Mixing genome annotation methods in a comparative analysis inflates the apparent number of lineage-specific genes Article Swipe
 YOU?
·
· 2022
· Open Access
·
· DOI: https://doi.org/10.1101/2022.01.13.476251
YOU?
·
· 2022
· Open Access
·
· DOI: https://doi.org/10.1101/2022.01.13.476251
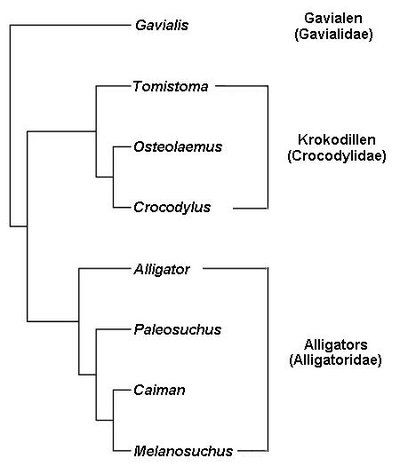
Summary Comparisons of genomes of different species are used to identify lineage-specific genes, those genes that appear unique to one species or clade. Lineage-specific genes are often thought to represent genetic novelty that underlies unique adaptations. Identification of these genes depends not only on genome sequences, but also on inferred gene annotations. Comparative analyses typically use available genomes that have been annotated using different methods, increasing the risk that orthologous DNA sequences may be erroneously annotated as a gene in one species but not another, appearing lineage-specific as a result. To evaluate the impact of such “annotation heterogeneity,” we identified four clades of species with sequenced genomes with more than one publicly available gene annotation, allowing us to compare the number of lineage-specific genes inferred when differing annotation methods are used to those resulting when annotation method is uniform across the clade. In these case studies, annotation heterogeneity increases the apparent number of lineage-specific genes by up to 15-fold, suggesting that annotation heterogeneity is a substantial source of potential artifact.
Related Topics
- Type
- preprint
- Language
- en
- Landing Page
- https://doi.org/10.1101/2022.01.13.476251
- https://www.biorxiv.org/content/biorxiv/early/2022/01/15/2022.01.13.476251.full.pdf
- OA Status
- green
- Cited By
- 9
- References
- 36
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4205528209
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4205528209Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1101/2022.01.13.476251Digital Object Identifier
- Title
-
Mixing genome annotation methods in a comparative analysis inflates the apparent number of lineage-specific genesWork title
- Type
-
preprintOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2022Year of publication
- Publication date
-
2022-01-15Full publication date if available
- Authors
-
Caroline M. Weisman, Andrew W. Murray, Sean R. EddyList of authors in order
- Landing page
-
https://doi.org/10.1101/2022.01.13.476251Publisher landing page
- PDF URL
-
https://www.biorxiv.org/content/biorxiv/early/2022/01/15/2022.01.13.476251.full.pdfDirect link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
greenOpen access status per OpenAlex
- OA URL
-
https://www.biorxiv.org/content/biorxiv/early/2022/01/15/2022.01.13.476251.full.pdfDirect OA link when available
- Concepts
-
Annotation, Genome, Biology, Lineage (genetic), Gene, Clade, Gene Annotation, Genetics, DNA sequencing, Genome project, Computational biology, Evolutionary biology, Comparative genomics, Genomics, PhylogeneticsTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
9Total citation count in OpenAlex
- Citations by year (recent)
-
2023: 4, 2022: 5Per-year citation counts (last 5 years)
- References (count)
-
36Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4205528209 |
|---|---|
| doi | https://doi.org/10.1101/2022.01.13.476251 |
| ids.doi | https://doi.org/10.1101/2022.01.13.476251 |
| ids.openalex | https://openalex.org/W4205528209 |
| fwci | 1.10762242 |
| type | preprint |
| title | Mixing genome annotation methods in a comparative analysis inflates the apparent number of lineage-specific genes |
| biblio.issue | |
| biblio.volume | |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T10015 |
| topics[0].field.id | https://openalex.org/fields/13 |
| topics[0].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[0].score | 0.9998000264167786 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1312 |
| topics[0].subfield.display_name | Molecular Biology |
| topics[0].display_name | Genomics and Phylogenetic Studies |
| topics[1].id | https://openalex.org/T10012 |
| topics[1].field.id | https://openalex.org/fields/13 |
| topics[1].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[1].score | 0.9994999766349792 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1311 |
| topics[1].subfield.display_name | Genetics |
| topics[1].display_name | Genetic diversity and population structure |
| topics[2].id | https://openalex.org/T10434 |
| topics[2].field.id | https://openalex.org/fields/11 |
| topics[2].field.display_name | Agricultural and Biological Sciences |
| topics[2].score | 0.9943000078201294 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1110 |
| topics[2].subfield.display_name | Plant Science |
| topics[2].display_name | Chromosomal and Genetic Variations |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C2776321320 |
| concepts[0].level | 2 |
| concepts[0].score | 0.8120704889297485 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q857525 |
| concepts[0].display_name | Annotation |
| concepts[1].id | https://openalex.org/C141231307 |
| concepts[1].level | 3 |
| concepts[1].score | 0.7855792045593262 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q7020 |
| concepts[1].display_name | Genome |
| concepts[2].id | https://openalex.org/C86803240 |
| concepts[2].level | 0 |
| concepts[2].score | 0.758435845375061 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[2].display_name | Biology |
| concepts[3].id | https://openalex.org/C2776817793 |
| concepts[3].level | 3 |
| concepts[3].score | 0.7554060220718384 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q6553369 |
| concepts[3].display_name | Lineage (genetic) |
| concepts[4].id | https://openalex.org/C104317684 |
| concepts[4].level | 2 |
| concepts[4].score | 0.6596043109893799 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[4].display_name | Gene |
| concepts[5].id | https://openalex.org/C44465124 |
| concepts[5].level | 4 |
| concepts[5].score | 0.6373881697654724 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q713623 |
| concepts[5].display_name | Clade |
| concepts[6].id | https://openalex.org/C2908923196 |
| concepts[6].level | 4 |
| concepts[6].score | 0.5604009032249451 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q5205742 |
| concepts[6].display_name | Gene Annotation |
| concepts[7].id | https://openalex.org/C54355233 |
| concepts[7].level | 1 |
| concepts[7].score | 0.5015144348144531 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[7].display_name | Genetics |
| concepts[8].id | https://openalex.org/C51679486 |
| concepts[8].level | 3 |
| concepts[8].score | 0.5011763572692871 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q380546 |
| concepts[8].display_name | DNA sequencing |
| concepts[9].id | https://openalex.org/C89566754 |
| concepts[9].level | 4 |
| concepts[9].score | 0.49465295672416687 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q2273828 |
| concepts[9].display_name | Genome project |
| concepts[10].id | https://openalex.org/C70721500 |
| concepts[10].level | 1 |
| concepts[10].score | 0.4857788383960724 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q177005 |
| concepts[10].display_name | Computational biology |
| concepts[11].id | https://openalex.org/C78458016 |
| concepts[11].level | 1 |
| concepts[11].score | 0.47118404507637024 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q840400 |
| concepts[11].display_name | Evolutionary biology |
| concepts[12].id | https://openalex.org/C105176652 |
| concepts[12].level | 5 |
| concepts[12].score | 0.42361193895339966 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q1147112 |
| concepts[12].display_name | Comparative genomics |
| concepts[13].id | https://openalex.org/C189206191 |
| concepts[13].level | 4 |
| concepts[13].score | 0.41212207078933716 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q222046 |
| concepts[13].display_name | Genomics |
| concepts[14].id | https://openalex.org/C90132467 |
| concepts[14].level | 3 |
| concepts[14].score | 0.3427683115005493 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q171184 |
| concepts[14].display_name | Phylogenetics |
| keywords[0].id | https://openalex.org/keywords/annotation |
| keywords[0].score | 0.8120704889297485 |
| keywords[0].display_name | Annotation |
| keywords[1].id | https://openalex.org/keywords/genome |
| keywords[1].score | 0.7855792045593262 |
| keywords[1].display_name | Genome |
| keywords[2].id | https://openalex.org/keywords/biology |
| keywords[2].score | 0.758435845375061 |
| keywords[2].display_name | Biology |
| keywords[3].id | https://openalex.org/keywords/lineage |
| keywords[3].score | 0.7554060220718384 |
| keywords[3].display_name | Lineage (genetic) |
| keywords[4].id | https://openalex.org/keywords/gene |
| keywords[4].score | 0.6596043109893799 |
| keywords[4].display_name | Gene |
| keywords[5].id | https://openalex.org/keywords/clade |
| keywords[5].score | 0.6373881697654724 |
| keywords[5].display_name | Clade |
| keywords[6].id | https://openalex.org/keywords/gene-annotation |
| keywords[6].score | 0.5604009032249451 |
| keywords[6].display_name | Gene Annotation |
| keywords[7].id | https://openalex.org/keywords/genetics |
| keywords[7].score | 0.5015144348144531 |
| keywords[7].display_name | Genetics |
| keywords[8].id | https://openalex.org/keywords/dna-sequencing |
| keywords[8].score | 0.5011763572692871 |
| keywords[8].display_name | DNA sequencing |
| keywords[9].id | https://openalex.org/keywords/genome-project |
| keywords[9].score | 0.49465295672416687 |
| keywords[9].display_name | Genome project |
| keywords[10].id | https://openalex.org/keywords/computational-biology |
| keywords[10].score | 0.4857788383960724 |
| keywords[10].display_name | Computational biology |
| keywords[11].id | https://openalex.org/keywords/evolutionary-biology |
| keywords[11].score | 0.47118404507637024 |
| keywords[11].display_name | Evolutionary biology |
| keywords[12].id | https://openalex.org/keywords/comparative-genomics |
| keywords[12].score | 0.42361193895339966 |
| keywords[12].display_name | Comparative genomics |
| keywords[13].id | https://openalex.org/keywords/genomics |
| keywords[13].score | 0.41212207078933716 |
| keywords[13].display_name | Genomics |
| keywords[14].id | https://openalex.org/keywords/phylogenetics |
| keywords[14].score | 0.3427683115005493 |
| keywords[14].display_name | Phylogenetics |
| language | en |
| locations[0].id | doi:10.1101/2022.01.13.476251 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4306402567 |
| locations[0].source.issn | |
| locations[0].source.type | repository |
| locations[0].source.is_oa | False |
| locations[0].source.issn_l | |
| locations[0].source.is_core | False |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| locations[0].source.host_organization | https://openalex.org/I2750212522 |
| locations[0].source.host_organization_name | Cold Spring Harbor Laboratory |
| locations[0].source.host_organization_lineage | https://openalex.org/I2750212522 |
| locations[0].license | cc-by-nc-nd |
| locations[0].pdf_url | https://www.biorxiv.org/content/biorxiv/early/2022/01/15/2022.01.13.476251.full.pdf |
| locations[0].version | acceptedVersion |
| locations[0].raw_type | posted-content |
| locations[0].license_id | https://openalex.org/licenses/cc-by-nc-nd |
| locations[0].is_accepted | True |
| locations[0].is_published | False |
| locations[0].raw_source_name | |
| locations[0].landing_page_url | https://doi.org/10.1101/2022.01.13.476251 |
| indexed_in | crossref |
| authorships[0].author.id | https://openalex.org/A5004241972 |
| authorships[0].author.orcid | https://orcid.org/0000-0002-1648-6776 |
| authorships[0].author.display_name | Caroline M. Weisman |
| authorships[0].countries | US |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I20089843 |
| authorships[0].affiliations[0].raw_affiliation_string | Lewis-Sigler Institute for Integrative Genomics, Princeton University, Princeton, NJ, USA |
| authorships[0].institutions[0].id | https://openalex.org/I20089843 |
| authorships[0].institutions[0].ror | https://ror.org/00hx57361 |
| authorships[0].institutions[0].type | education |
| authorships[0].institutions[0].lineage | https://openalex.org/I20089843 |
| authorships[0].institutions[0].country_code | US |
| authorships[0].institutions[0].display_name | Princeton University |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Caroline M. Weisman |
| authorships[0].is_corresponding | True |
| authorships[0].raw_affiliation_strings | Lewis-Sigler Institute for Integrative Genomics, Princeton University, Princeton, NJ, USA |
| authorships[1].author.id | https://openalex.org/A5020634412 |
| authorships[1].author.orcid | https://orcid.org/0000-0002-0868-6604 |
| authorships[1].author.display_name | Andrew W. Murray |
| authorships[1].countries | US |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I136199984 |
| authorships[1].affiliations[0].raw_affiliation_string | Department of Molecular & Cellular Biology, Harvard University, Cambridge MA, USA |
| authorships[1].institutions[0].id | https://openalex.org/I136199984 |
| authorships[1].institutions[0].ror | https://ror.org/03vek6s52 |
| authorships[1].institutions[0].type | education |
| authorships[1].institutions[0].lineage | https://openalex.org/I136199984 |
| authorships[1].institutions[0].country_code | US |
| authorships[1].institutions[0].display_name | Harvard University |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Andrew W. Murray |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | Department of Molecular & Cellular Biology, Harvard University, Cambridge MA, USA |
| authorships[2].author.id | https://openalex.org/A5020118397 |
| authorships[2].author.orcid | https://orcid.org/0000-0001-6676-4706 |
| authorships[2].author.display_name | Sean R. Eddy |
| authorships[2].countries | US |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I136199984 |
| authorships[2].affiliations[0].raw_affiliation_string | Department of Molecular & Cellular Biology, Harvard University, Cambridge MA, USA |
| authorships[2].affiliations[1].institution_ids | https://openalex.org/I136199984 |
| authorships[2].affiliations[1].raw_affiliation_string | John A. Paulson School of Engineering and Applied Sciences, Harvard University, Cambridge MA, USA |
| authorships[2].affiliations[2].institution_ids | https://openalex.org/I1344073410, https://openalex.org/I136199984 |
| authorships[2].affiliations[2].raw_affiliation_string | Howard Hughes Medical Institute, Harvard University, Cambridge MA, USA |
| authorships[2].institutions[0].id | https://openalex.org/I136199984 |
| authorships[2].institutions[0].ror | https://ror.org/03vek6s52 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I136199984 |
| authorships[2].institutions[0].country_code | US |
| authorships[2].institutions[0].display_name | Harvard University |
| authorships[2].institutions[1].id | https://openalex.org/I1344073410 |
| authorships[2].institutions[1].ror | https://ror.org/006w34k90 |
| authorships[2].institutions[1].type | facility |
| authorships[2].institutions[1].lineage | https://openalex.org/I1344073410 |
| authorships[2].institutions[1].country_code | US |
| authorships[2].institutions[1].display_name | Howard Hughes Medical Institute |
| authorships[2].author_position | last |
| authorships[2].raw_author_name | Sean R. Eddy |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | Department of Molecular & Cellular Biology, Harvard University, Cambridge MA, USA, Howard Hughes Medical Institute, Harvard University, Cambridge MA, USA, John A. Paulson School of Engineering and Applied Sciences, Harvard University, Cambridge MA, USA |
| has_content.pdf | True |
| has_content.grobid_xml | True |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://www.biorxiv.org/content/biorxiv/early/2022/01/15/2022.01.13.476251.full.pdf |
| open_access.oa_status | green |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Mixing genome annotation methods in a comparative analysis inflates the apparent number of lineage-specific genes |
| has_fulltext | True |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T10015 |
| primary_topic.field.id | https://openalex.org/fields/13 |
| primary_topic.field.display_name | Biochemistry, Genetics and Molecular Biology |
| primary_topic.score | 0.9998000264167786 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1312 |
| primary_topic.subfield.display_name | Molecular Biology |
| primary_topic.display_name | Genomics and Phylogenetic Studies |
| related_works | https://openalex.org/W2050399337, https://openalex.org/W4284881927, https://openalex.org/W4285093259, https://openalex.org/W1989890420, https://openalex.org/W2802715314, https://openalex.org/W1977692174, https://openalex.org/W2162782320, https://openalex.org/W2098218989, https://openalex.org/W2059565715, https://openalex.org/W2417531346 |
| cited_by_count | 9 |
| counts_by_year[0].year | 2023 |
| counts_by_year[0].cited_by_count | 4 |
| counts_by_year[1].year | 2022 |
| counts_by_year[1].cited_by_count | 5 |
| locations_count | 1 |
| best_oa_location.id | doi:10.1101/2022.01.13.476251 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4306402567 |
| best_oa_location.source.issn | |
| best_oa_location.source.type | repository |
| best_oa_location.source.is_oa | False |
| best_oa_location.source.issn_l | |
| best_oa_location.source.is_core | False |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| best_oa_location.source.host_organization | https://openalex.org/I2750212522 |
| best_oa_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| best_oa_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| best_oa_location.license | cc-by-nc-nd |
| best_oa_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2022/01/15/2022.01.13.476251.full.pdf |
| best_oa_location.version | acceptedVersion |
| best_oa_location.raw_type | posted-content |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by-nc-nd |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | False |
| best_oa_location.raw_source_name | |
| best_oa_location.landing_page_url | https://doi.org/10.1101/2022.01.13.476251 |
| primary_location.id | doi:10.1101/2022.01.13.476251 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4306402567 |
| primary_location.source.issn | |
| primary_location.source.type | repository |
| primary_location.source.is_oa | False |
| primary_location.source.issn_l | |
| primary_location.source.is_core | False |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| primary_location.source.host_organization | https://openalex.org/I2750212522 |
| primary_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| primary_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| primary_location.license | cc-by-nc-nd |
| primary_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2022/01/15/2022.01.13.476251.full.pdf |
| primary_location.version | acceptedVersion |
| primary_location.raw_type | posted-content |
| primary_location.license_id | https://openalex.org/licenses/cc-by-nc-nd |
| primary_location.is_accepted | True |
| primary_location.is_published | False |
| primary_location.raw_source_name | |
| primary_location.landing_page_url | https://doi.org/10.1101/2022.01.13.476251 |
| publication_date | 2022-01-15 |
| publication_year | 2022 |
| referenced_works | https://openalex.org/W2146729343, https://openalex.org/W2059223767, https://openalex.org/W2055777573, https://openalex.org/W2120507610, https://openalex.org/W2058587250, https://openalex.org/W2051418786, https://openalex.org/W2479687413, https://openalex.org/W3105122235, https://openalex.org/W2896689896, https://openalex.org/W3037321979, https://openalex.org/W2608157918, https://openalex.org/W2127673162, https://openalex.org/W2774045279, https://openalex.org/W2999484417, https://openalex.org/W2161153002, https://openalex.org/W2799953103, https://openalex.org/W2728408188, https://openalex.org/W4205247356, https://openalex.org/W2402793346, https://openalex.org/W1974342825, https://openalex.org/W2053551770, https://openalex.org/W2168292723, https://openalex.org/W2108108770, https://openalex.org/W4236236547, https://openalex.org/W3033528024, https://openalex.org/W2945454784, https://openalex.org/W2462408723, https://openalex.org/W3014506894, https://openalex.org/W3120435377, https://openalex.org/W3086082703, https://openalex.org/W2233603983, https://openalex.org/W2047348172, https://openalex.org/W2056200352, https://openalex.org/W2098675519, https://openalex.org/W3097658527, https://openalex.org/W2235483646 |
| referenced_works_count | 36 |
| abstract_inverted_index.a | 77, 88, 164 |
| abstract_inverted_index.In | 142 |
| abstract_inverted_index.To | 90 |
| abstract_inverted_index.as | 76, 87 |
| abstract_inverted_index.be | 73 |
| abstract_inverted_index.by | 155 |
| abstract_inverted_index.in | 79 |
| abstract_inverted_index.is | 137, 163 |
| abstract_inverted_index.of | 2, 4, 37, 94, 102, 121, 152, 167 |
| abstract_inverted_index.on | 43, 48 |
| abstract_inverted_index.or | 21 |
| abstract_inverted_index.to | 9, 18, 28, 117, 131, 157 |
| abstract_inverted_index.up | 156 |
| abstract_inverted_index.us | 116 |
| abstract_inverted_index.we | 98 |
| abstract_inverted_index.DNA | 70 |
| abstract_inverted_index.are | 7, 25, 129 |
| abstract_inverted_index.but | 46, 82 |
| abstract_inverted_index.may | 72 |
| abstract_inverted_index.not | 41, 83 |
| abstract_inverted_index.one | 19, 80, 110 |
| abstract_inverted_index.the | 66, 92, 119, 140, 149 |
| abstract_inverted_index.use | 55 |
| abstract_inverted_index.also | 47 |
| abstract_inverted_index.been | 60 |
| abstract_inverted_index.case | 144 |
| abstract_inverted_index.four | 100 |
| abstract_inverted_index.gene | 50, 78, 113 |
| abstract_inverted_index.have | 59 |
| abstract_inverted_index.more | 108 |
| abstract_inverted_index.only | 42 |
| abstract_inverted_index.risk | 67 |
| abstract_inverted_index.such | 95 |
| abstract_inverted_index.than | 109 |
| abstract_inverted_index.that | 15, 32, 58, 68, 160 |
| abstract_inverted_index.used | 8, 130 |
| abstract_inverted_index.when | 125, 134 |
| abstract_inverted_index.with | 104, 107 |
| abstract_inverted_index.genes | 14, 24, 39, 123, 154 |
| abstract_inverted_index.often | 26 |
| abstract_inverted_index.these | 38, 143 |
| abstract_inverted_index.those | 13, 132 |
| abstract_inverted_index.using | 62 |
| abstract_inverted_index.across | 139 |
| abstract_inverted_index.appear | 16 |
| abstract_inverted_index.clade. | 22, 141 |
| abstract_inverted_index.clades | 101 |
| abstract_inverted_index.genes, | 12 |
| abstract_inverted_index.genome | 44 |
| abstract_inverted_index.impact | 93 |
| abstract_inverted_index.method | 136 |
| abstract_inverted_index.number | 120, 151 |
| abstract_inverted_index.source | 166 |
| abstract_inverted_index.unique | 17, 34 |
| abstract_inverted_index.Summary | 0 |
| abstract_inverted_index.compare | 118 |
| abstract_inverted_index.depends | 40 |
| abstract_inverted_index.genetic | 30 |
| abstract_inverted_index.genomes | 3, 57, 106 |
| abstract_inverted_index.methods | 128 |
| abstract_inverted_index.novelty | 31 |
| abstract_inverted_index.result. | 89 |
| abstract_inverted_index.species | 6, 20, 81, 103 |
| abstract_inverted_index.thought | 27 |
| abstract_inverted_index.uniform | 138 |
| abstract_inverted_index.15-fold, | 158 |
| abstract_inverted_index.allowing | 115 |
| abstract_inverted_index.analyses | 53 |
| abstract_inverted_index.another, | 84 |
| abstract_inverted_index.apparent | 150 |
| abstract_inverted_index.evaluate | 91 |
| abstract_inverted_index.identify | 10 |
| abstract_inverted_index.inferred | 49, 124 |
| abstract_inverted_index.methods, | 64 |
| abstract_inverted_index.publicly | 111 |
| abstract_inverted_index.studies, | 145 |
| abstract_inverted_index.annotated | 61, 75 |
| abstract_inverted_index.appearing | 85 |
| abstract_inverted_index.artifact. | 169 |
| abstract_inverted_index.available | 56, 112 |
| abstract_inverted_index.different | 5, 63 |
| abstract_inverted_index.differing | 126 |
| abstract_inverted_index.increases | 148 |
| abstract_inverted_index.potential | 168 |
| abstract_inverted_index.represent | 29 |
| abstract_inverted_index.resulting | 133 |
| abstract_inverted_index.sequenced | 105 |
| abstract_inverted_index.sequences | 71 |
| abstract_inverted_index.typically | 54 |
| abstract_inverted_index.underlies | 33 |
| abstract_inverted_index.annotation | 127, 135, 146, 161 |
| abstract_inverted_index.identified | 99 |
| abstract_inverted_index.increasing | 65 |
| abstract_inverted_index.sequences, | 45 |
| abstract_inverted_index.suggesting | 159 |
| abstract_inverted_index.Comparative | 52 |
| abstract_inverted_index.Comparisons | 1 |
| abstract_inverted_index.annotation, | 114 |
| abstract_inverted_index.erroneously | 74 |
| abstract_inverted_index.orthologous | 69 |
| abstract_inverted_index.substantial | 165 |
| abstract_inverted_index.adaptations. | 35 |
| abstract_inverted_index.annotations. | 51 |
| abstract_inverted_index.heterogeneity | 147, 162 |
| abstract_inverted_index.“annotation | 96 |
| abstract_inverted_index.Identification | 36 |
| abstract_inverted_index.Lineage-specific | 23 |
| abstract_inverted_index.lineage-specific | 11, 86, 122, 153 |
| abstract_inverted_index.heterogeneity,” | 97 |
| cited_by_percentile_year.max | 98 |
| cited_by_percentile_year.min | 97 |
| corresponding_author_ids | https://openalex.org/A5004241972 |
| countries_distinct_count | 1 |
| institutions_distinct_count | 3 |
| corresponding_institution_ids | https://openalex.org/I20089843 |
| sustainable_development_goals[0].id | https://metadata.un.org/sdg/15 |
| sustainable_development_goals[0].score | 0.4099999964237213 |
| sustainable_development_goals[0].display_name | Life in Land |
| citation_normalized_percentile.value | 0.70353366 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |