Multi-ancestry genome-wide association study reveals novel genetic signals for lung function decline Article Swipe
 YOU?
·
· 2024
· Open Access
·
· DOI: https://doi.org/10.1101/2024.11.25.24317794
YOU?
·
· 2024
· Open Access
·
· DOI: https://doi.org/10.1101/2024.11.25.24317794
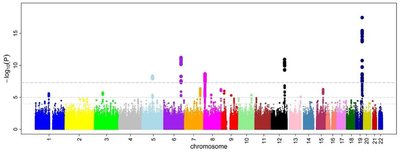
Rationale Accelerated decline in lung function contributes to the development of chronic respiratory disease. Despite evidence for a genetic component, few genetic associations with lung function decline have been identified. Objectives To evaluate genome-wide associations and putative downstream functionality of genetic variants with lung function decline in diverse general population cohorts. Methods We conducted genome-wide association study (GWAS) analyses of decline in the forced expiratory volume in the first second (FEV 1 ), forced vital capacity (FVC), and their ratio (FEV 1 /FVC) in participants across six cohorts in the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium and the UK Biobank. Genotypes were imputed to TOPMed (CHARGE cohorts) or Haplotype Reference Consortium (HRC) (UK Biobank) reference panels, and GWAS analyses used generalized estimating equation models with robust standard error. Models were stratified by cohort, ancestry, and sex, and adjusted for important lung function confounders and genotype principal components. Results were combined in cross-ancestry and ancestry-specific meta-analyses. Selected top variants were tested for replication in two independent COPD-enriched cohorts. Measurements and Main Results Our discovery analyses included 52,056 self-reported White (N=44,988), Black (N=5,788), Hispanic (N=550), and Chinese American (N=730) participants with a mean of 2.3 spirometry measurements and 8.6 years of follow-up. Functional mapping of GWAS meta-analysis results identified 361 distinct genome-wide significant (p<5E-08) variants in one or more of the FEV 1 , FVC, and FEV 1 /FVC decline phenotypes, which overlapped with previously reported genetic signals for several related pulmonary traits. Of these, 8 variants, or 20.5% of the variant set available for replication testing, were nominally associated (p<0.05) with at least one decline phenotype in COPD-enriched cohorts (White [N=4,778] and Black [N=1,118]). Using the GWAS results, gene-level analysis implicated 38 genes, including eight ( XIRP2 , GRIN2D , SATB1 , MARCHF4 , SIPA1L2 , ANO5 , H2BC10 , and FAF2 ) with consistent associations across ancestries or decline phenotypes. Annotation class analysis revealed significant enrichment of several regulatory processes for corticosteroid biosynthesis and metabolism. Drug repurposing analysis identified 43 approved compounds targeting eight of the implicated 38 genes. Conclusions Our multi-ancestry GWAS meta-analyses identified numerous genetic loci associated with lung function decline. These findings contribute knowledge to the genetic architecture of lung function decline, provide evidence for a role of endogenous corticosteroids in the etiology of lung function decline, and identify drug targets that merit further study for potential repurposing to slow lung function decline and treat lung disease.
Related Topics
- Type
- preprint
- Language
- en
- Landing Page
- https://doi.org/10.1101/2024.11.25.24317794
- OA Status
- green
- Cited By
- 1
- References
- 55
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4404788147
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4404788147Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1101/2024.11.25.24317794Digital Object Identifier
- Title
-
Multi-ancestry genome-wide association study reveals novel genetic signals for lung function declineWork title
- Type
-
preprintOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2024Year of publication
- Publication date
-
2024-11-27Full publication date if available
- Authors
-
Bonnie Patchen, Jingwen Zhang, Nathan Gaddis, Traci M. Bartz, Jing Chen, Catherine L. Debban, Hampton L. Leonard, Ngoc Quynh Nguyen, Ji Heui Seo, Courtney Tern, Richard J. Allen, Dawn L. DeMeo, Myriam Fornage, Carl Melbourne, Melyssa S. Minto, Matthew Moll, George O'connor, Tess D. Pottinger, Bruce M. Psaty, Stephen S. Rich, Laura M. Raffield, Edwin K. Silverman, Jeran K. Stratford, R. Graham Barr, Michael H. Cho, Sina A. Gharib, Ani Manichaikul, Kari North, Elizabeth C. Oelsner, Eleanor M. Simonsick, Martin D. Tobin, Bing Yu, Seung Hoan Choi, Josée Dupuis, Patricia A. Cassano, Dana B. HancockList of authors in order
- Landing page
-
https://doi.org/10.1101/2024.11.25.24317794Publisher landing page
- Open access
-
YesWhether a free full text is available
- OA status
-
greenOpen access status per OpenAlex
- OA URL
-
https://www.ncbi.nlm.nih.gov/pmc/articles/11623738Direct OA link when available
- Concepts
-
Genome-wide association study, Lung function, Association (psychology), Genetic association, Biology, Function (biology), Evolutionary biology, Computational biology, Genome, Genetic genealogy, Genetics, Medicine, Single-nucleotide polymorphism, Genotype, Lung, Environmental health, Internal medicine, Psychology, Gene, Population, PsychotherapistTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
1Total citation count in OpenAlex
- Citations by year (recent)
-
2025: 1Per-year citation counts (last 5 years)
- References (count)
-
55Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4404788147 |
|---|---|
| doi | https://doi.org/10.1101/2024.11.25.24317794 |
| ids.doi | https://doi.org/10.1101/2024.11.25.24317794 |
| ids.pmid | https://pubmed.ncbi.nlm.nih.gov/39649580 |
| ids.openalex | https://openalex.org/W4404788147 |
| fwci | 0.86262314 |
| type | preprint |
| title | Multi-ancestry genome-wide association study reveals novel genetic signals for lung function decline |
| biblio.issue | |
| biblio.volume | |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T10143 |
| topics[0].field.id | https://openalex.org/fields/27 |
| topics[0].field.display_name | Medicine |
| topics[0].score | 0.9959999918937683 |
| topics[0].domain.id | https://openalex.org/domains/4 |
| topics[0].domain.display_name | Health Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/2740 |
| topics[0].subfield.display_name | Pulmonary and Respiratory Medicine |
| topics[0].display_name | Chronic Obstructive Pulmonary Disease (COPD) Research |
| topics[1].id | https://openalex.org/T10051 |
| topics[1].field.id | https://openalex.org/fields/27 |
| topics[1].field.display_name | Medicine |
| topics[1].score | 0.982699990272522 |
| topics[1].domain.id | https://openalex.org/domains/4 |
| topics[1].domain.display_name | Health Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/2737 |
| topics[1].subfield.display_name | Physiology |
| topics[1].display_name | Asthma and respiratory diseases |
| topics[2].id | https://openalex.org/T10261 |
| topics[2].field.id | https://openalex.org/fields/13 |
| topics[2].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[2].score | 0.9772999882698059 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1311 |
| topics[2].subfield.display_name | Genetics |
| topics[2].display_name | Genetic Associations and Epidemiology |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C106208931 |
| concepts[0].level | 5 |
| concepts[0].score | 0.7700222730636597 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q1098876 |
| concepts[0].display_name | Genome-wide association study |
| concepts[1].id | https://openalex.org/C3018587741 |
| concepts[1].level | 3 |
| concepts[1].score | 0.5970984101295471 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q7886 |
| concepts[1].display_name | Lung function |
| concepts[2].id | https://openalex.org/C142853389 |
| concepts[2].level | 2 |
| concepts[2].score | 0.5868722796440125 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q744778 |
| concepts[2].display_name | Association (psychology) |
| concepts[3].id | https://openalex.org/C186413461 |
| concepts[3].level | 5 |
| concepts[3].score | 0.5729936957359314 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q744727 |
| concepts[3].display_name | Genetic association |
| concepts[4].id | https://openalex.org/C86803240 |
| concepts[4].level | 0 |
| concepts[4].score | 0.4681437611579895 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[4].display_name | Biology |
| concepts[5].id | https://openalex.org/C14036430 |
| concepts[5].level | 2 |
| concepts[5].score | 0.45641592144966125 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q3736076 |
| concepts[5].display_name | Function (biology) |
| concepts[6].id | https://openalex.org/C78458016 |
| concepts[6].level | 1 |
| concepts[6].score | 0.45107099413871765 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q840400 |
| concepts[6].display_name | Evolutionary biology |
| concepts[7].id | https://openalex.org/C70721500 |
| concepts[7].level | 1 |
| concepts[7].score | 0.42172789573669434 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q177005 |
| concepts[7].display_name | Computational biology |
| concepts[8].id | https://openalex.org/C141231307 |
| concepts[8].level | 3 |
| concepts[8].score | 0.41946378350257874 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q7020 |
| concepts[8].display_name | Genome |
| concepts[9].id | https://openalex.org/C75069973 |
| concepts[9].level | 3 |
| concepts[9].score | 0.41582080721855164 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q913584 |
| concepts[9].display_name | Genetic genealogy |
| concepts[10].id | https://openalex.org/C54355233 |
| concepts[10].level | 1 |
| concepts[10].score | 0.3813307285308838 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[10].display_name | Genetics |
| concepts[11].id | https://openalex.org/C71924100 |
| concepts[11].level | 0 |
| concepts[11].score | 0.2654328942298889 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q11190 |
| concepts[11].display_name | Medicine |
| concepts[12].id | https://openalex.org/C153209595 |
| concepts[12].level | 4 |
| concepts[12].score | 0.251570463180542 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q501128 |
| concepts[12].display_name | Single-nucleotide polymorphism |
| concepts[13].id | https://openalex.org/C135763542 |
| concepts[13].level | 3 |
| concepts[13].score | 0.22245478630065918 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q106016 |
| concepts[13].display_name | Genotype |
| concepts[14].id | https://openalex.org/C2777714996 |
| concepts[14].level | 2 |
| concepts[14].score | 0.21856674551963806 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q7886 |
| concepts[14].display_name | Lung |
| concepts[15].id | https://openalex.org/C99454951 |
| concepts[15].level | 1 |
| concepts[15].score | 0.13684159517288208 |
| concepts[15].wikidata | https://www.wikidata.org/wiki/Q932068 |
| concepts[15].display_name | Environmental health |
| concepts[16].id | https://openalex.org/C126322002 |
| concepts[16].level | 1 |
| concepts[16].score | 0.12375390529632568 |
| concepts[16].wikidata | https://www.wikidata.org/wiki/Q11180 |
| concepts[16].display_name | Internal medicine |
| concepts[17].id | https://openalex.org/C15744967 |
| concepts[17].level | 0 |
| concepts[17].score | 0.11901837587356567 |
| concepts[17].wikidata | https://www.wikidata.org/wiki/Q9418 |
| concepts[17].display_name | Psychology |
| concepts[18].id | https://openalex.org/C104317684 |
| concepts[18].level | 2 |
| concepts[18].score | 0.09317132830619812 |
| concepts[18].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[18].display_name | Gene |
| concepts[19].id | https://openalex.org/C2908647359 |
| concepts[19].level | 2 |
| concepts[19].score | 0.07192385196685791 |
| concepts[19].wikidata | https://www.wikidata.org/wiki/Q2625603 |
| concepts[19].display_name | Population |
| concepts[20].id | https://openalex.org/C542102704 |
| concepts[20].level | 1 |
| concepts[20].score | 0.0 |
| concepts[20].wikidata | https://www.wikidata.org/wiki/Q183257 |
| concepts[20].display_name | Psychotherapist |
| keywords[0].id | https://openalex.org/keywords/genome-wide-association-study |
| keywords[0].score | 0.7700222730636597 |
| keywords[0].display_name | Genome-wide association study |
| keywords[1].id | https://openalex.org/keywords/lung-function |
| keywords[1].score | 0.5970984101295471 |
| keywords[1].display_name | Lung function |
| keywords[2].id | https://openalex.org/keywords/association |
| keywords[2].score | 0.5868722796440125 |
| keywords[2].display_name | Association (psychology) |
| keywords[3].id | https://openalex.org/keywords/genetic-association |
| keywords[3].score | 0.5729936957359314 |
| keywords[3].display_name | Genetic association |
| keywords[4].id | https://openalex.org/keywords/biology |
| keywords[4].score | 0.4681437611579895 |
| keywords[4].display_name | Biology |
| keywords[5].id | https://openalex.org/keywords/function |
| keywords[5].score | 0.45641592144966125 |
| keywords[5].display_name | Function (biology) |
| keywords[6].id | https://openalex.org/keywords/evolutionary-biology |
| keywords[6].score | 0.45107099413871765 |
| keywords[6].display_name | Evolutionary biology |
| keywords[7].id | https://openalex.org/keywords/computational-biology |
| keywords[7].score | 0.42172789573669434 |
| keywords[7].display_name | Computational biology |
| keywords[8].id | https://openalex.org/keywords/genome |
| keywords[8].score | 0.41946378350257874 |
| keywords[8].display_name | Genome |
| keywords[9].id | https://openalex.org/keywords/genetic-genealogy |
| keywords[9].score | 0.41582080721855164 |
| keywords[9].display_name | Genetic genealogy |
| keywords[10].id | https://openalex.org/keywords/genetics |
| keywords[10].score | 0.3813307285308838 |
| keywords[10].display_name | Genetics |
| keywords[11].id | https://openalex.org/keywords/medicine |
| keywords[11].score | 0.2654328942298889 |
| keywords[11].display_name | Medicine |
| keywords[12].id | https://openalex.org/keywords/single-nucleotide-polymorphism |
| keywords[12].score | 0.251570463180542 |
| keywords[12].display_name | Single-nucleotide polymorphism |
| keywords[13].id | https://openalex.org/keywords/genotype |
| keywords[13].score | 0.22245478630065918 |
| keywords[13].display_name | Genotype |
| keywords[14].id | https://openalex.org/keywords/lung |
| keywords[14].score | 0.21856674551963806 |
| keywords[14].display_name | Lung |
| keywords[15].id | https://openalex.org/keywords/environmental-health |
| keywords[15].score | 0.13684159517288208 |
| keywords[15].display_name | Environmental health |
| keywords[16].id | https://openalex.org/keywords/internal-medicine |
| keywords[16].score | 0.12375390529632568 |
| keywords[16].display_name | Internal medicine |
| keywords[17].id | https://openalex.org/keywords/psychology |
| keywords[17].score | 0.11901837587356567 |
| keywords[17].display_name | Psychology |
| keywords[18].id | https://openalex.org/keywords/gene |
| keywords[18].score | 0.09317132830619812 |
| keywords[18].display_name | Gene |
| keywords[19].id | https://openalex.org/keywords/population |
| keywords[19].score | 0.07192385196685791 |
| keywords[19].display_name | Population |
| language | en |
| locations[0].id | doi:10.1101/2024.11.25.24317794 |
| locations[0].is_oa | False |
| locations[0].source.id | https://openalex.org/S4306402567 |
| locations[0].source.issn | |
| locations[0].source.type | repository |
| locations[0].source.is_oa | False |
| locations[0].source.issn_l | |
| locations[0].source.is_core | False |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| locations[0].source.host_organization | https://openalex.org/I2750212522 |
| locations[0].source.host_organization_name | Cold Spring Harbor Laboratory |
| locations[0].source.host_organization_lineage | https://openalex.org/I2750212522 |
| locations[0].license | |
| locations[0].pdf_url | |
| locations[0].version | acceptedVersion |
| locations[0].raw_type | posted-content |
| locations[0].license_id | |
| locations[0].is_accepted | True |
| locations[0].is_published | False |
| locations[0].raw_source_name | |
| locations[0].landing_page_url | https://doi.org/10.1101/2024.11.25.24317794 |
| locations[1].id | pmid:39649580 |
| locations[1].is_oa | False |
| locations[1].source.id | https://openalex.org/S4306525036 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | PubMed |
| locations[1].source.host_organization | https://openalex.org/I1299303238 |
| locations[1].source.host_organization_name | National Institutes of Health |
| locations[1].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[1].license | |
| locations[1].pdf_url | |
| locations[1].version | publishedVersion |
| locations[1].raw_type | |
| locations[1].license_id | |
| locations[1].is_accepted | True |
| locations[1].is_published | True |
| locations[1].raw_source_name | medRxiv : the preprint server for health sciences |
| locations[1].landing_page_url | https://pubmed.ncbi.nlm.nih.gov/39649580 |
| locations[2].id | pmh:oai:pubmedcentral.nih.gov:11623738 |
| locations[2].is_oa | True |
| locations[2].source.id | https://openalex.org/S2764455111 |
| locations[2].source.issn | |
| locations[2].source.type | repository |
| locations[2].source.is_oa | False |
| locations[2].source.issn_l | |
| locations[2].source.is_core | False |
| locations[2].source.is_in_doaj | False |
| locations[2].source.display_name | PubMed Central |
| locations[2].source.host_organization | https://openalex.org/I1299303238 |
| locations[2].source.host_organization_name | National Institutes of Health |
| locations[2].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[2].license | |
| locations[2].pdf_url | |
| locations[2].version | submittedVersion |
| locations[2].raw_type | Text |
| locations[2].license_id | |
| locations[2].is_accepted | False |
| locations[2].is_published | False |
| locations[2].raw_source_name | medRxiv |
| locations[2].landing_page_url | https://www.ncbi.nlm.nih.gov/pmc/articles/11623738 |
| indexed_in | crossref, pubmed |
| authorships[0].author.id | https://openalex.org/A5040316492 |
| authorships[0].author.orcid | https://orcid.org/0000-0003-3361-6744 |
| authorships[0].author.display_name | Bonnie Patchen |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Bonnie K. Patchen |
| authorships[0].is_corresponding | False |
| authorships[1].author.id | https://openalex.org/A5100435147 |
| authorships[1].author.orcid | |
| authorships[1].author.display_name | Jingwen Zhang |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Jingwen Zhang |
| authorships[1].is_corresponding | False |
| authorships[2].author.id | https://openalex.org/A5010195745 |
| authorships[2].author.orcid | https://orcid.org/0000-0001-5205-7138 |
| authorships[2].author.display_name | Nathan Gaddis |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Nathan Gaddis |
| authorships[2].is_corresponding | False |
| authorships[3].author.id | https://openalex.org/A5107467968 |
| authorships[3].author.orcid | |
| authorships[3].author.display_name | Traci M. Bartz |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Traci M. Bartz |
| authorships[3].is_corresponding | False |
| authorships[4].author.id | https://openalex.org/A5100786469 |
| authorships[4].author.orcid | https://orcid.org/0000-0003-1287-1930 |
| authorships[4].author.display_name | Jing Chen |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Jing Chen |
| authorships[4].is_corresponding | False |
| authorships[5].author.id | https://openalex.org/A5063472938 |
| authorships[5].author.orcid | |
| authorships[5].author.display_name | Catherine L. Debban |
| authorships[5].author_position | middle |
| authorships[5].raw_author_name | Catherine Debban |
| authorships[5].is_corresponding | False |
| authorships[6].author.id | https://openalex.org/A5033284534 |
| authorships[6].author.orcid | https://orcid.org/0000-0003-2390-8110 |
| authorships[6].author.display_name | Hampton L. Leonard |
| authorships[6].author_position | middle |
| authorships[6].raw_author_name | Hampton Leonard |
| authorships[6].is_corresponding | False |
| authorships[7].author.id | https://openalex.org/A5088351143 |
| authorships[7].author.orcid | |
| authorships[7].author.display_name | Ngoc Quynh Nguyen |
| authorships[7].author_position | middle |
| authorships[7].raw_author_name | Ngoc Quynh Nguyen |
| authorships[7].is_corresponding | False |
| authorships[8].author.id | https://openalex.org/A5112375876 |
| authorships[8].author.orcid | |
| authorships[8].author.display_name | Ji Heui Seo |
| authorships[8].author_position | middle |
| authorships[8].raw_author_name | Jungkun Seo |
| authorships[8].is_corresponding | False |
| authorships[9].author.id | https://openalex.org/A5012341524 |
| authorships[9].author.orcid | https://orcid.org/0000-0001-5029-595X |
| authorships[9].author.display_name | Courtney Tern |
| authorships[9].author_position | middle |
| authorships[9].raw_author_name | Courtney Tern |
| authorships[9].is_corresponding | False |
| authorships[10].author.id | https://openalex.org/A5066779629 |
| authorships[10].author.orcid | https://orcid.org/0000-0002-8450-3056 |
| authorships[10].author.display_name | Richard J. Allen |
| authorships[10].author_position | middle |
| authorships[10].raw_author_name | Richard Allen |
| authorships[10].is_corresponding | False |
| authorships[11].author.id | https://openalex.org/A5051093934 |
| authorships[11].author.orcid | https://orcid.org/0000-0001-9653-0636 |
| authorships[11].author.display_name | Dawn L. DeMeo |
| authorships[11].author_position | middle |
| authorships[11].raw_author_name | Dawn L. DeMeo |
| authorships[11].is_corresponding | False |
| authorships[12].author.id | https://openalex.org/A5100672057 |
| authorships[12].author.orcid | https://orcid.org/0000-0003-0677-8158 |
| authorships[12].author.display_name | Myriam Fornage |
| authorships[12].author_position | middle |
| authorships[12].raw_author_name | Myriam Fornage |
| authorships[12].is_corresponding | False |
| authorships[13].author.id | https://openalex.org/A5011026070 |
| authorships[13].author.orcid | https://orcid.org/0000-0001-7216-4547 |
| authorships[13].author.display_name | Carl Melbourne |
| authorships[13].author_position | middle |
| authorships[13].raw_author_name | Carl Melbourne |
| authorships[13].is_corresponding | False |
| authorships[14].author.id | https://openalex.org/A5048417722 |
| authorships[14].author.orcid | https://orcid.org/0000-0002-5438-7285 |
| authorships[14].author.display_name | Melyssa S. Minto |
| authorships[14].author_position | middle |
| authorships[14].raw_author_name | Melyssa Minto |
| authorships[14].is_corresponding | False |
| authorships[15].author.id | https://openalex.org/A5085242587 |
| authorships[15].author.orcid | https://orcid.org/0000-0002-0683-0975 |
| authorships[15].author.display_name | Matthew Moll |
| authorships[15].author_position | middle |
| authorships[15].raw_author_name | Matthew Moll |
| authorships[15].is_corresponding | False |
| authorships[16].author.id | https://openalex.org/A5059122240 |
| authorships[16].author.orcid | https://orcid.org/0000-0002-6476-3926 |
| authorships[16].author.display_name | George O'connor |
| authorships[16].author_position | middle |
| authorships[16].raw_author_name | George O’Connor |
| authorships[16].is_corresponding | False |
| authorships[17].author.id | https://openalex.org/A5024081090 |
| authorships[17].author.orcid | https://orcid.org/0000-0003-0647-5712 |
| authorships[17].author.display_name | Tess D. Pottinger |
| authorships[17].author_position | middle |
| authorships[17].raw_author_name | Tess Pottinger |
| authorships[17].is_corresponding | False |
| authorships[18].author.id | https://openalex.org/A5107858644 |
| authorships[18].author.orcid | |
| authorships[18].author.display_name | Bruce M. Psaty |
| authorships[18].author_position | middle |
| authorships[18].raw_author_name | Bruce M. Psaty |
| authorships[18].is_corresponding | False |
| authorships[19].author.id | https://openalex.org/A5070620956 |
| authorships[19].author.orcid | |
| authorships[19].author.display_name | Stephen S. Rich |
| authorships[19].author_position | middle |
| authorships[19].raw_author_name | Stephen S. Rich |
| authorships[19].is_corresponding | False |
| authorships[20].author.id | https://openalex.org/A5042360941 |
| authorships[20].author.orcid | https://orcid.org/0000-0002-7892-193X |
| authorships[20].author.display_name | Laura M. Raffield |
| authorships[20].author_position | middle |
| authorships[20].raw_author_name | Jerome I. Rotter |
| authorships[20].is_corresponding | False |
| authorships[21].author.id | https://openalex.org/A5035524361 |
| authorships[21].author.orcid | https://orcid.org/0000-0002-3641-3822 |
| authorships[21].author.display_name | Edwin K. Silverman |
| authorships[21].author_position | middle |
| authorships[21].raw_author_name | Edwin K. Silverman |
| authorships[21].is_corresponding | False |
| authorships[22].author.id | https://openalex.org/A5038774978 |
| authorships[22].author.orcid | https://orcid.org/0000-0003-4305-1468 |
| authorships[22].author.display_name | Jeran K. Stratford |
| authorships[22].author_position | middle |
| authorships[22].raw_author_name | Jeran Stratford |
| authorships[22].is_corresponding | False |
| authorships[23].author.id | https://openalex.org/A5103134292 |
| authorships[23].author.orcid | https://orcid.org/0000-0002-0743-2303 |
| authorships[23].author.display_name | R. Graham Barr |
| authorships[23].author_position | middle |
| authorships[23].raw_author_name | R. Graham Barr |
| authorships[23].is_corresponding | False |
| authorships[24].author.id | https://openalex.org/A5018495347 |
| authorships[24].author.orcid | https://orcid.org/0000-0002-4907-1657 |
| authorships[24].author.display_name | Michael H. Cho |
| authorships[24].author_position | middle |
| authorships[24].raw_author_name | Michael H. Cho |
| authorships[24].is_corresponding | False |
| authorships[25].author.id | https://openalex.org/A5084644058 |
| authorships[25].author.orcid | https://orcid.org/0000-0002-2480-4367 |
| authorships[25].author.display_name | Sina A. Gharib |
| authorships[25].author_position | middle |
| authorships[25].raw_author_name | Sina A. Gharib |
| authorships[25].is_corresponding | False |
| authorships[26].author.id | https://openalex.org/A5048424813 |
| authorships[26].author.orcid | |
| authorships[26].author.display_name | Ani Manichaikul |
| authorships[26].author_position | middle |
| authorships[26].raw_author_name | Ani Manichaikul |
| authorships[26].is_corresponding | False |
| authorships[27].author.id | https://openalex.org/A5025031281 |
| authorships[27].author.orcid | |
| authorships[27].author.display_name | Kari North |
| authorships[27].author_position | middle |
| authorships[27].raw_author_name | Kari North |
| authorships[27].is_corresponding | False |
| authorships[28].author.id | https://openalex.org/A5056182924 |
| authorships[28].author.orcid | https://orcid.org/0000-0002-7481-9671 |
| authorships[28].author.display_name | Elizabeth C. Oelsner |
| authorships[28].author_position | middle |
| authorships[28].raw_author_name | Elizabeth C. Oelsner |
| authorships[28].is_corresponding | False |
| authorships[29].author.id | https://openalex.org/A5090671996 |
| authorships[29].author.orcid | https://orcid.org/0000-0002-5460-3494 |
| authorships[29].author.display_name | Eleanor M. Simonsick |
| authorships[29].author_position | middle |
| authorships[29].raw_author_name | Eleanor M. Simonsick |
| authorships[29].is_corresponding | False |
| authorships[30].author.id | https://openalex.org/A5043513022 |
| authorships[30].author.orcid | https://orcid.org/0000-0002-3596-7874 |
| authorships[30].author.display_name | Martin D. Tobin |
| authorships[30].author_position | middle |
| authorships[30].raw_author_name | Martin D. Tobin |
| authorships[30].is_corresponding | False |
| authorships[31].author.id | https://openalex.org/A5042391667 |
| authorships[31].author.orcid | https://orcid.org/0000-0001-6013-5220 |
| authorships[31].author.display_name | Bing Yu |
| authorships[31].author_position | middle |
| authorships[31].raw_author_name | Bing Yu |
| authorships[31].is_corresponding | False |
| authorships[32].author.id | https://openalex.org/A5041586675 |
| authorships[32].author.orcid | https://orcid.org/0000-0002-2797-3190 |
| authorships[32].author.display_name | Seung Hoan Choi |
| authorships[32].author_position | middle |
| authorships[32].raw_author_name | Seung Hoan Choi |
| authorships[32].is_corresponding | False |
| authorships[33].author.id | https://openalex.org/A5053091216 |
| authorships[33].author.orcid | https://orcid.org/0000-0003-2871-3603 |
| authorships[33].author.display_name | Josée Dupuis |
| authorships[33].author_position | middle |
| authorships[33].raw_author_name | Josee Dupuis |
| authorships[33].is_corresponding | False |
| authorships[34].author.id | https://openalex.org/A5067632644 |
| authorships[34].author.orcid | https://orcid.org/0000-0003-4827-5073 |
| authorships[34].author.display_name | Patricia A. Cassano |
| authorships[34].author_position | middle |
| authorships[34].raw_author_name | Patricia A. Cassano |
| authorships[34].is_corresponding | False |
| authorships[35].author.id | https://openalex.org/A5019653549 |
| authorships[35].author.orcid | https://orcid.org/0000-0003-2240-3604 |
| authorships[35].author.display_name | Dana B. Hancock |
| authorships[35].author_position | last |
| authorships[35].raw_author_name | Dana B. Hancock |
| authorships[35].is_corresponding | False |
| has_content.pdf | False |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://www.ncbi.nlm.nih.gov/pmc/articles/11623738 |
| open_access.oa_status | green |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Multi-ancestry genome-wide association study reveals novel genetic signals for lung function decline |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T10143 |
| primary_topic.field.id | https://openalex.org/fields/27 |
| primary_topic.field.display_name | Medicine |
| primary_topic.score | 0.9959999918937683 |
| primary_topic.domain.id | https://openalex.org/domains/4 |
| primary_topic.domain.display_name | Health Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/2740 |
| primary_topic.subfield.display_name | Pulmonary and Respiratory Medicine |
| primary_topic.display_name | Chronic Obstructive Pulmonary Disease (COPD) Research |
| related_works | https://openalex.org/W2594299395, https://openalex.org/W2066467839, https://openalex.org/W2111842731, https://openalex.org/W2170506381, https://openalex.org/W2765794416, https://openalex.org/W2039666944, https://openalex.org/W1531630856, https://openalex.org/W1486217206, https://openalex.org/W1992310781, https://openalex.org/W2794350630 |
| cited_by_count | 1 |
| counts_by_year[0].year | 2025 |
| counts_by_year[0].cited_by_count | 1 |
| locations_count | 3 |
| best_oa_location.id | pmh:oai:pubmedcentral.nih.gov:11623738 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S2764455111 |
| best_oa_location.source.issn | |
| best_oa_location.source.type | repository |
| best_oa_location.source.is_oa | False |
| best_oa_location.source.issn_l | |
| best_oa_location.source.is_core | False |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | PubMed Central |
| best_oa_location.source.host_organization | https://openalex.org/I1299303238 |
| best_oa_location.source.host_organization_name | National Institutes of Health |
| best_oa_location.source.host_organization_lineage | https://openalex.org/I1299303238 |
| best_oa_location.license | |
| best_oa_location.pdf_url | |
| best_oa_location.version | submittedVersion |
| best_oa_location.raw_type | Text |
| best_oa_location.license_id | |
| best_oa_location.is_accepted | False |
| best_oa_location.is_published | False |
| best_oa_location.raw_source_name | medRxiv |
| best_oa_location.landing_page_url | https://www.ncbi.nlm.nih.gov/pmc/articles/11623738 |
| primary_location.id | doi:10.1101/2024.11.25.24317794 |
| primary_location.is_oa | False |
| primary_location.source.id | https://openalex.org/S4306402567 |
| primary_location.source.issn | |
| primary_location.source.type | repository |
| primary_location.source.is_oa | False |
| primary_location.source.issn_l | |
| primary_location.source.is_core | False |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| primary_location.source.host_organization | https://openalex.org/I2750212522 |
| primary_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| primary_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| primary_location.license | |
| primary_location.pdf_url | |
| primary_location.version | acceptedVersion |
| primary_location.raw_type | posted-content |
| primary_location.license_id | |
| primary_location.is_accepted | True |
| primary_location.is_published | False |
| primary_location.raw_source_name | |
| primary_location.landing_page_url | https://doi.org/10.1101/2024.11.25.24317794 |
| publication_date | 2024-11-27 |
| publication_year | 2024 |
| referenced_works | https://openalex.org/W2122590281, https://openalex.org/W2002726093, https://openalex.org/W2947236019, https://openalex.org/W4324045235, https://openalex.org/W2573109541, https://openalex.org/W2157361982, https://openalex.org/W4285733230, https://openalex.org/W2997704953, https://openalex.org/W2580815378, https://openalex.org/W2127818071, https://openalex.org/W2991133758, https://openalex.org/W2157186191, https://openalex.org/W2809787362, https://openalex.org/W2320254713, https://openalex.org/W1950067960, https://openalex.org/W2442409313, https://openalex.org/W2163184920, https://openalex.org/W2806078282, https://openalex.org/W2107134975, https://openalex.org/W1809582455, https://openalex.org/W2051314902, https://openalex.org/W2018838463, https://openalex.org/W2398690791, https://openalex.org/W2955177623, https://openalex.org/W3007352713, https://openalex.org/W2770092353, https://openalex.org/W1979594866, https://openalex.org/W4293702574, https://openalex.org/W3094762523, https://openalex.org/W2998400169, https://openalex.org/W3106143834, https://openalex.org/W3210331473, https://openalex.org/W2170146596, https://openalex.org/W2106811010, https://openalex.org/W2581166394, https://openalex.org/W4293168627, https://openalex.org/W4386116531, https://openalex.org/W4307824500, https://openalex.org/W2180085345, https://openalex.org/W967710558, https://openalex.org/W2343487858, https://openalex.org/W1972351108, https://openalex.org/W2008897742, https://openalex.org/W2147498958, https://openalex.org/W2164419379, https://openalex.org/W3190626600, https://openalex.org/W2122328984, https://openalex.org/W4306642258, https://openalex.org/W2023279935, https://openalex.org/W2982347822, https://openalex.org/W2591924740, https://openalex.org/W2169113398, https://openalex.org/W2982486303, https://openalex.org/W1969338627, https://openalex.org/W3205180240 |
| referenced_works_count | 55 |
| abstract_inverted_index.( | 290 |
| abstract_inverted_index.) | 307 |
| abstract_inverted_index., | 227, 292, 294, 296, 298, 300, 302, 304 |
| abstract_inverted_index.1 | 72, 82, 226, 231 |
| abstract_inverted_index.8 | 249 |
| abstract_inverted_index.a | 18, 195, 374 |
| abstract_inverted_index.), | 73 |
| abstract_inverted_index.38 | 286, 343 |
| abstract_inverted_index.43 | 335 |
| abstract_inverted_index.Of | 247 |
| abstract_inverted_index.To | 32 |
| abstract_inverted_index.UK | 104 |
| abstract_inverted_index.We | 53 |
| abstract_inverted_index.at | 266 |
| abstract_inverted_index.by | 137 |
| abstract_inverted_index.in | 4, 47, 62, 67, 84, 89, 97, 156, 168, 219, 271, 379 |
| abstract_inverted_index.of | 11, 40, 60, 197, 204, 208, 223, 253, 322, 340, 367, 376, 382 |
| abstract_inverted_index.or | 113, 221, 251, 313 |
| abstract_inverted_index.to | 8, 109, 363, 397 |
| abstract_inverted_index.(UK | 118 |
| abstract_inverted_index.2.3 | 198 |
| abstract_inverted_index.361 | 213 |
| abstract_inverted_index.8.6 | 202 |
| abstract_inverted_index.FEV | 225, 230 |
| abstract_inverted_index.Our | 177, 346 |
| abstract_inverted_index.and | 36, 78, 94, 102, 122, 140, 142, 149, 158, 174, 189, 201, 229, 276, 305, 329, 386, 402 |
| abstract_inverted_index.few | 21 |
| abstract_inverted_index.for | 17, 92, 144, 166, 242, 258, 326, 373, 394 |
| abstract_inverted_index.one | 220, 268 |
| abstract_inverted_index.set | 256 |
| abstract_inverted_index.six | 87 |
| abstract_inverted_index.the | 9, 63, 68, 90, 103, 224, 254, 280, 341, 364, 380 |
| abstract_inverted_index.top | 162 |
| abstract_inverted_index.two | 169 |
| abstract_inverted_index.(FEV | 71, 81 |
| abstract_inverted_index./FVC | 232 |
| abstract_inverted_index.ANO5 | 301 |
| abstract_inverted_index.Drug | 331 |
| abstract_inverted_index.FAF2 | 306 |
| abstract_inverted_index.FVC, | 228 |
| abstract_inverted_index.GWAS | 123, 209, 281, 348 |
| abstract_inverted_index.Main | 175 |
| abstract_inverted_index.been | 29 |
| abstract_inverted_index.drug | 388 |
| abstract_inverted_index.have | 28 |
| abstract_inverted_index.loci | 353 |
| abstract_inverted_index.lung | 5, 25, 44, 146, 356, 368, 383, 399, 404 |
| abstract_inverted_index.mean | 196 |
| abstract_inverted_index.more | 222 |
| abstract_inverted_index.role | 375 |
| abstract_inverted_index.sex, | 141 |
| abstract_inverted_index.slow | 398 |
| abstract_inverted_index.that | 390 |
| abstract_inverted_index.used | 125 |
| abstract_inverted_index.were | 107, 135, 154, 164, 261 |
| abstract_inverted_index.with | 24, 43, 130, 194, 237, 265, 308, 355 |
| abstract_inverted_index.(HRC) | 117 |
| abstract_inverted_index./FVC) | 83 |
| abstract_inverted_index.20.5% | 252 |
| abstract_inverted_index.Aging | 95 |
| abstract_inverted_index.Black | 185, 277 |
| abstract_inverted_index.Heart | 93 |
| abstract_inverted_index.SATB1 | 295 |
| abstract_inverted_index.These | 359 |
| abstract_inverted_index.Using | 279 |
| abstract_inverted_index.White | 183 |
| abstract_inverted_index.XIRP2 | 291 |
| abstract_inverted_index.class | 317 |
| abstract_inverted_index.eight | 289, 339 |
| abstract_inverted_index.first | 69 |
| abstract_inverted_index.least | 267 |
| abstract_inverted_index.merit | 391 |
| abstract_inverted_index.ratio | 80 |
| abstract_inverted_index.study | 57, 393 |
| abstract_inverted_index.their | 79 |
| abstract_inverted_index.treat | 403 |
| abstract_inverted_index.vital | 75 |
| abstract_inverted_index.which | 235 |
| abstract_inverted_index.years | 203 |
| abstract_inverted_index.(FVC), | 77 |
| abstract_inverted_index.(GWAS) | 58 |
| abstract_inverted_index.(White | 274 |
| abstract_inverted_index.52,056 | 181 |
| abstract_inverted_index.GRIN2D | 293 |
| abstract_inverted_index.H2BC10 | 303 |
| abstract_inverted_index.Models | 134 |
| abstract_inverted_index.TOPMed | 110 |
| abstract_inverted_index.across | 86, 311 |
| abstract_inverted_index.error. | 133 |
| abstract_inverted_index.forced | 64, 74 |
| abstract_inverted_index.genes, | 287 |
| abstract_inverted_index.genes. | 344 |
| abstract_inverted_index.models | 129 |
| abstract_inverted_index.robust | 131 |
| abstract_inverted_index.second | 70 |
| abstract_inverted_index.tested | 165 |
| abstract_inverted_index.these, | 248 |
| abstract_inverted_index.volume | 66 |
| abstract_inverted_index.(CHARGE | 111 |
| abstract_inverted_index.(N=730) | 192 |
| abstract_inverted_index.Chinese | 190 |
| abstract_inverted_index.Cohorts | 91 |
| abstract_inverted_index.Despite | 15 |
| abstract_inverted_index.Genomic | 98 |
| abstract_inverted_index.MARCHF4 | 297 |
| abstract_inverted_index.Methods | 52 |
| abstract_inverted_index.Results | 153, 176 |
| abstract_inverted_index.SIPA1L2 | 299 |
| abstract_inverted_index.chronic | 12 |
| abstract_inverted_index.cohort, | 138 |
| abstract_inverted_index.cohorts | 88, 273 |
| abstract_inverted_index.decline | 3, 27, 46, 61, 233, 269, 314, 401 |
| abstract_inverted_index.diverse | 48 |
| abstract_inverted_index.further | 392 |
| abstract_inverted_index.general | 49 |
| abstract_inverted_index.genetic | 19, 22, 41, 240, 352, 365 |
| abstract_inverted_index.imputed | 108 |
| abstract_inverted_index.mapping | 207 |
| abstract_inverted_index.panels, | 121 |
| abstract_inverted_index.provide | 371 |
| abstract_inverted_index.related | 244 |
| abstract_inverted_index.results | 211 |
| abstract_inverted_index.several | 243, 323 |
| abstract_inverted_index.signals | 241 |
| abstract_inverted_index.targets | 389 |
| abstract_inverted_index.traits. | 246 |
| abstract_inverted_index.variant | 255 |
| abstract_inverted_index.(CHARGE) | 100 |
| abstract_inverted_index.(N=550), | 188 |
| abstract_inverted_index.Abstract | 0 |
| abstract_inverted_index.American | 191 |
| abstract_inverted_index.Biobank) | 119 |
| abstract_inverted_index.Biobank. | 105 |
| abstract_inverted_index.Hispanic | 187 |
| abstract_inverted_index.Research | 96 |
| abstract_inverted_index.Selected | 161 |
| abstract_inverted_index.adjusted | 143 |
| abstract_inverted_index.analyses | 59, 124, 179 |
| abstract_inverted_index.analysis | 284, 318, 333 |
| abstract_inverted_index.approved | 336 |
| abstract_inverted_index.capacity | 76 |
| abstract_inverted_index.cohorts) | 112 |
| abstract_inverted_index.cohorts. | 51, 172 |
| abstract_inverted_index.combined | 155 |
| abstract_inverted_index.decline, | 370, 385 |
| abstract_inverted_index.decline. | 358 |
| abstract_inverted_index.disease. | 14, 405 |
| abstract_inverted_index.distinct | 214 |
| abstract_inverted_index.equation | 128 |
| abstract_inverted_index.etiology | 381 |
| abstract_inverted_index.evaluate | 33 |
| abstract_inverted_index.evidence | 16, 372 |
| abstract_inverted_index.findings | 360 |
| abstract_inverted_index.function | 6, 26, 45, 147, 357, 369, 384, 400 |
| abstract_inverted_index.genotype | 150 |
| abstract_inverted_index.identify | 387 |
| abstract_inverted_index.included | 180 |
| abstract_inverted_index.numerous | 351 |
| abstract_inverted_index.putative | 37 |
| abstract_inverted_index.reported | 239 |
| abstract_inverted_index.results, | 282 |
| abstract_inverted_index.revealed | 319 |
| abstract_inverted_index.standard | 132 |
| abstract_inverted_index.testing, | 260 |
| abstract_inverted_index.variants | 42, 163, 218 |
| abstract_inverted_index.Genotypes | 106 |
| abstract_inverted_index.Haplotype | 114 |
| abstract_inverted_index.Rationale | 1 |
| abstract_inverted_index.Reference | 115 |
| abstract_inverted_index.[N=4,778] | 275 |
| abstract_inverted_index.ancestry, | 139 |
| abstract_inverted_index.available | 257 |
| abstract_inverted_index.compounds | 337 |
| abstract_inverted_index.conducted | 54 |
| abstract_inverted_index.discovery | 178 |
| abstract_inverted_index.important | 145 |
| abstract_inverted_index.including | 288 |
| abstract_inverted_index.knowledge | 362 |
| abstract_inverted_index.nominally | 262 |
| abstract_inverted_index.phenotype | 270 |
| abstract_inverted_index.potential | 395 |
| abstract_inverted_index.principal | 151 |
| abstract_inverted_index.processes | 325 |
| abstract_inverted_index.pulmonary | 245 |
| abstract_inverted_index.reference | 120 |
| abstract_inverted_index.targeting | 338 |
| abstract_inverted_index.variants, | 250 |
| abstract_inverted_index.(N=5,788), | 186 |
| abstract_inverted_index.Annotation | 316 |
| abstract_inverted_index.Consortium | 116 |
| abstract_inverted_index.Functional | 206 |
| abstract_inverted_index.Objectives | 31 |
| abstract_inverted_index.ancestries | 312 |
| abstract_inverted_index.associated | 263, 354 |
| abstract_inverted_index.component, | 20 |
| abstract_inverted_index.consistent | 309 |
| abstract_inverted_index.consortium | 101 |
| abstract_inverted_index.contribute | 361 |
| abstract_inverted_index.downstream | 38 |
| abstract_inverted_index.endogenous | 377 |
| abstract_inverted_index.enrichment | 321 |
| abstract_inverted_index.estimating | 127 |
| abstract_inverted_index.expiratory | 65 |
| abstract_inverted_index.follow-up. | 205 |
| abstract_inverted_index.gene-level | 283 |
| abstract_inverted_index.identified | 212, 334, 350 |
| abstract_inverted_index.implicated | 285, 342 |
| abstract_inverted_index.overlapped | 236 |
| abstract_inverted_index.population | 50 |
| abstract_inverted_index.previously | 238 |
| abstract_inverted_index.regulatory | 324 |
| abstract_inverted_index.spirometry | 199 |
| abstract_inverted_index.stratified | 136 |
| abstract_inverted_index.(N=44,988), | 184 |
| abstract_inverted_index.(p<0.05) | 264 |
| abstract_inverted_index.Accelerated | 2 |
| abstract_inverted_index.Conclusions | 345 |
| abstract_inverted_index.[N=1,118]). | 278 |
| abstract_inverted_index.association | 56 |
| abstract_inverted_index.components. | 152 |
| abstract_inverted_index.confounders | 148 |
| abstract_inverted_index.contributes | 7 |
| abstract_inverted_index.development | 10 |
| abstract_inverted_index.generalized | 126 |
| abstract_inverted_index.genome-wide | 34, 55, 215 |
| abstract_inverted_index.identified. | 30 |
| abstract_inverted_index.independent | 170 |
| abstract_inverted_index.metabolism. | 330 |
| abstract_inverted_index.phenotypes, | 234 |
| abstract_inverted_index.phenotypes. | 315 |
| abstract_inverted_index.replication | 167, 259 |
| abstract_inverted_index.repurposing | 332, 396 |
| abstract_inverted_index.respiratory | 13 |
| abstract_inverted_index.significant | 216, 320 |
| abstract_inverted_index.(p<5E-08) | 217 |
| abstract_inverted_index.Epidemiology | 99 |
| abstract_inverted_index.Measurements | 173 |
| abstract_inverted_index.architecture | 366 |
| abstract_inverted_index.associations | 23, 35, 310 |
| abstract_inverted_index.biosynthesis | 328 |
| abstract_inverted_index.measurements | 200 |
| abstract_inverted_index.participants | 85, 193 |
| abstract_inverted_index.COPD-enriched | 171, 272 |
| abstract_inverted_index.functionality | 39 |
| abstract_inverted_index.meta-analyses | 349 |
| abstract_inverted_index.meta-analysis | 210 |
| abstract_inverted_index.self-reported | 182 |
| abstract_inverted_index.corticosteroid | 327 |
| abstract_inverted_index.cross-ancestry | 157 |
| abstract_inverted_index.meta-analyses. | 160 |
| abstract_inverted_index.multi-ancestry | 347 |
| abstract_inverted_index.corticosteroids | 378 |
| abstract_inverted_index.ancestry-specific | 159 |
| cited_by_percentile_year.max | 95 |
| cited_by_percentile_year.min | 91 |
| countries_distinct_count | 0 |
| institutions_distinct_count | 36 |
| sustainable_development_goals[0].id | https://metadata.un.org/sdg/3 |
| sustainable_development_goals[0].score | 0.8399999737739563 |
| sustainable_development_goals[0].display_name | Good health and well-being |
| citation_normalized_percentile.value | 0.73880644 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |