O-GlcNAcylation of boundary element associated factor (BEAF 32) in Drosophila melanogaster correlates with active histone marks at the promoters of its target genes Article Swipe
 YOU?
·
· 2017
· Open Access
·
· DOI: https://doi.org/10.1080/19491034.2017.1367887
YOU?
·
· 2017
· Open Access
·
· DOI: https://doi.org/10.1080/19491034.2017.1367887
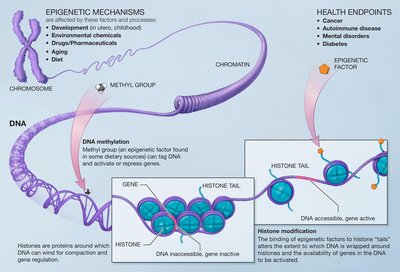
Boundary Element-Associated Factor 32 (BEAF 32) is a sequence specific DNA binding protein involved in functioning of chromatin domain boundaries in Drosophila. Several studies also show it to be involved in transcriptional regulation of a large number of genes, many of which are annotated to have cell cycle, development and differentiation related function. Since post-translational modifications (PTMs) of proteins add to their functional capacity, we investigated the PTMs on BEAF 32. The protein is known to be phosphorylated and O-GlcNAcylated. We mapped O-GlcNAc site at T91 of BEAF 32 and showed that it is linked to the deposition of active histone (H3K4me3) marks at transcription start site (TSS) of associated genes. Its role as a boundary associated factor, however, does not depend on this modification. Our study shows that by virtue of O-GlcNAcylation, BEAF 32 is linked to epigenetic mechanisms that activate a subset of associated genes.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.1080/19491034.2017.1367887
- https://www.tandfonline.com/doi/pdf/10.1080/19491034.2017.1367887?needAccess=true
- OA Status
- gold
- Cited By
- 3
- References
- 55
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W2755220396
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W2755220396Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1080/19491034.2017.1367887Digital Object Identifier
- Title
-
O-GlcNAcylation of boundary element associated factor (BEAF 32) in Drosophila melanogaster correlates with active histone marks at the promoters of its target genesWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2017Year of publication
- Publication date
-
2017-09-14Full publication date if available
- Authors
-
Debaditya De, Satish Kallappagoudar, Jae‐Min Lim, Rashmi U. Pathak, Rakesh K. MishraList of authors in order
- Landing page
-
https://doi.org/10.1080/19491034.2017.1367887Publisher landing page
- PDF URL
-
https://www.tandfonline.com/doi/pdf/10.1080/19491034.2017.1367887?needAccess=trueDirect link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
goldOpen access status per OpenAlex
- OA URL
-
https://www.tandfonline.com/doi/pdf/10.1080/19491034.2017.1367887?needAccess=trueDirect OA link when available
- Concepts
-
Chromatin, Histone, Biology, Genetics, Transcription factor, Drosophila melanogaster, Promoter, Gene, Cell biology, Epigenetics, Regulation of gene expression, Gene expressionTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
3Total citation count in OpenAlex
- Citations by year (recent)
-
2023: 1, 2022: 2Per-year citation counts (last 5 years)
- References (count)
-
55Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W2755220396 |
|---|---|
| doi | https://doi.org/10.1080/19491034.2017.1367887 |
| ids.doi | https://doi.org/10.1080/19491034.2017.1367887 |
| ids.mag | 2755220396 |
| ids.pmid | https://pubmed.ncbi.nlm.nih.gov/28910574 |
| ids.openalex | https://openalex.org/W2755220396 |
| fwci | 0.0 |
| mesh[0].qualifier_ui | Q000378 |
| mesh[0].descriptor_ui | D000117 |
| mesh[0].is_major_topic | False |
| mesh[0].qualifier_name | metabolism |
| mesh[0].descriptor_name | Acetylglucosamine |
| mesh[1].qualifier_ui | |
| mesh[1].descriptor_ui | D000818 |
| mesh[1].is_major_topic | False |
| mesh[1].qualifier_name | |
| mesh[1].descriptor_name | Animals |
| mesh[2].qualifier_ui | Q000737 |
| mesh[2].descriptor_ui | D004268 |
| mesh[2].is_major_topic | False |
| mesh[2].qualifier_name | chemistry |
| mesh[2].descriptor_name | DNA-Binding Proteins |
| mesh[3].qualifier_ui | Q000378 |
| mesh[3].descriptor_ui | D004268 |
| mesh[3].is_major_topic | False |
| mesh[3].qualifier_name | metabolism |
| mesh[3].descriptor_name | DNA-Binding Proteins |
| mesh[4].qualifier_ui | Q000737 |
| mesh[4].descriptor_ui | D029721 |
| mesh[4].is_major_topic | False |
| mesh[4].qualifier_name | chemistry |
| mesh[4].descriptor_name | Drosophila Proteins |
| mesh[5].qualifier_ui | Q000378 |
| mesh[5].descriptor_ui | D029721 |
| mesh[5].is_major_topic | False |
| mesh[5].qualifier_name | metabolism |
| mesh[5].descriptor_name | Drosophila Proteins |
| mesh[6].qualifier_ui | Q000235 |
| mesh[6].descriptor_ui | D004331 |
| mesh[6].is_major_topic | False |
| mesh[6].qualifier_name | genetics |
| mesh[6].descriptor_name | Drosophila melanogaster |
| mesh[7].qualifier_ui | Q000378 |
| mesh[7].descriptor_ui | D004331 |
| mesh[7].is_major_topic | False |
| mesh[7].qualifier_name | metabolism |
| mesh[7].descriptor_name | Drosophila melanogaster |
| mesh[8].qualifier_ui | Q000737 |
| mesh[8].descriptor_ui | D005136 |
| mesh[8].is_major_topic | False |
| mesh[8].qualifier_name | chemistry |
| mesh[8].descriptor_name | Eye Proteins |
| mesh[9].qualifier_ui | Q000378 |
| mesh[9].descriptor_ui | D005136 |
| mesh[9].is_major_topic | False |
| mesh[9].qualifier_name | metabolism |
| mesh[9].descriptor_name | Eye Proteins |
| mesh[10].qualifier_ui | Q000378 |
| mesh[10].descriptor_ui | D006657 |
| mesh[10].is_major_topic | False |
| mesh[10].qualifier_name | metabolism |
| mesh[10].descriptor_name | Histones |
| mesh[11].qualifier_ui | Q000235 |
| mesh[11].descriptor_ui | D011401 |
| mesh[11].is_major_topic | False |
| mesh[11].qualifier_name | genetics |
| mesh[11].descriptor_name | Promoter Regions, Genetic |
| mesh[12].qualifier_ui | Q000378 |
| mesh[12].descriptor_ui | D000117 |
| mesh[12].is_major_topic | False |
| mesh[12].qualifier_name | metabolism |
| mesh[12].descriptor_name | Acetylglucosamine |
| mesh[13].qualifier_ui | |
| mesh[13].descriptor_ui | D000818 |
| mesh[13].is_major_topic | False |
| mesh[13].qualifier_name | |
| mesh[13].descriptor_name | Animals |
| mesh[14].qualifier_ui | Q000737 |
| mesh[14].descriptor_ui | D004268 |
| mesh[14].is_major_topic | False |
| mesh[14].qualifier_name | chemistry |
| mesh[14].descriptor_name | DNA-Binding Proteins |
| mesh[15].qualifier_ui | Q000378 |
| mesh[15].descriptor_ui | D004268 |
| mesh[15].is_major_topic | False |
| mesh[15].qualifier_name | metabolism |
| mesh[15].descriptor_name | DNA-Binding Proteins |
| mesh[16].qualifier_ui | Q000737 |
| mesh[16].descriptor_ui | D029721 |
| mesh[16].is_major_topic | False |
| mesh[16].qualifier_name | chemistry |
| mesh[16].descriptor_name | Drosophila Proteins |
| mesh[17].qualifier_ui | Q000378 |
| mesh[17].descriptor_ui | D029721 |
| mesh[17].is_major_topic | False |
| mesh[17].qualifier_name | metabolism |
| mesh[17].descriptor_name | Drosophila Proteins |
| mesh[18].qualifier_ui | Q000235 |
| mesh[18].descriptor_ui | D004331 |
| mesh[18].is_major_topic | False |
| mesh[18].qualifier_name | genetics |
| mesh[18].descriptor_name | Drosophila melanogaster |
| mesh[19].qualifier_ui | Q000378 |
| mesh[19].descriptor_ui | D004331 |
| mesh[19].is_major_topic | False |
| mesh[19].qualifier_name | metabolism |
| mesh[19].descriptor_name | Drosophila melanogaster |
| mesh[20].qualifier_ui | Q000737 |
| mesh[20].descriptor_ui | D005136 |
| mesh[20].is_major_topic | False |
| mesh[20].qualifier_name | chemistry |
| mesh[20].descriptor_name | Eye Proteins |
| mesh[21].qualifier_ui | Q000378 |
| mesh[21].descriptor_ui | D005136 |
| mesh[21].is_major_topic | False |
| mesh[21].qualifier_name | metabolism |
| mesh[21].descriptor_name | Eye Proteins |
| mesh[22].qualifier_ui | Q000378 |
| mesh[22].descriptor_ui | D006657 |
| mesh[22].is_major_topic | False |
| mesh[22].qualifier_name | metabolism |
| mesh[22].descriptor_name | Histones |
| mesh[23].qualifier_ui | Q000235 |
| mesh[23].descriptor_ui | D011401 |
| mesh[23].is_major_topic | False |
| mesh[23].qualifier_name | genetics |
| mesh[23].descriptor_name | Promoter Regions, Genetic |
| type | article |
| title | O-GlcNAcylation of boundary element associated factor (BEAF 32) in Drosophila melanogaster correlates with active histone marks at the promoters of its target genes |
| biblio.issue | 1 |
| biblio.volume | 9 |
| biblio.last_page | 86 |
| biblio.first_page | 65 |
| topics[0].id | https://openalex.org/T10222 |
| topics[0].field.id | https://openalex.org/fields/13 |
| topics[0].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[0].score | 0.9993000030517578 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1312 |
| topics[0].subfield.display_name | Molecular Biology |
| topics[0].display_name | Genomics and Chromatin Dynamics |
| topics[1].id | https://openalex.org/T10602 |
| topics[1].field.id | https://openalex.org/fields/13 |
| topics[1].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[1].score | 0.9984999895095825 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1312 |
| topics[1].subfield.display_name | Molecular Biology |
| topics[1].display_name | Glycosylation and Glycoproteins Research |
| topics[2].id | https://openalex.org/T11041 |
| topics[2].field.id | https://openalex.org/fields/13 |
| topics[2].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[2].score | 0.9976999759674072 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1312 |
| topics[2].subfield.display_name | Molecular Biology |
| topics[2].display_name | Ubiquitin and proteasome pathways |
| is_xpac | False |
| apc_list.value | 1625 |
| apc_list.currency | GBP |
| apc_list.value_usd | 1993 |
| apc_paid.value | 1625 |
| apc_paid.currency | GBP |
| apc_paid.value_usd | 1993 |
| concepts[0].id | https://openalex.org/C83640560 |
| concepts[0].level | 3 |
| concepts[0].score | 0.6700718998908997 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q180951 |
| concepts[0].display_name | Chromatin |
| concepts[1].id | https://openalex.org/C64927066 |
| concepts[1].level | 3 |
| concepts[1].score | 0.6353249549865723 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q36293 |
| concepts[1].display_name | Histone |
| concepts[2].id | https://openalex.org/C86803240 |
| concepts[2].level | 0 |
| concepts[2].score | 0.5727463364601135 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[2].display_name | Biology |
| concepts[3].id | https://openalex.org/C54355233 |
| concepts[3].level | 1 |
| concepts[3].score | 0.562188446521759 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[3].display_name | Genetics |
| concepts[4].id | https://openalex.org/C86339819 |
| concepts[4].level | 3 |
| concepts[4].score | 0.537690281867981 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q407384 |
| concepts[4].display_name | Transcription factor |
| concepts[5].id | https://openalex.org/C2780104201 |
| concepts[5].level | 3 |
| concepts[5].score | 0.4989471435546875 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q130888 |
| concepts[5].display_name | Drosophila melanogaster |
| concepts[6].id | https://openalex.org/C101762097 |
| concepts[6].level | 4 |
| concepts[6].score | 0.4966338276863098 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q224093 |
| concepts[6].display_name | Promoter |
| concepts[7].id | https://openalex.org/C104317684 |
| concepts[7].level | 2 |
| concepts[7].score | 0.4963584542274475 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[7].display_name | Gene |
| concepts[8].id | https://openalex.org/C95444343 |
| concepts[8].level | 1 |
| concepts[8].score | 0.48787951469421387 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q7141 |
| concepts[8].display_name | Cell biology |
| concepts[9].id | https://openalex.org/C41091548 |
| concepts[9].level | 3 |
| concepts[9].score | 0.4530307352542877 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q26939 |
| concepts[9].display_name | Epigenetics |
| concepts[10].id | https://openalex.org/C165864922 |
| concepts[10].level | 3 |
| concepts[10].score | 0.43394702672958374 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q411391 |
| concepts[10].display_name | Regulation of gene expression |
| concepts[11].id | https://openalex.org/C150194340 |
| concepts[11].level | 3 |
| concepts[11].score | 0.2328580915927887 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q26972 |
| concepts[11].display_name | Gene expression |
| keywords[0].id | https://openalex.org/keywords/chromatin |
| keywords[0].score | 0.6700718998908997 |
| keywords[0].display_name | Chromatin |
| keywords[1].id | https://openalex.org/keywords/histone |
| keywords[1].score | 0.6353249549865723 |
| keywords[1].display_name | Histone |
| keywords[2].id | https://openalex.org/keywords/biology |
| keywords[2].score | 0.5727463364601135 |
| keywords[2].display_name | Biology |
| keywords[3].id | https://openalex.org/keywords/genetics |
| keywords[3].score | 0.562188446521759 |
| keywords[3].display_name | Genetics |
| keywords[4].id | https://openalex.org/keywords/transcription-factor |
| keywords[4].score | 0.537690281867981 |
| keywords[4].display_name | Transcription factor |
| keywords[5].id | https://openalex.org/keywords/drosophila-melanogaster |
| keywords[5].score | 0.4989471435546875 |
| keywords[5].display_name | Drosophila melanogaster |
| keywords[6].id | https://openalex.org/keywords/promoter |
| keywords[6].score | 0.4966338276863098 |
| keywords[6].display_name | Promoter |
| keywords[7].id | https://openalex.org/keywords/gene |
| keywords[7].score | 0.4963584542274475 |
| keywords[7].display_name | Gene |
| keywords[8].id | https://openalex.org/keywords/cell-biology |
| keywords[8].score | 0.48787951469421387 |
| keywords[8].display_name | Cell biology |
| keywords[9].id | https://openalex.org/keywords/epigenetics |
| keywords[9].score | 0.4530307352542877 |
| keywords[9].display_name | Epigenetics |
| keywords[10].id | https://openalex.org/keywords/regulation-of-gene-expression |
| keywords[10].score | 0.43394702672958374 |
| keywords[10].display_name | Regulation of gene expression |
| keywords[11].id | https://openalex.org/keywords/gene-expression |
| keywords[11].score | 0.2328580915927887 |
| keywords[11].display_name | Gene expression |
| language | en |
| locations[0].id | doi:10.1080/19491034.2017.1367887 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S200139231 |
| locations[0].source.issn | 1949-1034, 1949-1042 |
| locations[0].source.type | journal |
| locations[0].source.is_oa | True |
| locations[0].source.issn_l | 1949-1034 |
| locations[0].source.is_core | True |
| locations[0].source.is_in_doaj | True |
| locations[0].source.display_name | Nucleus |
| locations[0].source.host_organization | https://openalex.org/P4310320547 |
| locations[0].source.host_organization_name | Taylor & Francis |
| locations[0].source.host_organization_lineage | https://openalex.org/P4310320547 |
| locations[0].source.host_organization_lineage_names | Taylor & Francis |
| locations[0].license | |
| locations[0].pdf_url | https://www.tandfonline.com/doi/pdf/10.1080/19491034.2017.1367887?needAccess=true |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | Nucleus |
| locations[0].landing_page_url | https://doi.org/10.1080/19491034.2017.1367887 |
| locations[1].id | pmid:28910574 |
| locations[1].is_oa | False |
| locations[1].source.id | https://openalex.org/S4306525036 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | PubMed |
| locations[1].source.host_organization | https://openalex.org/I1299303238 |
| locations[1].source.host_organization_name | National Institutes of Health |
| locations[1].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[1].license | |
| locations[1].pdf_url | |
| locations[1].version | publishedVersion |
| locations[1].raw_type | |
| locations[1].license_id | |
| locations[1].is_accepted | True |
| locations[1].is_published | True |
| locations[1].raw_source_name | Nucleus (Austin, Tex.) |
| locations[1].landing_page_url | https://pubmed.ncbi.nlm.nih.gov/28910574 |
| locations[2].id | pmh:oai:pubmedcentral.nih.gov:5973196 |
| locations[2].is_oa | True |
| locations[2].source.id | https://openalex.org/S2764455111 |
| locations[2].source.issn | |
| locations[2].source.type | repository |
| locations[2].source.is_oa | False |
| locations[2].source.issn_l | |
| locations[2].source.is_core | False |
| locations[2].source.is_in_doaj | False |
| locations[2].source.display_name | PubMed Central |
| locations[2].source.host_organization | https://openalex.org/I1299303238 |
| locations[2].source.host_organization_name | National Institutes of Health |
| locations[2].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[2].license | cc-by |
| locations[2].pdf_url | |
| locations[2].version | submittedVersion |
| locations[2].raw_type | Text |
| locations[2].license_id | https://openalex.org/licenses/cc-by |
| locations[2].is_accepted | False |
| locations[2].is_published | False |
| locations[2].raw_source_name | Nucleus |
| locations[2].landing_page_url | https://www.ncbi.nlm.nih.gov/pmc/articles/5973196 |
| indexed_in | crossref, doaj, pubmed |
| authorships[0].author.id | https://openalex.org/A5030569374 |
| authorships[0].author.orcid | |
| authorships[0].author.display_name | Debaditya De |
| authorships[0].countries | IN |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I185638415 |
| authorships[0].affiliations[0].raw_affiliation_string | CSIR-Centre for Cellular and Molecular Biology, Hyderabad, India |
| authorships[0].institutions[0].id | https://openalex.org/I185638415 |
| authorships[0].institutions[0].ror | https://ror.org/05shq4n12 |
| authorships[0].institutions[0].type | facility |
| authorships[0].institutions[0].lineage | https://openalex.org/I185638415, https://openalex.org/I2799351866, https://openalex.org/I4210134808, https://openalex.org/I66760702 |
| authorships[0].institutions[0].country_code | IN |
| authorships[0].institutions[0].display_name | Centre for Cellular and Molecular Biology |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Debaditya De |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | CSIR-Centre for Cellular and Molecular Biology, Hyderabad, India |
| authorships[1].author.id | https://openalex.org/A5007934754 |
| authorships[1].author.orcid | https://orcid.org/0000-0001-9557-176X |
| authorships[1].author.display_name | Satish Kallappagoudar |
| authorships[1].countries | IN |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I185638415 |
| authorships[1].affiliations[0].raw_affiliation_string | CSIR-Centre for Cellular and Molecular Biology, Hyderabad, India |
| authorships[1].institutions[0].id | https://openalex.org/I185638415 |
| authorships[1].institutions[0].ror | https://ror.org/05shq4n12 |
| authorships[1].institutions[0].type | facility |
| authorships[1].institutions[0].lineage | https://openalex.org/I185638415, https://openalex.org/I2799351866, https://openalex.org/I4210134808, https://openalex.org/I66760702 |
| authorships[1].institutions[0].country_code | IN |
| authorships[1].institutions[0].display_name | Centre for Cellular and Molecular Biology |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Satish Kallappagoudar |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | CSIR-Centre for Cellular and Molecular Biology, Hyderabad, India |
| authorships[2].author.id | https://openalex.org/A5085947482 |
| authorships[2].author.orcid | https://orcid.org/0000-0002-7153-0194 |
| authorships[2].author.display_name | Jae‐Min Lim |
| authorships[2].countries | KR |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I174101054 |
| authorships[2].affiliations[0].raw_affiliation_string | Department of Chemistry, Changwon National University, Changwon, Gyeongnam, South Korea |
| authorships[2].institutions[0].id | https://openalex.org/I174101054 |
| authorships[2].institutions[0].ror | https://ror.org/04ts4qa58 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I174101054 |
| authorships[2].institutions[0].country_code | KR |
| authorships[2].institutions[0].display_name | Changwon National University |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Jae-Min Lim |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | Department of Chemistry, Changwon National University, Changwon, Gyeongnam, South Korea |
| authorships[3].author.id | https://openalex.org/A5068728283 |
| authorships[3].author.orcid | https://orcid.org/0000-0002-0328-9300 |
| authorships[3].author.display_name | Rashmi U. Pathak |
| authorships[3].countries | IN |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I185638415 |
| authorships[3].affiliations[0].raw_affiliation_string | CSIR-Centre for Cellular and Molecular Biology, Hyderabad, India |
| authorships[3].institutions[0].id | https://openalex.org/I185638415 |
| authorships[3].institutions[0].ror | https://ror.org/05shq4n12 |
| authorships[3].institutions[0].type | facility |
| authorships[3].institutions[0].lineage | https://openalex.org/I185638415, https://openalex.org/I2799351866, https://openalex.org/I4210134808, https://openalex.org/I66760702 |
| authorships[3].institutions[0].country_code | IN |
| authorships[3].institutions[0].display_name | Centre for Cellular and Molecular Biology |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Rashmi U. Pathak |
| authorships[3].is_corresponding | True |
| authorships[3].raw_affiliation_strings | CSIR-Centre for Cellular and Molecular Biology, Hyderabad, India |
| authorships[4].author.id | https://openalex.org/A5100674488 |
| authorships[4].author.orcid | https://orcid.org/0000-0001-6636-7380 |
| authorships[4].author.display_name | Rakesh K. Mishra |
| authorships[4].countries | IN |
| authorships[4].affiliations[0].institution_ids | https://openalex.org/I185638415 |
| authorships[4].affiliations[0].raw_affiliation_string | CSIR-Centre for Cellular and Molecular Biology, Hyderabad, India |
| authorships[4].institutions[0].id | https://openalex.org/I185638415 |
| authorships[4].institutions[0].ror | https://ror.org/05shq4n12 |
| authorships[4].institutions[0].type | facility |
| authorships[4].institutions[0].lineage | https://openalex.org/I185638415, https://openalex.org/I2799351866, https://openalex.org/I4210134808, https://openalex.org/I66760702 |
| authorships[4].institutions[0].country_code | IN |
| authorships[4].institutions[0].display_name | Centre for Cellular and Molecular Biology |
| authorships[4].author_position | last |
| authorships[4].raw_author_name | Rakesh K. Mishra |
| authorships[4].is_corresponding | True |
| authorships[4].raw_affiliation_strings | CSIR-Centre for Cellular and Molecular Biology, Hyderabad, India |
| has_content.pdf | True |
| has_content.grobid_xml | True |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://www.tandfonline.com/doi/pdf/10.1080/19491034.2017.1367887?needAccess=true |
| open_access.oa_status | gold |
| open_access.any_repository_has_fulltext | False |
| created_date | 2017-09-25T00:00:00 |
| display_name | O-GlcNAcylation of boundary element associated factor (BEAF 32) in Drosophila melanogaster correlates with active histone marks at the promoters of its target genes |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T10222 |
| primary_topic.field.id | https://openalex.org/fields/13 |
| primary_topic.field.display_name | Biochemistry, Genetics and Molecular Biology |
| primary_topic.score | 0.9993000030517578 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1312 |
| primary_topic.subfield.display_name | Molecular Biology |
| primary_topic.display_name | Genomics and Chromatin Dynamics |
| related_works | https://openalex.org/W2587366952, https://openalex.org/W2393920775, https://openalex.org/W1969771092, https://openalex.org/W2014093629, https://openalex.org/W98379584, https://openalex.org/W2413824894, https://openalex.org/W2030159793, https://openalex.org/W3145419749, https://openalex.org/W3038565614, https://openalex.org/W2581702729 |
| cited_by_count | 3 |
| counts_by_year[0].year | 2023 |
| counts_by_year[0].cited_by_count | 1 |
| counts_by_year[1].year | 2022 |
| counts_by_year[1].cited_by_count | 2 |
| locations_count | 3 |
| best_oa_location.id | doi:10.1080/19491034.2017.1367887 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S200139231 |
| best_oa_location.source.issn | 1949-1034, 1949-1042 |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | True |
| best_oa_location.source.issn_l | 1949-1034 |
| best_oa_location.source.is_core | True |
| best_oa_location.source.is_in_doaj | True |
| best_oa_location.source.display_name | Nucleus |
| best_oa_location.source.host_organization | https://openalex.org/P4310320547 |
| best_oa_location.source.host_organization_name | Taylor & Francis |
| best_oa_location.source.host_organization_lineage | https://openalex.org/P4310320547 |
| best_oa_location.source.host_organization_lineage_names | Taylor & Francis |
| best_oa_location.license | |
| best_oa_location.pdf_url | https://www.tandfonline.com/doi/pdf/10.1080/19491034.2017.1367887?needAccess=true |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | journal-article |
| best_oa_location.license_id | |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | Nucleus |
| best_oa_location.landing_page_url | https://doi.org/10.1080/19491034.2017.1367887 |
| primary_location.id | doi:10.1080/19491034.2017.1367887 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S200139231 |
| primary_location.source.issn | 1949-1034, 1949-1042 |
| primary_location.source.type | journal |
| primary_location.source.is_oa | True |
| primary_location.source.issn_l | 1949-1034 |
| primary_location.source.is_core | True |
| primary_location.source.is_in_doaj | True |
| primary_location.source.display_name | Nucleus |
| primary_location.source.host_organization | https://openalex.org/P4310320547 |
| primary_location.source.host_organization_name | Taylor & Francis |
| primary_location.source.host_organization_lineage | https://openalex.org/P4310320547 |
| primary_location.source.host_organization_lineage_names | Taylor & Francis |
| primary_location.license | |
| primary_location.pdf_url | https://www.tandfonline.com/doi/pdf/10.1080/19491034.2017.1367887?needAccess=true |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | Nucleus |
| primary_location.landing_page_url | https://doi.org/10.1080/19491034.2017.1367887 |
| publication_date | 2017-09-14 |
| publication_year | 2017 |
| referenced_works | https://openalex.org/W2159192458, https://openalex.org/W1600994582, https://openalex.org/W1832452463, https://openalex.org/W2161638097, https://openalex.org/W2157382629, https://openalex.org/W2006669822, https://openalex.org/W1970321798, https://openalex.org/W2042500803, https://openalex.org/W2070021921, https://openalex.org/W2056482755, https://openalex.org/W2157576490, https://openalex.org/W2172400873, https://openalex.org/W2088016816, https://openalex.org/W2029722028, https://openalex.org/W1989373989, https://openalex.org/W2106895159, https://openalex.org/W2095400582, https://openalex.org/W2136583745, https://openalex.org/W2137769752, https://openalex.org/W2145420651, https://openalex.org/W2123017585, https://openalex.org/W2146743044, https://openalex.org/W2331889471, https://openalex.org/W2116942623, https://openalex.org/W2099104943, https://openalex.org/W2087243573, https://openalex.org/W2171125622, https://openalex.org/W2161559093, https://openalex.org/W2102282402, https://openalex.org/W2087172939, https://openalex.org/W2105427468, https://openalex.org/W1994598776, https://openalex.org/W2157954574, https://openalex.org/W2165299990, https://openalex.org/W2011609862, https://openalex.org/W2120176673, https://openalex.org/W2018658338, https://openalex.org/W2077309544, https://openalex.org/W2105816640, https://openalex.org/W1996786134, https://openalex.org/W2016172184, https://openalex.org/W2065955997, https://openalex.org/W2061011762, https://openalex.org/W2099766227, https://openalex.org/W2035837644, https://openalex.org/W2067423895, https://openalex.org/W2053017792, https://openalex.org/W2016015885, https://openalex.org/W1522904209, https://openalex.org/W2035281138, https://openalex.org/W2108966929, https://openalex.org/W2002644225, https://openalex.org/W2167671063, https://openalex.org/W2107279676, https://openalex.org/W2112551809 |
| referenced_works_count | 55 |
| abstract_inverted_index.a | 7, 34, 114, 142 |
| abstract_inverted_index.32 | 3, 88, 134 |
| abstract_inverted_index.We | 80 |
| abstract_inverted_index.as | 113 |
| abstract_inverted_index.at | 84, 103 |
| abstract_inverted_index.be | 28, 76 |
| abstract_inverted_index.by | 129 |
| abstract_inverted_index.in | 14, 20, 30 |
| abstract_inverted_index.is | 6, 73, 93, 135 |
| abstract_inverted_index.it | 26, 92 |
| abstract_inverted_index.of | 16, 33, 37, 40, 57, 86, 98, 108, 131, 144 |
| abstract_inverted_index.on | 68, 122 |
| abstract_inverted_index.to | 27, 44, 60, 75, 95, 137 |
| abstract_inverted_index.we | 64 |
| abstract_inverted_index.32) | 5 |
| abstract_inverted_index.32. | 70 |
| abstract_inverted_index.DNA | 10 |
| abstract_inverted_index.Its | 111 |
| abstract_inverted_index.Our | 125 |
| abstract_inverted_index.T91 | 85 |
| abstract_inverted_index.The | 71 |
| abstract_inverted_index.add | 59 |
| abstract_inverted_index.and | 49, 78, 89 |
| abstract_inverted_index.are | 42 |
| abstract_inverted_index.not | 120 |
| abstract_inverted_index.the | 66, 96 |
| abstract_inverted_index.BEAF | 69, 87, 133 |
| abstract_inverted_index.PTMs | 67 |
| abstract_inverted_index.also | 24 |
| abstract_inverted_index.cell | 46 |
| abstract_inverted_index.does | 119 |
| abstract_inverted_index.have | 45 |
| abstract_inverted_index.many | 39 |
| abstract_inverted_index.role | 112 |
| abstract_inverted_index.show | 25 |
| abstract_inverted_index.site | 83, 106 |
| abstract_inverted_index.that | 91, 128, 140 |
| abstract_inverted_index.this | 123 |
| abstract_inverted_index.(BEAF | 4 |
| abstract_inverted_index.(TSS) | 107 |
| abstract_inverted_index.Since | 53 |
| abstract_inverted_index.known | 74 |
| abstract_inverted_index.large | 35 |
| abstract_inverted_index.marks | 102 |
| abstract_inverted_index.shows | 127 |
| abstract_inverted_index.start | 105 |
| abstract_inverted_index.study | 126 |
| abstract_inverted_index.their | 61 |
| abstract_inverted_index.which | 41 |
| abstract_inverted_index.(PTMs) | 56 |
| abstract_inverted_index.Factor | 2 |
| abstract_inverted_index.active | 99 |
| abstract_inverted_index.cycle, | 47 |
| abstract_inverted_index.depend | 121 |
| abstract_inverted_index.domain | 18 |
| abstract_inverted_index.genes, | 38 |
| abstract_inverted_index.genes. | 110, 146 |
| abstract_inverted_index.linked | 94, 136 |
| abstract_inverted_index.mapped | 81 |
| abstract_inverted_index.number | 36 |
| abstract_inverted_index.showed | 90 |
| abstract_inverted_index.subset | 143 |
| abstract_inverted_index.virtue | 130 |
| abstract_inverted_index.Several | 22 |
| abstract_inverted_index.binding | 11 |
| abstract_inverted_index.factor, | 117 |
| abstract_inverted_index.histone | 100 |
| abstract_inverted_index.protein | 12, 72 |
| abstract_inverted_index.related | 51 |
| abstract_inverted_index.studies | 23 |
| abstract_inverted_index.Boundary | 0 |
| abstract_inverted_index.O-GlcNAc | 82 |
| abstract_inverted_index.activate | 141 |
| abstract_inverted_index.boundary | 115 |
| abstract_inverted_index.however, | 118 |
| abstract_inverted_index.involved | 13, 29 |
| abstract_inverted_index.proteins | 58 |
| abstract_inverted_index.sequence | 8 |
| abstract_inverted_index.specific | 9 |
| abstract_inverted_index.(H3K4me3) | 101 |
| abstract_inverted_index.annotated | 43 |
| abstract_inverted_index.capacity, | 63 |
| abstract_inverted_index.chromatin | 17 |
| abstract_inverted_index.function. | 52 |
| abstract_inverted_index.associated | 109, 116, 145 |
| abstract_inverted_index.boundaries | 19 |
| abstract_inverted_index.deposition | 97 |
| abstract_inverted_index.epigenetic | 138 |
| abstract_inverted_index.functional | 62 |
| abstract_inverted_index.mechanisms | 139 |
| abstract_inverted_index.regulation | 32 |
| abstract_inverted_index.Drosophila. | 21 |
| abstract_inverted_index.development | 48 |
| abstract_inverted_index.functioning | 15 |
| abstract_inverted_index.investigated | 65 |
| abstract_inverted_index.modification. | 124 |
| abstract_inverted_index.modifications | 55 |
| abstract_inverted_index.transcription | 104 |
| abstract_inverted_index.phosphorylated | 77 |
| abstract_inverted_index.O-GlcNAcylated. | 79 |
| abstract_inverted_index.differentiation | 50 |
| abstract_inverted_index.transcriptional | 31 |
| abstract_inverted_index.O-GlcNAcylation, | 132 |
| abstract_inverted_index.Element-Associated | 1 |
| abstract_inverted_index.post-translational | 54 |
| cited_by_percentile_year.max | 96 |
| cited_by_percentile_year.min | 89 |
| corresponding_author_ids | https://openalex.org/A5100674488, https://openalex.org/A5068728283 |
| countries_distinct_count | 2 |
| institutions_distinct_count | 5 |
| corresponding_institution_ids | https://openalex.org/I185638415 |
| citation_normalized_percentile.value | 0.10283271 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |