Mining for Potent Inhibitors through Artificial Intelligence and Physics: A Unified Methodology for Ligand Based and Structure Based Drug Design Article Swipe
 YOU?
·
· 2024
· Open Access
·
· DOI: https://doi.org/10.1021/acs.jcim.4c00634
YOU?
·
· 2024
· Open Access
·
· DOI: https://doi.org/10.1021/acs.jcim.4c00634
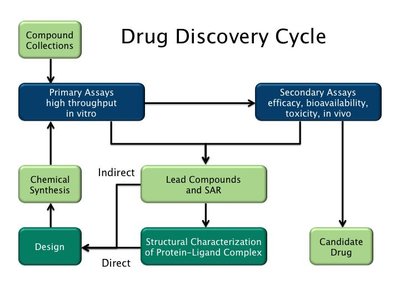
Determining the viability of a new drug molecule is a time- and resource-intensive task that makes computer-aided assessments a vital approach to rapid drug discovery. Here we develop a machine learning algorithm, iMiner, that generates novel inhibitor molecules for target proteins by combining deep reinforcement learning with real-time 3D molecular docking using AutoDock Vina, thereby simultaneously creating chemical novelty while constraining molecules for shape and molecular compatibility with target active sites. Moreover, through the use of various types of reward functions, we have introduced novelty in generative tasks for new molecules such as chemical similarity to a target ligand, molecules grown from known protein bound fragments, and creation of molecules that enforce interactions with target residues in the protein active site. The iMiner algorithm is embedded in a composite workflow that filters out Pan-assay interference compounds, Lipinski rule violations, uncommon structures in medicinal chemistry, and poor synthetic accessibility with options for cross-validation against other docking scoring functions and automation of a molecular dynamics simulation to measure pose stability. We also allow users to define a set of rules for the structures they would like to exclude during the training process and postfiltering steps. Because our approach relies only on the structure of the target protein, iMiner can be easily adapted for the future development of other inhibitors or small molecule therapeutics of any target protein.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.1021/acs.jcim.4c00634
- https://pubs.acs.org/doi/pdf/10.1021/acs.jcim.4c00634
- OA Status
- hybrid
- Cited By
- 4
- References
- 67
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4399393786
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4399393786Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1021/acs.jcim.4c00634Digital Object Identifier
- Title
-
Mining for Potent Inhibitors through Artificial Intelligence and Physics: A Unified Methodology for Ligand Based and Structure Based Drug DesignWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2024Year of publication
- Publication date
-
2024-06-06Full publication date if available
- Authors
-
Jie Li, Oufan Zhang, Kunyang Sun, Yingze Wang, Xingyi Guan, Dorian Bagni, Mojtaba Haghighatlari, Fiona L. Kearns, conor parks, Rommie E. Amaro, Teresa Head‐GordonList of authors in order
- Landing page
-
https://doi.org/10.1021/acs.jcim.4c00634Publisher landing page
- PDF URL
-
https://pubs.acs.org/doi/pdf/10.1021/acs.jcim.4c00634Direct link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
hybridOpen access status per OpenAlex
- OA URL
-
https://pubs.acs.org/doi/pdf/10.1021/acs.jcim.4c00634Direct OA link when available
- Concepts
-
Computer science, Drug discovery, Artificial intelligence, Novelty, DrugBank, AutoDock, Virtual screening, Docking (animal), Lipinski's rule of five, In silico, Machine learning, Computational biology, Chemistry, Bioinformatics, Drug, Biology, Philosophy, Nursing, Pharmacology, Medicine, Gene, Biochemistry, TheologyTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
4Total citation count in OpenAlex
- Citations by year (recent)
-
2025: 3, 2024: 1Per-year citation counts (last 5 years)
- References (count)
-
67Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4399393786 |
|---|---|
| doi | https://doi.org/10.1021/acs.jcim.4c00634 |
| ids.doi | https://doi.org/10.1021/acs.jcim.4c00634 |
| ids.pmid | https://pubmed.ncbi.nlm.nih.gov/38843070 |
| ids.openalex | https://openalex.org/W4399393786 |
| fwci | 3.1605423 |
| mesh[0].qualifier_ui | |
| mesh[0].descriptor_ui | D008024 |
| mesh[0].is_major_topic | False |
| mesh[0].qualifier_name | |
| mesh[0].descriptor_name | Ligands |
| mesh[1].qualifier_ui | |
| mesh[1].descriptor_ui | D015195 |
| mesh[1].is_major_topic | True |
| mesh[1].qualifier_name | |
| mesh[1].descriptor_name | Drug Design |
| mesh[2].qualifier_ui | |
| mesh[2].descriptor_ui | D062105 |
| mesh[2].is_major_topic | True |
| mesh[2].qualifier_name | |
| mesh[2].descriptor_name | Molecular Docking Simulation |
| mesh[3].qualifier_ui | |
| mesh[3].descriptor_ui | D001185 |
| mesh[3].is_major_topic | True |
| mesh[3].qualifier_name | |
| mesh[3].descriptor_name | Artificial Intelligence |
| mesh[4].qualifier_ui | |
| mesh[4].descriptor_ui | D056004 |
| mesh[4].is_major_topic | False |
| mesh[4].qualifier_name | |
| mesh[4].descriptor_name | Molecular Dynamics Simulation |
| mesh[5].qualifier_ui | |
| mesh[5].descriptor_ui | D057225 |
| mesh[5].is_major_topic | False |
| mesh[5].qualifier_name | |
| mesh[5].descriptor_name | Data Mining |
| mesh[6].qualifier_ui | |
| mesh[6].descriptor_ui | D000465 |
| mesh[6].is_major_topic | False |
| mesh[6].qualifier_name | |
| mesh[6].descriptor_name | Algorithms |
| mesh[7].qualifier_ui | Q000737 |
| mesh[7].descriptor_ui | D011506 |
| mesh[7].is_major_topic | False |
| mesh[7].qualifier_name | chemistry |
| mesh[7].descriptor_name | Proteins |
| mesh[8].qualifier_ui | Q000037 |
| mesh[8].descriptor_ui | D011506 |
| mesh[8].is_major_topic | False |
| mesh[8].qualifier_name | antagonists & inhibitors |
| mesh[8].descriptor_name | Proteins |
| mesh[9].qualifier_ui | Q000378 |
| mesh[9].descriptor_ui | D011506 |
| mesh[9].is_major_topic | False |
| mesh[9].qualifier_name | metabolism |
| mesh[9].descriptor_name | Proteins |
| mesh[10].qualifier_ui | |
| mesh[10].descriptor_ui | D000069550 |
| mesh[10].is_major_topic | False |
| mesh[10].qualifier_name | |
| mesh[10].descriptor_name | Machine Learning |
| mesh[11].qualifier_ui | |
| mesh[11].descriptor_ui | D008024 |
| mesh[11].is_major_topic | False |
| mesh[11].qualifier_name | |
| mesh[11].descriptor_name | Ligands |
| mesh[12].qualifier_ui | |
| mesh[12].descriptor_ui | D015195 |
| mesh[12].is_major_topic | True |
| mesh[12].qualifier_name | |
| mesh[12].descriptor_name | Drug Design |
| mesh[13].qualifier_ui | |
| mesh[13].descriptor_ui | D062105 |
| mesh[13].is_major_topic | True |
| mesh[13].qualifier_name | |
| mesh[13].descriptor_name | Molecular Docking Simulation |
| mesh[14].qualifier_ui | |
| mesh[14].descriptor_ui | D001185 |
| mesh[14].is_major_topic | True |
| mesh[14].qualifier_name | |
| mesh[14].descriptor_name | Artificial Intelligence |
| mesh[15].qualifier_ui | |
| mesh[15].descriptor_ui | D056004 |
| mesh[15].is_major_topic | False |
| mesh[15].qualifier_name | |
| mesh[15].descriptor_name | Molecular Dynamics Simulation |
| mesh[16].qualifier_ui | |
| mesh[16].descriptor_ui | D057225 |
| mesh[16].is_major_topic | False |
| mesh[16].qualifier_name | |
| mesh[16].descriptor_name | Data Mining |
| mesh[17].qualifier_ui | |
| mesh[17].descriptor_ui | D000465 |
| mesh[17].is_major_topic | False |
| mesh[17].qualifier_name | |
| mesh[17].descriptor_name | Algorithms |
| mesh[18].qualifier_ui | Q000737 |
| mesh[18].descriptor_ui | D011506 |
| mesh[18].is_major_topic | False |
| mesh[18].qualifier_name | chemistry |
| mesh[18].descriptor_name | Proteins |
| mesh[19].qualifier_ui | Q000037 |
| mesh[19].descriptor_ui | D011506 |
| mesh[19].is_major_topic | False |
| mesh[19].qualifier_name | antagonists & inhibitors |
| mesh[19].descriptor_name | Proteins |
| mesh[20].qualifier_ui | Q000378 |
| mesh[20].descriptor_ui | D011506 |
| mesh[20].is_major_topic | False |
| mesh[20].qualifier_name | metabolism |
| mesh[20].descriptor_name | Proteins |
| mesh[21].qualifier_ui | |
| mesh[21].descriptor_ui | D000069550 |
| mesh[21].is_major_topic | False |
| mesh[21].qualifier_name | |
| mesh[21].descriptor_name | Machine Learning |
| mesh[22].qualifier_ui | |
| mesh[22].descriptor_ui | D008024 |
| mesh[22].is_major_topic | False |
| mesh[22].qualifier_name | |
| mesh[22].descriptor_name | Ligands |
| mesh[23].qualifier_ui | |
| mesh[23].descriptor_ui | D015195 |
| mesh[23].is_major_topic | True |
| mesh[23].qualifier_name | |
| mesh[23].descriptor_name | Drug Design |
| mesh[24].qualifier_ui | |
| mesh[24].descriptor_ui | D062105 |
| mesh[24].is_major_topic | True |
| mesh[24].qualifier_name | |
| mesh[24].descriptor_name | Molecular Docking Simulation |
| mesh[25].qualifier_ui | |
| mesh[25].descriptor_ui | D001185 |
| mesh[25].is_major_topic | True |
| mesh[25].qualifier_name | |
| mesh[25].descriptor_name | Artificial Intelligence |
| mesh[26].qualifier_ui | |
| mesh[26].descriptor_ui | D056004 |
| mesh[26].is_major_topic | False |
| mesh[26].qualifier_name | |
| mesh[26].descriptor_name | Molecular Dynamics Simulation |
| mesh[27].qualifier_ui | |
| mesh[27].descriptor_ui | D057225 |
| mesh[27].is_major_topic | False |
| mesh[27].qualifier_name | |
| mesh[27].descriptor_name | Data Mining |
| mesh[28].qualifier_ui | |
| mesh[28].descriptor_ui | D000465 |
| mesh[28].is_major_topic | False |
| mesh[28].qualifier_name | |
| mesh[28].descriptor_name | Algorithms |
| mesh[29].qualifier_ui | Q000737 |
| mesh[29].descriptor_ui | D011506 |
| mesh[29].is_major_topic | False |
| mesh[29].qualifier_name | chemistry |
| mesh[29].descriptor_name | Proteins |
| mesh[30].qualifier_ui | Q000037 |
| mesh[30].descriptor_ui | D011506 |
| mesh[30].is_major_topic | False |
| mesh[30].qualifier_name | antagonists & inhibitors |
| mesh[30].descriptor_name | Proteins |
| mesh[31].qualifier_ui | Q000378 |
| mesh[31].descriptor_ui | D011506 |
| mesh[31].is_major_topic | False |
| mesh[31].qualifier_name | metabolism |
| mesh[31].descriptor_name | Proteins |
| mesh[32].qualifier_ui | |
| mesh[32].descriptor_ui | D000069550 |
| mesh[32].is_major_topic | False |
| mesh[32].qualifier_name | |
| mesh[32].descriptor_name | Machine Learning |
| type | article |
| title | Mining for Potent Inhibitors through Artificial Intelligence and Physics: A Unified Methodology for Ligand Based and Structure Based Drug Design |
| awards[0].id | https://openalex.org/G474321178 |
| awards[0].funder_id | https://openalex.org/F4320337355 |
| awards[0].display_name | |
| awards[0].funder_award_id | U19-AI171954 |
| awards[0].funder_display_name | National Institute of Allergy and Infectious Diseases |
| biblio.issue | 24 |
| biblio.volume | 64 |
| biblio.last_page | 9097 |
| biblio.first_page | 9082 |
| topics[0].id | https://openalex.org/T10211 |
| topics[0].field.id | https://openalex.org/fields/17 |
| topics[0].field.display_name | Computer Science |
| topics[0].score | 0.9998999834060669 |
| topics[0].domain.id | https://openalex.org/domains/3 |
| topics[0].domain.display_name | Physical Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1703 |
| topics[0].subfield.display_name | Computational Theory and Mathematics |
| topics[0].display_name | Computational Drug Discovery Methods |
| topics[1].id | https://openalex.org/T11948 |
| topics[1].field.id | https://openalex.org/fields/25 |
| topics[1].field.display_name | Materials Science |
| topics[1].score | 0.9609000086784363 |
| topics[1].domain.id | https://openalex.org/domains/3 |
| topics[1].domain.display_name | Physical Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/2505 |
| topics[1].subfield.display_name | Materials Chemistry |
| topics[1].display_name | Machine Learning in Materials Science |
| topics[2].id | https://openalex.org/T10044 |
| topics[2].field.id | https://openalex.org/fields/13 |
| topics[2].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[2].score | 0.9336000084877014 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1312 |
| topics[2].subfield.display_name | Molecular Biology |
| topics[2].display_name | Protein Structure and Dynamics |
| funders[0].id | https://openalex.org/F4320337355 |
| funders[0].ror | https://ror.org/043z4tv69 |
| funders[0].display_name | National Institute of Allergy and Infectious Diseases |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C41008148 |
| concepts[0].level | 0 |
| concepts[0].score | 0.6376495361328125 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q21198 |
| concepts[0].display_name | Computer science |
| concepts[1].id | https://openalex.org/C74187038 |
| concepts[1].level | 2 |
| concepts[1].score | 0.5702682137489319 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q1418791 |
| concepts[1].display_name | Drug discovery |
| concepts[2].id | https://openalex.org/C154945302 |
| concepts[2].level | 1 |
| concepts[2].score | 0.5520074963569641 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q11660 |
| concepts[2].display_name | Artificial intelligence |
| concepts[3].id | https://openalex.org/C2778738651 |
| concepts[3].level | 2 |
| concepts[3].score | 0.527379035949707 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q16546687 |
| concepts[3].display_name | Novelty |
| concepts[4].id | https://openalex.org/C155261790 |
| concepts[4].level | 3 |
| concepts[4].score | 0.49793553352355957 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q1122544 |
| concepts[4].display_name | DrugBank |
| concepts[5].id | https://openalex.org/C2780152424 |
| concepts[5].level | 4 |
| concepts[5].score | 0.4896295666694641 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q4826062 |
| concepts[5].display_name | AutoDock |
| concepts[6].id | https://openalex.org/C103697762 |
| concepts[6].level | 3 |
| concepts[6].score | 0.4866112172603607 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q4112105 |
| concepts[6].display_name | Virtual screening |
| concepts[7].id | https://openalex.org/C41685203 |
| concepts[7].level | 2 |
| concepts[7].score | 0.46555712819099426 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q1974042 |
| concepts[7].display_name | Docking (animal) |
| concepts[8].id | https://openalex.org/C41847442 |
| concepts[8].level | 4 |
| concepts[8].score | 0.4428609609603882 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q916723 |
| concepts[8].display_name | Lipinski's rule of five |
| concepts[9].id | https://openalex.org/C2775905019 |
| concepts[9].level | 3 |
| concepts[9].score | 0.4119924306869507 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q192572 |
| concepts[9].display_name | In silico |
| concepts[10].id | https://openalex.org/C119857082 |
| concepts[10].level | 1 |
| concepts[10].score | 0.4030681550502777 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q2539 |
| concepts[10].display_name | Machine learning |
| concepts[11].id | https://openalex.org/C70721500 |
| concepts[11].level | 1 |
| concepts[11].score | 0.39412355422973633 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q177005 |
| concepts[11].display_name | Computational biology |
| concepts[12].id | https://openalex.org/C185592680 |
| concepts[12].level | 0 |
| concepts[12].score | 0.2747400104999542 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q2329 |
| concepts[12].display_name | Chemistry |
| concepts[13].id | https://openalex.org/C60644358 |
| concepts[13].level | 1 |
| concepts[13].score | 0.23261916637420654 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q128570 |
| concepts[13].display_name | Bioinformatics |
| concepts[14].id | https://openalex.org/C2780035454 |
| concepts[14].level | 2 |
| concepts[14].score | 0.16095709800720215 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q8386 |
| concepts[14].display_name | Drug |
| concepts[15].id | https://openalex.org/C86803240 |
| concepts[15].level | 0 |
| concepts[15].score | 0.12871089577674866 |
| concepts[15].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[15].display_name | Biology |
| concepts[16].id | https://openalex.org/C138885662 |
| concepts[16].level | 0 |
| concepts[16].score | 0.0 |
| concepts[16].wikidata | https://www.wikidata.org/wiki/Q5891 |
| concepts[16].display_name | Philosophy |
| concepts[17].id | https://openalex.org/C159110408 |
| concepts[17].level | 1 |
| concepts[17].score | 0.0 |
| concepts[17].wikidata | https://www.wikidata.org/wiki/Q121176 |
| concepts[17].display_name | Nursing |
| concepts[18].id | https://openalex.org/C98274493 |
| concepts[18].level | 1 |
| concepts[18].score | 0.0 |
| concepts[18].wikidata | https://www.wikidata.org/wiki/Q128406 |
| concepts[18].display_name | Pharmacology |
| concepts[19].id | https://openalex.org/C71924100 |
| concepts[19].level | 0 |
| concepts[19].score | 0.0 |
| concepts[19].wikidata | https://www.wikidata.org/wiki/Q11190 |
| concepts[19].display_name | Medicine |
| concepts[20].id | https://openalex.org/C104317684 |
| concepts[20].level | 2 |
| concepts[20].score | 0.0 |
| concepts[20].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[20].display_name | Gene |
| concepts[21].id | https://openalex.org/C55493867 |
| concepts[21].level | 1 |
| concepts[21].score | 0.0 |
| concepts[21].wikidata | https://www.wikidata.org/wiki/Q7094 |
| concepts[21].display_name | Biochemistry |
| concepts[22].id | https://openalex.org/C27206212 |
| concepts[22].level | 1 |
| concepts[22].score | 0.0 |
| concepts[22].wikidata | https://www.wikidata.org/wiki/Q34178 |
| concepts[22].display_name | Theology |
| keywords[0].id | https://openalex.org/keywords/computer-science |
| keywords[0].score | 0.6376495361328125 |
| keywords[0].display_name | Computer science |
| keywords[1].id | https://openalex.org/keywords/drug-discovery |
| keywords[1].score | 0.5702682137489319 |
| keywords[1].display_name | Drug discovery |
| keywords[2].id | https://openalex.org/keywords/artificial-intelligence |
| keywords[2].score | 0.5520074963569641 |
| keywords[2].display_name | Artificial intelligence |
| keywords[3].id | https://openalex.org/keywords/novelty |
| keywords[3].score | 0.527379035949707 |
| keywords[3].display_name | Novelty |
| keywords[4].id | https://openalex.org/keywords/drugbank |
| keywords[4].score | 0.49793553352355957 |
| keywords[4].display_name | DrugBank |
| keywords[5].id | https://openalex.org/keywords/autodock |
| keywords[5].score | 0.4896295666694641 |
| keywords[5].display_name | AutoDock |
| keywords[6].id | https://openalex.org/keywords/virtual-screening |
| keywords[6].score | 0.4866112172603607 |
| keywords[6].display_name | Virtual screening |
| keywords[7].id | https://openalex.org/keywords/docking |
| keywords[7].score | 0.46555712819099426 |
| keywords[7].display_name | Docking (animal) |
| keywords[8].id | https://openalex.org/keywords/lipinskis-rule-of-five |
| keywords[8].score | 0.4428609609603882 |
| keywords[8].display_name | Lipinski's rule of five |
| keywords[9].id | https://openalex.org/keywords/in-silico |
| keywords[9].score | 0.4119924306869507 |
| keywords[9].display_name | In silico |
| keywords[10].id | https://openalex.org/keywords/machine-learning |
| keywords[10].score | 0.4030681550502777 |
| keywords[10].display_name | Machine learning |
| keywords[11].id | https://openalex.org/keywords/computational-biology |
| keywords[11].score | 0.39412355422973633 |
| keywords[11].display_name | Computational biology |
| keywords[12].id | https://openalex.org/keywords/chemistry |
| keywords[12].score | 0.2747400104999542 |
| keywords[12].display_name | Chemistry |
| keywords[13].id | https://openalex.org/keywords/bioinformatics |
| keywords[13].score | 0.23261916637420654 |
| keywords[13].display_name | Bioinformatics |
| keywords[14].id | https://openalex.org/keywords/drug |
| keywords[14].score | 0.16095709800720215 |
| keywords[14].display_name | Drug |
| keywords[15].id | https://openalex.org/keywords/biology |
| keywords[15].score | 0.12871089577674866 |
| keywords[15].display_name | Biology |
| language | en |
| locations[0].id | doi:10.1021/acs.jcim.4c00634 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S167262187 |
| locations[0].source.issn | 1549-9596, 1549-960X |
| locations[0].source.type | journal |
| locations[0].source.is_oa | False |
| locations[0].source.issn_l | 1549-9596 |
| locations[0].source.is_core | True |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | Journal of Chemical Information and Modeling |
| locations[0].source.host_organization | https://openalex.org/P4310320006 |
| locations[0].source.host_organization_name | American Chemical Society |
| locations[0].source.host_organization_lineage | https://openalex.org/P4310320006 |
| locations[0].source.host_organization_lineage_names | American Chemical Society |
| locations[0].license | cc-by |
| locations[0].pdf_url | https://pubs.acs.org/doi/pdf/10.1021/acs.jcim.4c00634 |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | Journal of Chemical Information and Modeling |
| locations[0].landing_page_url | https://doi.org/10.1021/acs.jcim.4c00634 |
| locations[1].id | pmid:38843070 |
| locations[1].is_oa | False |
| locations[1].source.id | https://openalex.org/S4306525036 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | PubMed |
| locations[1].source.host_organization | https://openalex.org/I1299303238 |
| locations[1].source.host_organization_name | National Institutes of Health |
| locations[1].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[1].license | |
| locations[1].pdf_url | |
| locations[1].version | publishedVersion |
| locations[1].raw_type | |
| locations[1].license_id | |
| locations[1].is_accepted | True |
| locations[1].is_published | True |
| locations[1].raw_source_name | Journal of chemical information and modeling |
| locations[1].landing_page_url | https://pubmed.ncbi.nlm.nih.gov/38843070 |
| locations[2].id | pmh:oai:escholarship.org:ark:/13030/qt99t2x783 |
| locations[2].is_oa | True |
| locations[2].source | |
| locations[2].license | cc-by |
| locations[2].pdf_url | |
| locations[2].version | submittedVersion |
| locations[2].raw_type | article |
| locations[2].license_id | https://openalex.org/licenses/cc-by |
| locations[2].is_accepted | False |
| locations[2].is_published | False |
| locations[2].raw_source_name | Journal of chemical information and computer sciences, vol 64, iss 24 |
| locations[2].landing_page_url | https://escholarship.org/uc/item/99t2x783 |
| locations[3].id | pmh:oai:pubmedcentral.nih.gov:11683870 |
| locations[3].is_oa | True |
| locations[3].source.id | https://openalex.org/S2764455111 |
| locations[3].source.issn | |
| locations[3].source.type | repository |
| locations[3].source.is_oa | False |
| locations[3].source.issn_l | |
| locations[3].source.is_core | False |
| locations[3].source.is_in_doaj | False |
| locations[3].source.display_name | PubMed Central |
| locations[3].source.host_organization | https://openalex.org/I1299303238 |
| locations[3].source.host_organization_name | National Institutes of Health |
| locations[3].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[3].license | cc-by |
| locations[3].pdf_url | |
| locations[3].version | submittedVersion |
| locations[3].raw_type | Text |
| locations[3].license_id | https://openalex.org/licenses/cc-by |
| locations[3].is_accepted | False |
| locations[3].is_published | False |
| locations[3].raw_source_name | J Chem Inf Model |
| locations[3].landing_page_url | https://www.ncbi.nlm.nih.gov/pmc/articles/11683870 |
| indexed_in | crossref, pubmed |
| authorships[0].author.id | https://openalex.org/A5100428251 |
| authorships[0].author.orcid | https://orcid.org/0000-0002-4727-1786 |
| authorships[0].author.display_name | Jie Li |
| authorships[0].countries | US |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I95457486 |
| authorships[0].affiliations[0].raw_affiliation_string | Pitzer Center for Theoretical Chemistry, Department of Chemistry, University of California, Berkeley, California 94720, United States |
| authorships[0].institutions[0].id | https://openalex.org/I95457486 |
| authorships[0].institutions[0].ror | https://ror.org/01an7q238 |
| authorships[0].institutions[0].type | education |
| authorships[0].institutions[0].lineage | https://openalex.org/I95457486 |
| authorships[0].institutions[0].country_code | US |
| authorships[0].institutions[0].display_name | University of California, Berkeley |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Jie Li |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Pitzer Center for Theoretical Chemistry, Department of Chemistry, University of California, Berkeley, California 94720, United States |
| authorships[1].author.id | https://openalex.org/A5062154585 |
| authorships[1].author.orcid | https://orcid.org/0009-0003-2782-3998 |
| authorships[1].author.display_name | Oufan Zhang |
| authorships[1].countries | US |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I95457486 |
| authorships[1].affiliations[0].raw_affiliation_string | Pitzer Center for Theoretical Chemistry, Department of Chemistry, University of California, Berkeley, California 94720, United States |
| authorships[1].institutions[0].id | https://openalex.org/I95457486 |
| authorships[1].institutions[0].ror | https://ror.org/01an7q238 |
| authorships[1].institutions[0].type | education |
| authorships[1].institutions[0].lineage | https://openalex.org/I95457486 |
| authorships[1].institutions[0].country_code | US |
| authorships[1].institutions[0].display_name | University of California, Berkeley |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Oufan Zhang |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | Pitzer Center for Theoretical Chemistry, Department of Chemistry, University of California, Berkeley, California 94720, United States |
| authorships[2].author.id | https://openalex.org/A5060511972 |
| authorships[2].author.orcid | https://orcid.org/0000-0001-6472-1665 |
| authorships[2].author.display_name | Kunyang Sun |
| authorships[2].countries | US |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I95457486 |
| authorships[2].affiliations[0].raw_affiliation_string | Pitzer Center for Theoretical Chemistry, Department of Chemistry, University of California, Berkeley, California 94720, United States |
| authorships[2].institutions[0].id | https://openalex.org/I95457486 |
| authorships[2].institutions[0].ror | https://ror.org/01an7q238 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I95457486 |
| authorships[2].institutions[0].country_code | US |
| authorships[2].institutions[0].display_name | University of California, Berkeley |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Kunyang Sun |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | Pitzer Center for Theoretical Chemistry, Department of Chemistry, University of California, Berkeley, California 94720, United States |
| authorships[3].author.id | https://openalex.org/A5101958743 |
| authorships[3].author.orcid | https://orcid.org/0000-0002-1706-3791 |
| authorships[3].author.display_name | Yingze Wang |
| authorships[3].countries | US |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I95457486 |
| authorships[3].affiliations[0].raw_affiliation_string | Pitzer Center for Theoretical Chemistry, Department of Chemistry, University of California, Berkeley, California 94720, United States |
| authorships[3].institutions[0].id | https://openalex.org/I95457486 |
| authorships[3].institutions[0].ror | https://ror.org/01an7q238 |
| authorships[3].institutions[0].type | education |
| authorships[3].institutions[0].lineage | https://openalex.org/I95457486 |
| authorships[3].institutions[0].country_code | US |
| authorships[3].institutions[0].display_name | University of California, Berkeley |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Yingze Wang |
| authorships[3].is_corresponding | False |
| authorships[3].raw_affiliation_strings | Pitzer Center for Theoretical Chemistry, Department of Chemistry, University of California, Berkeley, California 94720, United States |
| authorships[4].author.id | https://openalex.org/A5019347904 |
| authorships[4].author.orcid | https://orcid.org/0000-0001-7623-4012 |
| authorships[4].author.display_name | Xingyi Guan |
| authorships[4].countries | US |
| authorships[4].affiliations[0].institution_ids | https://openalex.org/I95457486 |
| authorships[4].affiliations[0].raw_affiliation_string | Pitzer Center for Theoretical Chemistry, Department of Chemistry, University of California, Berkeley, California 94720, United States |
| authorships[4].institutions[0].id | https://openalex.org/I95457486 |
| authorships[4].institutions[0].ror | https://ror.org/01an7q238 |
| authorships[4].institutions[0].type | education |
| authorships[4].institutions[0].lineage | https://openalex.org/I95457486 |
| authorships[4].institutions[0].country_code | US |
| authorships[4].institutions[0].display_name | University of California, Berkeley |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Xingyi Guan |
| authorships[4].is_corresponding | False |
| authorships[4].raw_affiliation_strings | Pitzer Center for Theoretical Chemistry, Department of Chemistry, University of California, Berkeley, California 94720, United States |
| authorships[5].author.id | https://openalex.org/A5112984665 |
| authorships[5].author.orcid | |
| authorships[5].author.display_name | Dorian Bagni |
| authorships[5].countries | US |
| authorships[5].affiliations[0].institution_ids | https://openalex.org/I95457486 |
| authorships[5].affiliations[0].raw_affiliation_string | Pitzer Center for Theoretical Chemistry, Department of Chemistry, University of California, Berkeley, California 94720, United States |
| authorships[5].institutions[0].id | https://openalex.org/I95457486 |
| authorships[5].institutions[0].ror | https://ror.org/01an7q238 |
| authorships[5].institutions[0].type | education |
| authorships[5].institutions[0].lineage | https://openalex.org/I95457486 |
| authorships[5].institutions[0].country_code | US |
| authorships[5].institutions[0].display_name | University of California, Berkeley |
| authorships[5].author_position | middle |
| authorships[5].raw_author_name | Dorian Bagni |
| authorships[5].is_corresponding | False |
| authorships[5].raw_affiliation_strings | Pitzer Center for Theoretical Chemistry, Department of Chemistry, University of California, Berkeley, California 94720, United States |
| authorships[6].author.id | https://openalex.org/A5000952998 |
| authorships[6].author.orcid | https://orcid.org/0000-0002-3779-2246 |
| authorships[6].author.display_name | Mojtaba Haghighatlari |
| authorships[6].countries | US |
| authorships[6].affiliations[0].institution_ids | https://openalex.org/I95457486 |
| authorships[6].affiliations[0].raw_affiliation_string | Pitzer Center for Theoretical Chemistry, Department of Chemistry, University of California, Berkeley, California 94720, United States |
| authorships[6].institutions[0].id | https://openalex.org/I95457486 |
| authorships[6].institutions[0].ror | https://ror.org/01an7q238 |
| authorships[6].institutions[0].type | education |
| authorships[6].institutions[0].lineage | https://openalex.org/I95457486 |
| authorships[6].institutions[0].country_code | US |
| authorships[6].institutions[0].display_name | University of California, Berkeley |
| authorships[6].author_position | middle |
| authorships[6].raw_author_name | Mojtaba Haghighatlari |
| authorships[6].is_corresponding | False |
| authorships[6].raw_affiliation_strings | Pitzer Center for Theoretical Chemistry, Department of Chemistry, University of California, Berkeley, California 94720, United States |
| authorships[7].author.id | https://openalex.org/A5080498765 |
| authorships[7].author.orcid | https://orcid.org/0000-0002-5469-9035 |
| authorships[7].author.display_name | Fiona L. Kearns |
| authorships[7].countries | US |
| authorships[7].affiliations[0].institution_ids | https://openalex.org/I36258959 |
| authorships[7].affiliations[0].raw_affiliation_string | Department of Chemistry and Biochemistry, University of California, San Diego, La Jolla, California 92093, United States |
| authorships[7].institutions[0].id | https://openalex.org/I36258959 |
| authorships[7].institutions[0].ror | https://ror.org/0168r3w48 |
| authorships[7].institutions[0].type | education |
| authorships[7].institutions[0].lineage | https://openalex.org/I36258959 |
| authorships[7].institutions[0].country_code | US |
| authorships[7].institutions[0].display_name | University of California, San Diego |
| authorships[7].author_position | middle |
| authorships[7].raw_author_name | Fiona L. Kearns |
| authorships[7].is_corresponding | False |
| authorships[7].raw_affiliation_strings | Department of Chemistry and Biochemistry, University of California, San Diego, La Jolla, California 92093, United States |
| authorships[8].author.id | https://openalex.org/A5014201274 |
| authorships[8].author.orcid | https://orcid.org/0000-0001-8158-5116 |
| authorships[8].author.display_name | conor parks |
| authorships[8].countries | US |
| authorships[8].affiliations[0].institution_ids | https://openalex.org/I36258959 |
| authorships[8].affiliations[0].raw_affiliation_string | Department of Chemistry and Biochemistry, University of California, San Diego, La Jolla, California 92093, United States |
| authorships[8].institutions[0].id | https://openalex.org/I36258959 |
| authorships[8].institutions[0].ror | https://ror.org/0168r3w48 |
| authorships[8].institutions[0].type | education |
| authorships[8].institutions[0].lineage | https://openalex.org/I36258959 |
| authorships[8].institutions[0].country_code | US |
| authorships[8].institutions[0].display_name | University of California, San Diego |
| authorships[8].author_position | middle |
| authorships[8].raw_author_name | Conor Parks |
| authorships[8].is_corresponding | False |
| authorships[8].raw_affiliation_strings | Department of Chemistry and Biochemistry, University of California, San Diego, La Jolla, California 92093, United States |
| authorships[9].author.id | https://openalex.org/A5081468273 |
| authorships[9].author.orcid | https://orcid.org/0000-0002-9275-9553 |
| authorships[9].author.display_name | Rommie E. Amaro |
| authorships[9].countries | US |
| authorships[9].affiliations[0].institution_ids | https://openalex.org/I36258959 |
| authorships[9].affiliations[0].raw_affiliation_string | Department of Chemistry and Biochemistry, University of California, San Diego, La Jolla, California 92093, United States |
| authorships[9].institutions[0].id | https://openalex.org/I36258959 |
| authorships[9].institutions[0].ror | https://ror.org/0168r3w48 |
| authorships[9].institutions[0].type | education |
| authorships[9].institutions[0].lineage | https://openalex.org/I36258959 |
| authorships[9].institutions[0].country_code | US |
| authorships[9].institutions[0].display_name | University of California, San Diego |
| authorships[9].author_position | middle |
| authorships[9].raw_author_name | Rommie E. Amaro |
| authorships[9].is_corresponding | False |
| authorships[9].raw_affiliation_strings | Department of Chemistry and Biochemistry, University of California, San Diego, La Jolla, California 92093, United States |
| authorships[10].author.id | https://openalex.org/A5056814590 |
| authorships[10].author.orcid | https://orcid.org/0000-0003-0025-8987 |
| authorships[10].author.display_name | Teresa Head‐Gordon |
| authorships[10].countries | US |
| authorships[10].affiliations[0].institution_ids | https://openalex.org/I95457486 |
| authorships[10].affiliations[0].raw_affiliation_string | Departments of Bioengineering and Chemical and Biomolecular Engineering, University of California, Berkeley, California 94720, United States |
| authorships[10].affiliations[1].institution_ids | https://openalex.org/I95457486 |
| authorships[10].affiliations[1].raw_affiliation_string | Pitzer Center for Theoretical Chemistry, Department of Chemistry, University of California, Berkeley, California 94720, United States |
| authorships[10].institutions[0].id | https://openalex.org/I95457486 |
| authorships[10].institutions[0].ror | https://ror.org/01an7q238 |
| authorships[10].institutions[0].type | education |
| authorships[10].institutions[0].lineage | https://openalex.org/I95457486 |
| authorships[10].institutions[0].country_code | US |
| authorships[10].institutions[0].display_name | University of California, Berkeley |
| authorships[10].author_position | last |
| authorships[10].raw_author_name | Teresa Head-Gordon |
| authorships[10].is_corresponding | True |
| authorships[10].raw_affiliation_strings | Departments of Bioengineering and Chemical and Biomolecular Engineering, University of California, Berkeley, California 94720, United States, Pitzer Center for Theoretical Chemistry, Department of Chemistry, University of California, Berkeley, California 94720, United States |
| has_content.pdf | True |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://pubs.acs.org/doi/pdf/10.1021/acs.jcim.4c00634 |
| open_access.oa_status | hybrid |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Mining for Potent Inhibitors through Artificial Intelligence and Physics: A Unified Methodology for Ligand Based and Structure Based Drug Design |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T10211 |
| primary_topic.field.id | https://openalex.org/fields/17 |
| primary_topic.field.display_name | Computer Science |
| primary_topic.score | 0.9998999834060669 |
| primary_topic.domain.id | https://openalex.org/domains/3 |
| primary_topic.domain.display_name | Physical Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1703 |
| primary_topic.subfield.display_name | Computational Theory and Mathematics |
| primary_topic.display_name | Computational Drug Discovery Methods |
| related_works | https://openalex.org/W2089188080, https://openalex.org/W3134751040, https://openalex.org/W4280640169, https://openalex.org/W2058674645, https://openalex.org/W2766845742, https://openalex.org/W1594539819, https://openalex.org/W4224235344, https://openalex.org/W2007257620, https://openalex.org/W2130867670, https://openalex.org/W2336040088 |
| cited_by_count | 4 |
| counts_by_year[0].year | 2025 |
| counts_by_year[0].cited_by_count | 3 |
| counts_by_year[1].year | 2024 |
| counts_by_year[1].cited_by_count | 1 |
| locations_count | 4 |
| best_oa_location.id | doi:10.1021/acs.jcim.4c00634 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S167262187 |
| best_oa_location.source.issn | 1549-9596, 1549-960X |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | False |
| best_oa_location.source.issn_l | 1549-9596 |
| best_oa_location.source.is_core | True |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | Journal of Chemical Information and Modeling |
| best_oa_location.source.host_organization | https://openalex.org/P4310320006 |
| best_oa_location.source.host_organization_name | American Chemical Society |
| best_oa_location.source.host_organization_lineage | https://openalex.org/P4310320006 |
| best_oa_location.source.host_organization_lineage_names | American Chemical Society |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | https://pubs.acs.org/doi/pdf/10.1021/acs.jcim.4c00634 |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | journal-article |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | Journal of Chemical Information and Modeling |
| best_oa_location.landing_page_url | https://doi.org/10.1021/acs.jcim.4c00634 |
| primary_location.id | doi:10.1021/acs.jcim.4c00634 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S167262187 |
| primary_location.source.issn | 1549-9596, 1549-960X |
| primary_location.source.type | journal |
| primary_location.source.is_oa | False |
| primary_location.source.issn_l | 1549-9596 |
| primary_location.source.is_core | True |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | Journal of Chemical Information and Modeling |
| primary_location.source.host_organization | https://openalex.org/P4310320006 |
| primary_location.source.host_organization_name | American Chemical Society |
| primary_location.source.host_organization_lineage | https://openalex.org/P4310320006 |
| primary_location.source.host_organization_lineage_names | American Chemical Society |
| primary_location.license | cc-by |
| primary_location.pdf_url | https://pubs.acs.org/doi/pdf/10.1021/acs.jcim.4c00634 |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | Journal of Chemical Information and Modeling |
| primary_location.landing_page_url | https://doi.org/10.1021/acs.jcim.4c00634 |
| publication_date | 2024-06-06 |
| publication_year | 2024 |
| referenced_works | https://openalex.org/W2901719664, https://openalex.org/W2948831670, https://openalex.org/W2152630148, https://openalex.org/W2798017727, https://openalex.org/W2096541451, https://openalex.org/W3097145107, https://openalex.org/W1757990252, https://openalex.org/W4235946121, https://openalex.org/W2023818227, https://openalex.org/W2005481380, https://openalex.org/W2065263819, https://openalex.org/W2883583109, https://openalex.org/W2786565076, https://openalex.org/W2610148085, https://openalex.org/W2773987374, https://openalex.org/W2971690404, https://openalex.org/W4206579729, https://openalex.org/W3090171308, https://openalex.org/W3130227682, https://openalex.org/W4284898351, https://openalex.org/W4381470101, https://openalex.org/W3111168552, https://openalex.org/W4303431765, https://openalex.org/W4386498912, https://openalex.org/W4307416138, https://openalex.org/W3009321976, https://openalex.org/W2134967712, https://openalex.org/W2069506489, https://openalex.org/W2332896222, https://openalex.org/W3015608194, https://openalex.org/W3130155181, https://openalex.org/W3111988956, https://openalex.org/W2066273100, https://openalex.org/W2113364934, https://openalex.org/W2189116376, https://openalex.org/W4280550952, https://openalex.org/W2736601468, https://openalex.org/W2383406194, https://openalex.org/W2034549041, https://openalex.org/W2022476850, https://openalex.org/W2038287971, https://openalex.org/W2025816743, https://openalex.org/W4248107770, https://openalex.org/W2593436234, https://openalex.org/W2052666656, https://openalex.org/W4200139236, https://openalex.org/W3013426051, https://openalex.org/W3120469917, https://openalex.org/W2900694120, https://openalex.org/W2887447356, https://openalex.org/W3013468691, https://openalex.org/W4381996511, https://openalex.org/W2950124390, https://openalex.org/W3027630905, https://openalex.org/W4225286967, https://openalex.org/W4211050935, https://openalex.org/W2102469876, https://openalex.org/W4361305322, https://openalex.org/W2086161653, https://openalex.org/W3006436762, https://openalex.org/W2964121744, https://openalex.org/W2963587345, https://openalex.org/W4241811150, https://openalex.org/W3100751385, https://openalex.org/W2990138404, https://openalex.org/W3045928028, https://openalex.org/W3098350627 |
| referenced_works_count | 67 |
| abstract_inverted_index.a | 4, 9, 18, 28, 96, 127, 160, 174 |
| abstract_inverted_index.3D | 48 |
| abstract_inverted_index.We | 168 |
| abstract_inverted_index.as | 92 |
| abstract_inverted_index.be | 207 |
| abstract_inverted_index.by | 41 |
| abstract_inverted_index.in | 85, 116, 126, 141 |
| abstract_inverted_index.is | 8, 124 |
| abstract_inverted_index.of | 3, 75, 78, 108, 159, 176, 201, 214, 221 |
| abstract_inverted_index.on | 198 |
| abstract_inverted_index.or | 217 |
| abstract_inverted_index.to | 21, 95, 164, 172, 184 |
| abstract_inverted_index.we | 26, 81 |
| abstract_inverted_index.The | 121 |
| abstract_inverted_index.and | 11, 64, 106, 144, 157, 190 |
| abstract_inverted_index.any | 222 |
| abstract_inverted_index.can | 206 |
| abstract_inverted_index.for | 38, 62, 88, 150, 178, 210 |
| abstract_inverted_index.new | 5, 89 |
| abstract_inverted_index.our | 194 |
| abstract_inverted_index.out | 132 |
| abstract_inverted_index.set | 175 |
| abstract_inverted_index.the | 1, 73, 117, 179, 187, 199, 202, 211 |
| abstract_inverted_index.use | 74 |
| abstract_inverted_index.Here | 25 |
| abstract_inverted_index.also | 169 |
| abstract_inverted_index.deep | 43 |
| abstract_inverted_index.drug | 6, 23 |
| abstract_inverted_index.from | 101 |
| abstract_inverted_index.have | 82 |
| abstract_inverted_index.like | 183 |
| abstract_inverted_index.only | 197 |
| abstract_inverted_index.poor | 145 |
| abstract_inverted_index.pose | 166 |
| abstract_inverted_index.rule | 137 |
| abstract_inverted_index.such | 91 |
| abstract_inverted_index.task | 13 |
| abstract_inverted_index.that | 14, 33, 110, 130 |
| abstract_inverted_index.they | 181 |
| abstract_inverted_index.with | 46, 67, 113, 148 |
| abstract_inverted_index.Vina, | 53 |
| abstract_inverted_index.allow | 170 |
| abstract_inverted_index.bound | 104 |
| abstract_inverted_index.grown | 100 |
| abstract_inverted_index.known | 102 |
| abstract_inverted_index.makes | 15 |
| abstract_inverted_index.novel | 35 |
| abstract_inverted_index.other | 153, 215 |
| abstract_inverted_index.rapid | 22 |
| abstract_inverted_index.rules | 177 |
| abstract_inverted_index.shape | 63 |
| abstract_inverted_index.site. | 120 |
| abstract_inverted_index.small | 218 |
| abstract_inverted_index.tasks | 87 |
| abstract_inverted_index.time- | 10 |
| abstract_inverted_index.types | 77 |
| abstract_inverted_index.users | 171 |
| abstract_inverted_index.using | 51 |
| abstract_inverted_index.vital | 19 |
| abstract_inverted_index.while | 59 |
| abstract_inverted_index.would | 182 |
| abstract_inverted_index.active | 69, 119 |
| abstract_inverted_index.define | 173 |
| abstract_inverted_index.during | 186 |
| abstract_inverted_index.easily | 208 |
| abstract_inverted_index.future | 212 |
| abstract_inverted_index.iMiner | 122, 205 |
| abstract_inverted_index.relies | 196 |
| abstract_inverted_index.reward | 79 |
| abstract_inverted_index.sites. | 70 |
| abstract_inverted_index.steps. | 192 |
| abstract_inverted_index.target | 39, 68, 97, 114, 203, 223 |
| abstract_inverted_index.Because | 193 |
| abstract_inverted_index.adapted | 209 |
| abstract_inverted_index.against | 152 |
| abstract_inverted_index.develop | 27 |
| abstract_inverted_index.docking | 50, 154 |
| abstract_inverted_index.enforce | 111 |
| abstract_inverted_index.exclude | 185 |
| abstract_inverted_index.filters | 131 |
| abstract_inverted_index.iMiner, | 32 |
| abstract_inverted_index.ligand, | 98 |
| abstract_inverted_index.machine | 29 |
| abstract_inverted_index.measure | 165 |
| abstract_inverted_index.novelty | 58, 84 |
| abstract_inverted_index.options | 149 |
| abstract_inverted_index.process | 189 |
| abstract_inverted_index.protein | 103, 118 |
| abstract_inverted_index.scoring | 155 |
| abstract_inverted_index.thereby | 54 |
| abstract_inverted_index.through | 72 |
| abstract_inverted_index.various | 76 |
| abstract_inverted_index.AutoDock | 52 |
| abstract_inverted_index.Lipinski | 136 |
| abstract_inverted_index.approach | 20, 195 |
| abstract_inverted_index.chemical | 57, 93 |
| abstract_inverted_index.creating | 56 |
| abstract_inverted_index.creation | 107 |
| abstract_inverted_index.dynamics | 162 |
| abstract_inverted_index.embedded | 125 |
| abstract_inverted_index.learning | 30, 45 |
| abstract_inverted_index.molecule | 7, 219 |
| abstract_inverted_index.protein, | 204 |
| abstract_inverted_index.protein. | 224 |
| abstract_inverted_index.proteins | 40 |
| abstract_inverted_index.residues | 115 |
| abstract_inverted_index.training | 188 |
| abstract_inverted_index.uncommon | 139 |
| abstract_inverted_index.workflow | 129 |
| abstract_inverted_index.Moreover, | 71 |
| abstract_inverted_index.Pan-assay | 133 |
| abstract_inverted_index.algorithm | 123 |
| abstract_inverted_index.combining | 42 |
| abstract_inverted_index.composite | 128 |
| abstract_inverted_index.functions | 156 |
| abstract_inverted_index.generates | 34 |
| abstract_inverted_index.inhibitor | 36 |
| abstract_inverted_index.medicinal | 142 |
| abstract_inverted_index.molecular | 49, 65, 161 |
| abstract_inverted_index.molecules | 37, 61, 90, 99, 109 |
| abstract_inverted_index.real-time | 47 |
| abstract_inverted_index.structure | 200 |
| abstract_inverted_index.synthetic | 146 |
| abstract_inverted_index.viability | 2 |
| abstract_inverted_index.algorithm, | 31 |
| abstract_inverted_index.automation | 158 |
| abstract_inverted_index.chemistry, | 143 |
| abstract_inverted_index.compounds, | 135 |
| abstract_inverted_index.discovery. | 24 |
| abstract_inverted_index.fragments, | 105 |
| abstract_inverted_index.functions, | 80 |
| abstract_inverted_index.generative | 86 |
| abstract_inverted_index.inhibitors | 216 |
| abstract_inverted_index.introduced | 83 |
| abstract_inverted_index.similarity | 94 |
| abstract_inverted_index.simulation | 163 |
| abstract_inverted_index.stability. | 167 |
| abstract_inverted_index.structures | 140, 180 |
| abstract_inverted_index.Determining | 0 |
| abstract_inverted_index.assessments | 17 |
| abstract_inverted_index.development | 213 |
| abstract_inverted_index.violations, | 138 |
| abstract_inverted_index.constraining | 60 |
| abstract_inverted_index.interactions | 112 |
| abstract_inverted_index.interference | 134 |
| abstract_inverted_index.therapeutics | 220 |
| abstract_inverted_index.accessibility | 147 |
| abstract_inverted_index.compatibility | 66 |
| abstract_inverted_index.postfiltering | 191 |
| abstract_inverted_index.reinforcement | 44 |
| abstract_inverted_index.computer-aided | 16 |
| abstract_inverted_index.simultaneously | 55 |
| abstract_inverted_index.cross-validation | 151 |
| abstract_inverted_index.resource-intensive | 12 |
| cited_by_percentile_year.max | 97 |
| cited_by_percentile_year.min | 90 |
| corresponding_author_ids | https://openalex.org/A5056814590 |
| countries_distinct_count | 1 |
| institutions_distinct_count | 11 |
| corresponding_institution_ids | https://openalex.org/I95457486 |
| citation_normalized_percentile.value | 0.87165328 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | True |