 206 |
| abstract_inverted_index.our | 155 |
| abstract_inverted_index.set | 230 |
| abstract_inverted_index.sum | 34 |
| abstract_inverted_index.the | 33, 42, 62, 69, 95, 102, 122, 151, 176, 190 |
| abstract_inverted_index.two | 65 |
| abstract_inverted_index.χ2 | 36 |
| abstract_inverted_index.(LD) | 72 |
| abstract_inverted_index.GWAS | 166 |
| abstract_inverted_index.SNPs | 29, 56, 232 |
| abstract_inverted_index.body | 159 |
| abstract_inverted_index.find | 112 |
| abstract_inverted_index.gene | 11, 218 |
| abstract_inverted_index.into | 7 |
| abstract_inverted_index.loss | 52 |
| abstract_inverted_index.mass | 160 |
| abstract_inverted_index.only | 179, 203 |
| abstract_inverted_index.real | 109 |
| abstract_inverted_index.sets | 8 |
| abstract_inverted_index.show | 127, 170 |
| abstract_inverted_index.test | 82 |
| abstract_inverted_index.than | 87 |
| abstract_inverted_index.that | 83, 113, 140, 171 |
| abstract_inverted_index.this | 39 |
| abstract_inverted_index.used | 16 |
| abstract_inverted_index.when | 61 |
| abstract_inverted_index.with | 57, 164, 182, 193, 241 |
| abstract_inverted_index.4,273 | 136 |
| abstract_inverted_index.95.7% | 174 |
| abstract_inverted_index.Here, | 76 |
| abstract_inverted_index.blood | 116 |
| abstract_inverted_index.could | 48 |
| abstract_inverted_index.data. | 110, 220 |
| abstract_inverted_index.e.g., | 60 |
| abstract_inverted_index.genes | 177, 201 |
| abstract_inverted_index.other | 88 |
| abstract_inverted_index.power | 54, 153 |
| abstract_inverted_index.sizes | 168, 185 |
| abstract_inverted_index.tests | 2, 25 |
| abstract_inverted_index.total | 134 |
| abstract_inverted_index.urine | 118 |
| abstract_inverted_index.which | 47 |
| abstract_inverted_index.Eleven | 200 |
| abstract_inverted_index.better | 85 |
| abstract_inverted_index.common | 21, 144 |
| abstract_inverted_index.detect | 91 |
| abstract_inverted_index.hidden | 237 |
| abstract_inverted_index.index, | 161 |
| abstract_inverted_index.method | 103, 156 |
| abstract_inverted_index.pairs, | 138 |
| abstract_inverted_index.refine | 234 |
| abstract_inverted_index.result | 49 |
| abstract_inverted_index.sample | 167, 184, 198 |
| abstract_inverted_index.sizes. | 199 |
| abstract_inverted_index.traits | 120, 126 |
| abstract_inverted_index.widely | 15 |
| abstract_inverted_index.applied | 227 |
| abstract_inverted_index.average | 173 |
| abstract_inverted_index.combine | 28 |
| abstract_inverted_index.complex | 146, 242 |
| abstract_inverted_index.context | 96 |
| abstract_inverted_index.defined | 9 |
| abstract_inverted_index.effects | 67, 131, 142 |
| abstract_inverted_index.further | 149 |
| abstract_inverted_index.genomic | 239 |
| abstract_inverted_index.height, | 158 |
| abstract_inverted_index.ignores | 41 |
| abstract_inverted_index.linkage | 70 |
| abstract_inverted_index.masking | 58, 98, 130, 141 |
| abstract_inverted_index.methods | 89 |
| abstract_inverted_index.powered | 86 |
| abstract_inverted_index.product | 63 |
| abstract_inverted_index.regions | 240 |
| abstract_inverted_index.signals | 236 |
| abstract_inverted_index.smaller | 183 |
| abstract_inverted_index.through | 104 |
| abstract_inverted_index.traits. | 147 |
| abstract_inverted_index.1.7-fold | 195 |
| abstract_inverted_index.Biobank, | 124 |
| abstract_inverted_index.However, | 38 |
| abstract_inverted_index.approach | 22, 192 |
| abstract_inverted_index.detected | 178 |
| abstract_inverted_index.effects, | 46, 59 |
| abstract_inverted_index.effects. | 99 |
| abstract_inverted_index.evidence | 128 |
| abstract_inverted_index.improved | 152 |
| abstract_inverted_index.increase | 196 |
| abstract_inverted_index.informed | 212 |
| abstract_inverted_index.multiple | 4 |
| abstract_inverted_index.strategy | 40 |
| abstract_inverted_index.validate | 101, 150 |
| abstract_inverted_index.Mendelian | 215 |
| abstract_inverted_index.SNP-trait | 5 |
| abstract_inverted_index.aggregate | 3 |
| abstract_inverted_index.analysis. | 19 |
| abstract_inverted_index.biomarker | 119 |
| abstract_inverted_index.computing | 32 |
| abstract_inverted_index.confirmed | 209 |
| abstract_inverted_index.different | 165 |
| abstract_inverted_index.framework | 222 |
| abstract_inverted_index.introduce | 78 |
| abstract_inverted_index.multi-SNP | 92 |
| abstract_inverted_index.negative. | 75 |
| abstract_inverted_index.post-GWAS | 18 |
| abstract_inverted_index.set-based | 81 |
| abstract_inverted_index.Gene-based | 0 |
| abstract_inverted_index.boundaries | 12 |
| abstract_inverted_index.directions | 43 |
| abstract_inverted_index.expression | 219 |
| abstract_inverted_index.gene-based | 24 |
| abstract_inverted_index.gene-trait | 137 |
| abstract_inverted_index.identified | 188 |
| abstract_inverted_index.indicating | 139 |
| abstract_inverted_index.mBAT-combo | 181, 205, 224 |
| abstract_inverted_index.single-SNP | 191 |
| abstract_inverted_index.association | 1 |
| abstract_inverted_index.correlation | 73 |
| abstract_inverted_index.integrating | 217 |
| abstract_inverted_index.significant | 202 |
| abstract_inverted_index.simulations | 105 |
| abstract_inverted_index.statistics. | 37 |
| abstract_inverted_index.structures. | 244 |
| abstract_inverted_index.applications | 107 |
| abstract_inverted_index.associations | 6, 30, 93 |
| abstract_inverted_index.fine-mapping | 213 |
| abstract_inverted_index.functionally | 211 |
| abstract_inverted_index.randomization | 216 |
| abstract_inverted_index.schizophrenia | 163, 207 |
| abstract_inverted_index.disequilibrium | 71 |
| abstract_inverted_index.trait-association | 235 |
| abstract_inverted_index.“mBAT-combo,” | 79 |
| cited_by_percentile_year | |
| countries_distinct_count | 0 |
| institutions_distinct_count | 11 |
| citation_normalized_percentile.value | 0.13215577 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |