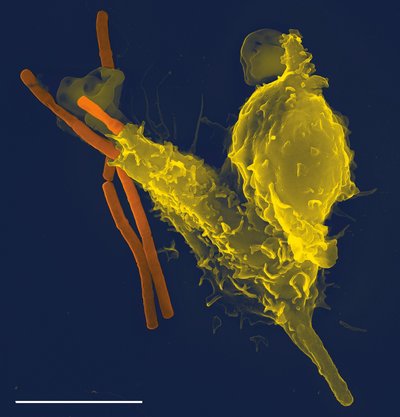
Molecular, Metabolic, and Subcellular Mapping of the Tumor Immune Microenvironment via 3D Targeted and Non-Targeted Multiplex Multi-Omics Analyses Article Swipe
 YOU?
·
· 2024
· Open Access
·
· DOI: https://doi.org/10.17615/v28r-q380
YOU?
·
· 2024
· Open Access
·
· DOI: https://doi.org/10.17615/v28r-q380
Most platforms used for the molecular reconstruction of the tumor-immune microenvironment (TIME) of a solid tumor fail to explore the spatial context of the three-dimensional (3D) space of the tumor at a single-cell resolution, and thus lack information about cell-cell or cell-extracellular matrix (ECM) interactions. To address this issue, a pipeline which integrated multiplex spatially resolved multi-omics platforms was developed to identify crosstalk signaling networks among various cell types and the ECM in the 3D TIME of two FFPE (formalin-fixed paraffin embedded) gynecologic tumor samples. These platforms include non-targeted mass spectrometry imaging (glycans, metabolites, and peptides) and Stereo-seq (spatial transcriptomics) and targeted seqIF (IHC proteomics). The spatially resolved imaging data in a two- and three-dimensional space demonstrated various cellular neighborhoods in both samples. The collection of spatially resolved analytes in a voxel (3D pixel) across serial sections of the tissue was also demonstrated. Data collected from this analytical pipeline were used to construct spatial 3D maps with single-cell resolution, which revealed cell identity, activation, and energized status. These maps will provide not only insights into the molecular basis of spatial cell heterogeneity in the TIME, but also novel predictive biomarkers and therapeutic targets, which can improve patient survival rates.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.17615/v28r-q380
- OA Status
- green
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4403710662
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4403710662Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.17615/v28r-q380Digital Object Identifier
- Title
-
Molecular, Metabolic, and Subcellular Mapping of the Tumor Immune Microenvironment via 3D Targeted and Non-Targeted Multiplex Multi-Omics AnalysesWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2024Year of publication
- Publication date
-
2024-10-23Full publication date if available
- Authors
-
Sammy Ferri‐Borgogno, Jared K. Burks, Erin H. Seeley, Trevor D. McKee, Danielle L. Stolley, Akshay Basi, Javier A. Gomez, Basant T. Gamal, Shamini Ayyadhury, Barrett C. Lawson, Melinda S. Yates, Michael J. Birrer, Karen H. Lu, Samuel C. MokList of authors in order
- Landing page
-
https://doi.org/10.17615/v28r-q380Publisher landing page
- Open access
-
YesWhether a free full text is available
- OA status
-
greenOpen access status per OpenAlex
- OA URL
-
https://doi.org/10.17615/v28r-q380Direct OA link when available
- Concepts
-
Multiplex, Computational biology, Immune system, Omics, Biology, Tumor microenvironment, Bioinformatics, GeneticsTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
0Total citation count in OpenAlex
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4403710662 |
|---|---|
| doi | https://doi.org/10.17615/v28r-q380 |
| ids.doi | https://doi.org/10.17615/v28r-q380 |
| ids.openalex | https://openalex.org/W4403710662 |
| fwci | 0.0 |
| type | article |
| title | Molecular, Metabolic, and Subcellular Mapping of the Tumor Immune Microenvironment via 3D Targeted and Non-Targeted Multiplex Multi-Omics Analyses |
| biblio.issue | |
| biblio.volume | |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T11313 |
| topics[0].field.id | https://openalex.org/fields/24 |
| topics[0].field.display_name | Immunology and Microbiology |
| topics[0].score | 0.9930999875068665 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/2403 |
| topics[0].subfield.display_name | Immunology |
| topics[0].display_name | Immune cells in cancer |
| topics[1].id | https://openalex.org/T12996 |
| topics[1].field.id | https://openalex.org/fields/13 |
| topics[1].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[1].score | 0.9872999787330627 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1305 |
| topics[1].subfield.display_name | Biotechnology |
| topics[1].display_name | Cancer Research and Treatments |
| topics[2].id | https://openalex.org/T10158 |
| topics[2].field.id | https://openalex.org/fields/27 |
| topics[2].field.display_name | Medicine |
| topics[2].score | 0.9847000241279602 |
| topics[2].domain.id | https://openalex.org/domains/4 |
| topics[2].domain.display_name | Health Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/2730 |
| topics[2].subfield.display_name | Oncology |
| topics[2].display_name | Cancer Immunotherapy and Biomarkers |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C2781188995 |
| concepts[0].level | 2 |
| concepts[0].score | 0.87071293592453 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q6934982 |
| concepts[0].display_name | Multiplex |
| concepts[1].id | https://openalex.org/C70721500 |
| concepts[1].level | 1 |
| concepts[1].score | 0.6675289869308472 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q177005 |
| concepts[1].display_name | Computational biology |
| concepts[2].id | https://openalex.org/C8891405 |
| concepts[2].level | 2 |
| concepts[2].score | 0.5838205218315125 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q1059 |
| concepts[2].display_name | Immune system |
| concepts[3].id | https://openalex.org/C157585117 |
| concepts[3].level | 2 |
| concepts[3].score | 0.5495341420173645 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q158666 |
| concepts[3].display_name | Omics |
| concepts[4].id | https://openalex.org/C86803240 |
| concepts[4].level | 0 |
| concepts[4].score | 0.5156192183494568 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[4].display_name | Biology |
| concepts[5].id | https://openalex.org/C2776107976 |
| concepts[5].level | 3 |
| concepts[5].score | 0.42359471321105957 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q1786433 |
| concepts[5].display_name | Tumor microenvironment |
| concepts[6].id | https://openalex.org/C60644358 |
| concepts[6].level | 1 |
| concepts[6].score | 0.3070160150527954 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q128570 |
| concepts[6].display_name | Bioinformatics |
| concepts[7].id | https://openalex.org/C54355233 |
| concepts[7].level | 1 |
| concepts[7].score | 0.206889808177948 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[7].display_name | Genetics |
| keywords[0].id | https://openalex.org/keywords/multiplex |
| keywords[0].score | 0.87071293592453 |
| keywords[0].display_name | Multiplex |
| keywords[1].id | https://openalex.org/keywords/computational-biology |
| keywords[1].score | 0.6675289869308472 |
| keywords[1].display_name | Computational biology |
| keywords[2].id | https://openalex.org/keywords/immune-system |
| keywords[2].score | 0.5838205218315125 |
| keywords[2].display_name | Immune system |
| keywords[3].id | https://openalex.org/keywords/omics |
| keywords[3].score | 0.5495341420173645 |
| keywords[3].display_name | Omics |
| keywords[4].id | https://openalex.org/keywords/biology |
| keywords[4].score | 0.5156192183494568 |
| keywords[4].display_name | Biology |
| keywords[5].id | https://openalex.org/keywords/tumor-microenvironment |
| keywords[5].score | 0.42359471321105957 |
| keywords[5].display_name | Tumor microenvironment |
| keywords[6].id | https://openalex.org/keywords/bioinformatics |
| keywords[6].score | 0.3070160150527954 |
| keywords[6].display_name | Bioinformatics |
| keywords[7].id | https://openalex.org/keywords/genetics |
| keywords[7].score | 0.206889808177948 |
| keywords[7].display_name | Genetics |
| language | en |
| locations[0].id | doi:10.17615/v28r-q380 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S7407051488 |
| locations[0].source.type | repository |
| locations[0].source.is_oa | False |
| locations[0].source.issn_l | |
| locations[0].source.is_core | False |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | UNC Libraries |
| locations[0].source.host_organization | |
| locations[0].source.host_organization_name | |
| locations[0].license | |
| locations[0].pdf_url | |
| locations[0].version | |
| locations[0].raw_type | article-journal |
| locations[0].license_id | |
| locations[0].is_accepted | False |
| locations[0].is_published | |
| locations[0].raw_source_name | |
| locations[0].landing_page_url | https://doi.org/10.17615/v28r-q380 |
| indexed_in | datacite |
| authorships[0].author.id | https://openalex.org/A5060541999 |
| authorships[0].author.orcid | https://orcid.org/0000-0002-8270-8963 |
| authorships[0].author.display_name | Sammy Ferri‐Borgogno |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Ferri-Borgogno, Sammy |
| authorships[0].is_corresponding | False |
| authorships[1].author.id | https://openalex.org/A5008665039 |
| authorships[1].author.orcid | https://orcid.org/0000-0002-6173-9074 |
| authorships[1].author.display_name | Jared K. Burks |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Burks, Jared K. |
| authorships[1].is_corresponding | False |
| authorships[2].author.id | https://openalex.org/A5086658186 |
| authorships[2].author.orcid | https://orcid.org/0000-0002-8000-5754 |
| authorships[2].author.display_name | Erin H. Seeley |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Seeley, Erin H. |
| authorships[2].is_corresponding | False |
| authorships[3].author.id | https://openalex.org/A5090391995 |
| authorships[3].author.orcid | https://orcid.org/0000-0002-2195-6146 |
| authorships[3].author.display_name | Trevor D. McKee |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | McKee, Trevor D. |
| authorships[3].is_corresponding | False |
| authorships[4].author.id | https://openalex.org/A5066645015 |
| authorships[4].author.orcid | https://orcid.org/0000-0003-0479-7902 |
| authorships[4].author.display_name | Danielle L. Stolley |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Stolley, Danielle L. |
| authorships[4].is_corresponding | False |
| authorships[5].author.id | https://openalex.org/A5092110230 |
| authorships[5].author.orcid | |
| authorships[5].author.display_name | Akshay Basi |
| authorships[5].author_position | middle |
| authorships[5].raw_author_name | Basi, Akshay V. |
| authorships[5].is_corresponding | False |
| authorships[6].author.id | https://openalex.org/A5069469851 |
| authorships[6].author.orcid | https://orcid.org/0000-0002-6579-744X |
| authorships[6].author.display_name | Javier A. Gomez |
| authorships[6].author_position | middle |
| authorships[6].raw_author_name | Gomez, Javier A. |
| authorships[6].is_corresponding | False |
| authorships[7].author.id | https://openalex.org/A5093960572 |
| authorships[7].author.orcid | |
| authorships[7].author.display_name | Basant T. Gamal |
| authorships[7].author_position | middle |
| authorships[7].raw_author_name | Gamal, Basant T. |
| authorships[7].is_corresponding | False |
| authorships[8].author.id | https://openalex.org/A5025784364 |
| authorships[8].author.orcid | https://orcid.org/0000-0002-8717-637X |
| authorships[8].author.display_name | Shamini Ayyadhury |
| authorships[8].author_position | middle |
| authorships[8].raw_author_name | Ayyadhury, Shamini |
| authorships[8].is_corresponding | False |
| authorships[9].author.id | https://openalex.org/A5038063176 |
| authorships[9].author.orcid | https://orcid.org/0000-0003-2485-7087 |
| authorships[9].author.display_name | Barrett C. Lawson |
| authorships[9].author_position | middle |
| authorships[9].raw_author_name | Lawson, Barrett C. |
| authorships[9].is_corresponding | False |
| authorships[10].author.id | https://openalex.org/A5001744690 |
| authorships[10].author.orcid | https://orcid.org/0000-0001-9450-5517 |
| authorships[10].author.display_name | Melinda S. Yates |
| authorships[10].author_position | middle |
| authorships[10].raw_author_name | Yates, Melinda S. |
| authorships[10].is_corresponding | False |
| authorships[11].author.id | https://openalex.org/A5103220093 |
| authorships[11].author.orcid | https://orcid.org/0000-0002-3861-4521 |
| authorships[11].author.display_name | Michael J. Birrer |
| authorships[11].author_position | middle |
| authorships[11].raw_author_name | Birrer, Michael J. |
| authorships[11].is_corresponding | False |
| authorships[12].author.id | https://openalex.org/A5078692182 |
| authorships[12].author.orcid | https://orcid.org/0000-0002-5317-9927 |
| authorships[12].author.display_name | Karen H. Lu |
| authorships[12].author_position | middle |
| authorships[12].raw_author_name | Lu, Karen H. |
| authorships[12].is_corresponding | False |
| authorships[13].author.id | https://openalex.org/A5010693778 |
| authorships[13].author.orcid | https://orcid.org/0000-0001-7013-1805 |
| authorships[13].author.display_name | Samuel C. Mok |
| authorships[13].author_position | last |
| authorships[13].raw_author_name | Mok, Samuel C. |
| authorships[13].is_corresponding | False |
| has_content.pdf | False |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://doi.org/10.17615/v28r-q380 |
| open_access.oa_status | green |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Molecular, Metabolic, and Subcellular Mapping of the Tumor Immune Microenvironment via 3D Targeted and Non-Targeted Multiplex Multi-Omics Analyses |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-06T06:51:31.235846 |
| primary_topic.id | https://openalex.org/T11313 |
| primary_topic.field.id | https://openalex.org/fields/24 |
| primary_topic.field.display_name | Immunology and Microbiology |
| primary_topic.score | 0.9930999875068665 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/2403 |
| primary_topic.subfield.display_name | Immunology |
| primary_topic.display_name | Immune cells in cancer |
| related_works | https://openalex.org/W3006199032, https://openalex.org/W1963826946, https://openalex.org/W177137871, https://openalex.org/W2087421209, https://openalex.org/W2547948671, https://openalex.org/W4403685181, https://openalex.org/W4292302930, https://openalex.org/W4281783318, https://openalex.org/W3195112886, https://openalex.org/W2590543013 |
| cited_by_count | 0 |
| locations_count | 1 |
| best_oa_location.id | doi:10.17615/v28r-q380 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S7407051488 |
| best_oa_location.source.type | repository |
| best_oa_location.source.is_oa | False |
| best_oa_location.source.issn_l | |
| best_oa_location.source.is_core | False |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | UNC Libraries |
| best_oa_location.source.host_organization | |
| best_oa_location.source.host_organization_name | |
| best_oa_location.license | |
| best_oa_location.pdf_url | |
| best_oa_location.version | |
| best_oa_location.raw_type | article-journal |
| best_oa_location.license_id | |
| best_oa_location.is_accepted | False |
| best_oa_location.is_published | False |
| best_oa_location.raw_source_name | |
| best_oa_location.landing_page_url | https://doi.org/10.17615/v28r-q380 |
| primary_location.id | doi:10.17615/v28r-q380 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S7407051488 |
| primary_location.source.type | repository |
| primary_location.source.is_oa | False |
| primary_location.source.issn_l | |
| primary_location.source.is_core | False |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | UNC Libraries |
| primary_location.source.host_organization | |
| primary_location.source.host_organization_name | |
| primary_location.license | |
| primary_location.pdf_url | |
| primary_location.version | |
| primary_location.raw_type | article-journal |
| primary_location.license_id | |
| primary_location.is_accepted | False |
| primary_location.is_published | False |
| primary_location.raw_source_name | |
| primary_location.landing_page_url | https://doi.org/10.17615/v28r-q380 |
| publication_date | 2024-10-23 |
| publication_year | 2024 |
| referenced_works_count | 0 |
| abstract_inverted_index.a | 13, 31, 49, 111, 130 |
| abstract_inverted_index.3D | 74, 154 |
| abstract_inverted_index.To | 45 |
| abstract_inverted_index.at | 30 |
| abstract_inverted_index.in | 72, 110, 120, 129, 182 |
| abstract_inverted_index.of | 7, 12, 22, 27, 76, 125, 137, 178 |
| abstract_inverted_index.or | 40 |
| abstract_inverted_index.to | 17, 60, 151 |
| abstract_inverted_index.(3D | 132 |
| abstract_inverted_index.ECM | 71 |
| abstract_inverted_index.The | 105, 123 |
| abstract_inverted_index.and | 34, 69, 94, 96, 100, 113, 164, 190 |
| abstract_inverted_index.but | 185 |
| abstract_inverted_index.can | 194 |
| abstract_inverted_index.for | 3 |
| abstract_inverted_index.not | 171 |
| abstract_inverted_index.the | 4, 8, 19, 23, 28, 70, 73, 138, 175, 183 |
| abstract_inverted_index.two | 77 |
| abstract_inverted_index.was | 58, 140 |
| abstract_inverted_index.(3D) | 25 |
| abstract_inverted_index.(IHC | 103 |
| abstract_inverted_index.Data | 143 |
| abstract_inverted_index.FFPE | 78 |
| abstract_inverted_index.Most | 0 |
| abstract_inverted_index.TIME | 75 |
| abstract_inverted_index.also | 141, 186 |
| abstract_inverted_index.both | 121 |
| abstract_inverted_index.cell | 67, 161, 180 |
| abstract_inverted_index.data | 109 |
| abstract_inverted_index.fail | 16 |
| abstract_inverted_index.from | 145 |
| abstract_inverted_index.into | 174 |
| abstract_inverted_index.lack | 36 |
| abstract_inverted_index.maps | 155, 168 |
| abstract_inverted_index.mass | 89 |
| abstract_inverted_index.only | 172 |
| abstract_inverted_index.this | 47, 146 |
| abstract_inverted_index.thus | 35 |
| abstract_inverted_index.two- | 112 |
| abstract_inverted_index.used | 2, 150 |
| abstract_inverted_index.were | 149 |
| abstract_inverted_index.will | 169 |
| abstract_inverted_index.with | 156 |
| abstract_inverted_index.(ECM) | 43 |
| abstract_inverted_index.TIME, | 184 |
| abstract_inverted_index.These | 85, 167 |
| abstract_inverted_index.about | 38 |
| abstract_inverted_index.among | 65 |
| abstract_inverted_index.basis | 177 |
| abstract_inverted_index.novel | 187 |
| abstract_inverted_index.seqIF | 102 |
| abstract_inverted_index.solid | 14 |
| abstract_inverted_index.space | 26, 115 |
| abstract_inverted_index.tumor | 15, 29, 83 |
| abstract_inverted_index.types | 68 |
| abstract_inverted_index.voxel | 131 |
| abstract_inverted_index.which | 51, 159, 193 |
| abstract_inverted_index.(TIME) | 11 |
| abstract_inverted_index.across | 134 |
| abstract_inverted_index.issue, | 48 |
| abstract_inverted_index.matrix | 42 |
| abstract_inverted_index.pixel) | 133 |
| abstract_inverted_index.rates. | 198 |
| abstract_inverted_index.serial | 135 |
| abstract_inverted_index.tissue | 139 |
| abstract_inverted_index.address | 46 |
| abstract_inverted_index.context | 21 |
| abstract_inverted_index.explore | 18 |
| abstract_inverted_index.imaging | 91, 108 |
| abstract_inverted_index.improve | 195 |
| abstract_inverted_index.include | 87 |
| abstract_inverted_index.patient | 196 |
| abstract_inverted_index.provide | 170 |
| abstract_inverted_index.spatial | 20, 153, 179 |
| abstract_inverted_index.status. | 166 |
| abstract_inverted_index.various | 66, 117 |
| abstract_inverted_index.(spatial | 98 |
| abstract_inverted_index.analytes | 128 |
| abstract_inverted_index.cellular | 118 |
| abstract_inverted_index.identify | 61 |
| abstract_inverted_index.insights | 173 |
| abstract_inverted_index.networks | 64 |
| abstract_inverted_index.paraffin | 80 |
| abstract_inverted_index.pipeline | 50, 148 |
| abstract_inverted_index.resolved | 55, 107, 127 |
| abstract_inverted_index.revealed | 160 |
| abstract_inverted_index.samples. | 84, 122 |
| abstract_inverted_index.sections | 136 |
| abstract_inverted_index.survival | 197 |
| abstract_inverted_index.targeted | 101 |
| abstract_inverted_index.targets, | 192 |
| abstract_inverted_index.(glycans, | 92 |
| abstract_inverted_index.cell-cell | 39 |
| abstract_inverted_index.collected | 144 |
| abstract_inverted_index.construct | 152 |
| abstract_inverted_index.crosstalk | 62 |
| abstract_inverted_index.developed | 59 |
| abstract_inverted_index.embedded) | 81 |
| abstract_inverted_index.energized | 165 |
| abstract_inverted_index.identity, | 162 |
| abstract_inverted_index.molecular | 5, 176 |
| abstract_inverted_index.multiplex | 53 |
| abstract_inverted_index.peptides) | 95 |
| abstract_inverted_index.platforms | 1, 57, 86 |
| abstract_inverted_index.signaling | 63 |
| abstract_inverted_index.spatially | 54, 106, 126 |
| abstract_inverted_index.Stereo-seq | 97 |
| abstract_inverted_index.analytical | 147 |
| abstract_inverted_index.biomarkers | 189 |
| abstract_inverted_index.collection | 124 |
| abstract_inverted_index.integrated | 52 |
| abstract_inverted_index.predictive | 188 |
| abstract_inverted_index.activation, | 163 |
| abstract_inverted_index.gynecologic | 82 |
| abstract_inverted_index.information | 37 |
| abstract_inverted_index.multi-omics | 56 |
| abstract_inverted_index.resolution, | 33, 158 |
| abstract_inverted_index.single-cell | 32, 157 |
| abstract_inverted_index.therapeutic | 191 |
| abstract_inverted_index.demonstrated | 116 |
| abstract_inverted_index.metabolites, | 93 |
| abstract_inverted_index.non-targeted | 88 |
| abstract_inverted_index.proteomics). | 104 |
| abstract_inverted_index.spectrometry | 90 |
| abstract_inverted_index.tumor-immune | 9 |
| abstract_inverted_index.demonstrated. | 142 |
| abstract_inverted_index.heterogeneity | 181 |
| abstract_inverted_index.interactions. | 44 |
| abstract_inverted_index.neighborhoods | 119 |
| abstract_inverted_index.reconstruction | 6 |
| abstract_inverted_index.(formalin-fixed | 79 |
| abstract_inverted_index.microenvironment | 10 |
| abstract_inverted_index.transcriptomics) | 99 |
| abstract_inverted_index.three-dimensional | 24, 114 |
| abstract_inverted_index.cell-extracellular | 41 |
| cited_by_percentile_year | |
| countries_distinct_count | 0 |
| institutions_distinct_count | 14 |
| sustainable_development_goals[0].id | https://metadata.un.org/sdg/2 |
| sustainable_development_goals[0].score | 0.5 |
| sustainable_development_goals[0].display_name | Zero hunger |
| citation_normalized_percentile.value | 0.26944564 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |