PathwayKO: An integrated platform for deciphering the systems-level signaling pathways Article Swipe
 YOU?
·
· 2023
· Open Access
·
· DOI: https://doi.org/10.3389/fimmu.2023.1103392
YOU?
·
· 2023
· Open Access
·
· DOI: https://doi.org/10.3389/fimmu.2023.1103392
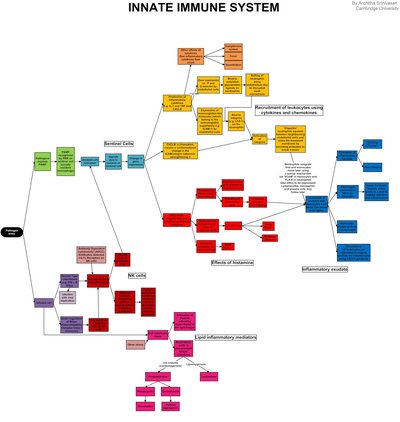
Systems characterization of immune landscapes in health, disease and clinical intervention cases is a priority in modern medicine. High-throughput transcriptomes accumulated from gene-knockout (KO) experiments are crucial for deciphering target KO signaling pathways that are impaired by KO genes at the systems-level. There is a demand for integrative platforms. This article describes the PathwayKO platform, which has integrated state-of-the-art methods of pathway enrichment analysis, statistics analysis, and visualizing analysis to conduct cutting-edge integrative pathway analysis in a pipeline fashion and decipher target KO signaling pathways at the systems-level. We focus on describing the methodology, principles and application features of PathwayKO. First, we demonstrate that the PathwayKO platform can be utilized to comprehensively analyze real-world mouse KO transcriptomes (GSE22873 and GSE24327), which reveal systemic mechanisms underlying the innate immune responses triggered by non-infectious extensive hepatectomy (2 hours after 85% liver resection surgery) and infectious CASP-model sepsis (12 hours after CASP-model surgery). Strikingly, our results indicate that both cases hit the same core set of 21 KO MyD88-associated signaling pathways, including the Toll-like receptor signaling pathway, the NFκB signaling pathway, the MAPK signaling pathway, and the PD-L1 expression and PD-1 checkpoint pathway in cancer, alongside the pathways of bacterial, viral and parasitic infections. These findings suggest common fundamental mechanisms between these immune responses and offer informative cues that warrant future experimental validation. Such mechanisms in mice may serve as models for humans and ultimately guide formulating the research paradigms and composite strategies to reduce the high mortality rates of patients in intensive care units who have undergone successful traumatic surgical treatments. Second, we demonstrate that the PathwayKO platform model-based assessments can effectively evaluate the performance difference of pathway analysis methods when benchmarked with a collection of proper transcriptomes. Together, such advances in methods for deciphering biological insights at the systems-level may benefit the fields of bioinformatics, systems immunology and beyond.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.3389/fimmu.2023.1103392
- OA Status
- gold
- References
- 49
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4360863139
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4360863139Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.3389/fimmu.2023.1103392Digital Object Identifier
- Title
-
PathwayKO: An integrated platform for deciphering the systems-level signaling pathwaysWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2023Year of publication
- Publication date
-
2023-03-23Full publication date if available
- Authors
-
Hannan Ai, Fanmei Meng, Yuncan AiList of authors in order
- Landing page
-
https://doi.org/10.3389/fimmu.2023.1103392Publisher landing page
- Open access
-
YesWhether a free full text is available
- OA status
-
goldOpen access status per OpenAlex
- OA URL
-
https://doi.org/10.3389/fimmu.2023.1103392Direct OA link when available
- Concepts
-
Signal transduction, Transcriptome, Computational biology, Biology, Systems biology, Innate immune system, Immune system, Alternative complement pathway, Biological pathway, Bioinformatics, Complement system, Medicine, Immunology, Gene, Cell biology, Genetics, Gene expressionTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
0Total citation count in OpenAlex
- References (count)
-
49Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4360863139 |
|---|---|
| doi | https://doi.org/10.3389/fimmu.2023.1103392 |
| ids.doi | https://doi.org/10.3389/fimmu.2023.1103392 |
| ids.pmid | https://pubmed.ncbi.nlm.nih.gov/37033947 |
| ids.openalex | https://openalex.org/W4360863139 |
| fwci | 0.0 |
| mesh[0].qualifier_ui | |
| mesh[0].descriptor_ui | D006801 |
| mesh[0].is_major_topic | False |
| mesh[0].qualifier_name | |
| mesh[0].descriptor_name | Humans |
| mesh[1].qualifier_ui | |
| mesh[1].descriptor_ui | D051379 |
| mesh[1].is_major_topic | False |
| mesh[1].qualifier_name | |
| mesh[1].descriptor_name | Mice |
| mesh[2].qualifier_ui | |
| mesh[2].descriptor_ui | D000818 |
| mesh[2].is_major_topic | False |
| mesh[2].qualifier_name | |
| mesh[2].descriptor_name | Animals |
| mesh[3].qualifier_ui | Q000235 |
| mesh[3].descriptor_ui | D015398 |
| mesh[3].is_major_topic | True |
| mesh[3].qualifier_name | genetics |
| mesh[3].descriptor_name | Signal Transduction |
| mesh[4].qualifier_ui | Q000378 |
| mesh[4].descriptor_ui | D008099 |
| mesh[4].is_major_topic | True |
| mesh[4].qualifier_name | metabolism |
| mesh[4].descriptor_name | Liver |
| mesh[5].qualifier_ui | Q000378 |
| mesh[5].descriptor_ui | D016328 |
| mesh[5].is_major_topic | False |
| mesh[5].qualifier_name | metabolism |
| mesh[5].descriptor_name | NF-kappa B |
| mesh[6].qualifier_ui | |
| mesh[6].descriptor_ui | D007113 |
| mesh[6].is_major_topic | False |
| mesh[6].qualifier_name | |
| mesh[6].descriptor_name | Immunity, Innate |
| mesh[7].qualifier_ui | |
| mesh[7].descriptor_ui | D059467 |
| mesh[7].is_major_topic | False |
| mesh[7].qualifier_name | |
| mesh[7].descriptor_name | Transcriptome |
| mesh[8].qualifier_ui | |
| mesh[8].descriptor_ui | D006801 |
| mesh[8].is_major_topic | False |
| mesh[8].qualifier_name | |
| mesh[8].descriptor_name | Humans |
| mesh[9].qualifier_ui | |
| mesh[9].descriptor_ui | D051379 |
| mesh[9].is_major_topic | False |
| mesh[9].qualifier_name | |
| mesh[9].descriptor_name | Mice |
| mesh[10].qualifier_ui | |
| mesh[10].descriptor_ui | D000818 |
| mesh[10].is_major_topic | False |
| mesh[10].qualifier_name | |
| mesh[10].descriptor_name | Animals |
| mesh[11].qualifier_ui | Q000235 |
| mesh[11].descriptor_ui | D015398 |
| mesh[11].is_major_topic | True |
| mesh[11].qualifier_name | genetics |
| mesh[11].descriptor_name | Signal Transduction |
| mesh[12].qualifier_ui | Q000378 |
| mesh[12].descriptor_ui | D008099 |
| mesh[12].is_major_topic | True |
| mesh[12].qualifier_name | metabolism |
| mesh[12].descriptor_name | Liver |
| mesh[13].qualifier_ui | Q000378 |
| mesh[13].descriptor_ui | D016328 |
| mesh[13].is_major_topic | False |
| mesh[13].qualifier_name | metabolism |
| mesh[13].descriptor_name | NF-kappa B |
| mesh[14].qualifier_ui | |
| mesh[14].descriptor_ui | D007113 |
| mesh[14].is_major_topic | False |
| mesh[14].qualifier_name | |
| mesh[14].descriptor_name | Immunity, Innate |
| mesh[15].qualifier_ui | |
| mesh[15].descriptor_ui | D059467 |
| mesh[15].is_major_topic | False |
| mesh[15].qualifier_name | |
| mesh[15].descriptor_name | Transcriptome |
| type | article |
| title | PathwayKO: An integrated platform for deciphering the systems-level signaling pathways |
| biblio.issue | |
| biblio.volume | 14 |
| biblio.last_page | 1103392 |
| biblio.first_page | 1103392 |
| topics[0].id | https://openalex.org/T10211 |
| topics[0].field.id | https://openalex.org/fields/17 |
| topics[0].field.display_name | Computer Science |
| topics[0].score | 0.9918000102043152 |
| topics[0].domain.id | https://openalex.org/domains/3 |
| topics[0].domain.display_name | Physical Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1703 |
| topics[0].subfield.display_name | Computational Theory and Mathematics |
| topics[0].display_name | Computational Drug Discovery Methods |
| topics[1].id | https://openalex.org/T10678 |
| topics[1].field.id | https://openalex.org/fields/27 |
| topics[1].field.display_name | Medicine |
| topics[1].score | 0.991100013256073 |
| topics[1].domain.id | https://openalex.org/domains/4 |
| topics[1].domain.display_name | Health Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/2736 |
| topics[1].subfield.display_name | Pharmacology |
| topics[1].display_name | Inflammatory mediators and NSAID effects |
| topics[2].id | https://openalex.org/T11591 |
| topics[2].field.id | https://openalex.org/fields/13 |
| topics[2].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[2].score | 0.9879999756813049 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1306 |
| topics[2].subfield.display_name | Cancer Research |
| topics[2].display_name | NF-κB Signaling Pathways |
| is_xpac | False |
| apc_list.value | 2950 |
| apc_list.currency | USD |
| apc_list.value_usd | 2950 |
| apc_paid.value | 2950 |
| apc_paid.currency | USD |
| apc_paid.value_usd | 2950 |
| concepts[0].id | https://openalex.org/C62478195 |
| concepts[0].level | 2 |
| concepts[0].score | 0.6626632213592529 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q828130 |
| concepts[0].display_name | Signal transduction |
| concepts[1].id | https://openalex.org/C162317418 |
| concepts[1].level | 4 |
| concepts[1].score | 0.5952986478805542 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q252857 |
| concepts[1].display_name | Transcriptome |
| concepts[2].id | https://openalex.org/C70721500 |
| concepts[2].level | 1 |
| concepts[2].score | 0.5785784125328064 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q177005 |
| concepts[2].display_name | Computational biology |
| concepts[3].id | https://openalex.org/C86803240 |
| concepts[3].level | 0 |
| concepts[3].score | 0.5516065359115601 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[3].display_name | Biology |
| concepts[4].id | https://openalex.org/C152662350 |
| concepts[4].level | 2 |
| concepts[4].score | 0.541509211063385 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q815297 |
| concepts[4].display_name | Systems biology |
| concepts[5].id | https://openalex.org/C136449434 |
| concepts[5].level | 3 |
| concepts[5].score | 0.4985694885253906 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q428253 |
| concepts[5].display_name | Innate immune system |
| concepts[6].id | https://openalex.org/C8891405 |
| concepts[6].level | 2 |
| concepts[6].score | 0.4535747766494751 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q1059 |
| concepts[6].display_name | Immune system |
| concepts[7].id | https://openalex.org/C189446657 |
| concepts[7].level | 4 |
| concepts[7].score | 0.45074185729026794 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q4736500 |
| concepts[7].display_name | Alternative complement pathway |
| concepts[8].id | https://openalex.org/C9927688 |
| concepts[8].level | 4 |
| concepts[8].score | 0.42500805854797363 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q4915012 |
| concepts[8].display_name | Biological pathway |
| concepts[9].id | https://openalex.org/C60644358 |
| concepts[9].level | 1 |
| concepts[9].score | 0.41033878922462463 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q128570 |
| concepts[9].display_name | Bioinformatics |
| concepts[10].id | https://openalex.org/C111684460 |
| concepts[10].level | 3 |
| concepts[10].score | 0.3918585777282715 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q334848 |
| concepts[10].display_name | Complement system |
| concepts[11].id | https://openalex.org/C71924100 |
| concepts[11].level | 0 |
| concepts[11].score | 0.33196157217025757 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q11190 |
| concepts[11].display_name | Medicine |
| concepts[12].id | https://openalex.org/C203014093 |
| concepts[12].level | 1 |
| concepts[12].score | 0.29818195104599 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q101929 |
| concepts[12].display_name | Immunology |
| concepts[13].id | https://openalex.org/C104317684 |
| concepts[13].level | 2 |
| concepts[13].score | 0.2548319101333618 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[13].display_name | Gene |
| concepts[14].id | https://openalex.org/C95444343 |
| concepts[14].level | 1 |
| concepts[14].score | 0.19973719120025635 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q7141 |
| concepts[14].display_name | Cell biology |
| concepts[15].id | https://openalex.org/C54355233 |
| concepts[15].level | 1 |
| concepts[15].score | 0.18328434228897095 |
| concepts[15].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[15].display_name | Genetics |
| concepts[16].id | https://openalex.org/C150194340 |
| concepts[16].level | 3 |
| concepts[16].score | 0.1492024064064026 |
| concepts[16].wikidata | https://www.wikidata.org/wiki/Q26972 |
| concepts[16].display_name | Gene expression |
| keywords[0].id | https://openalex.org/keywords/signal-transduction |
| keywords[0].score | 0.6626632213592529 |
| keywords[0].display_name | Signal transduction |
| keywords[1].id | https://openalex.org/keywords/transcriptome |
| keywords[1].score | 0.5952986478805542 |
| keywords[1].display_name | Transcriptome |
| keywords[2].id | https://openalex.org/keywords/computational-biology |
| keywords[2].score | 0.5785784125328064 |
| keywords[2].display_name | Computational biology |
| keywords[3].id | https://openalex.org/keywords/biology |
| keywords[3].score | 0.5516065359115601 |
| keywords[3].display_name | Biology |
| keywords[4].id | https://openalex.org/keywords/systems-biology |
| keywords[4].score | 0.541509211063385 |
| keywords[4].display_name | Systems biology |
| keywords[5].id | https://openalex.org/keywords/innate-immune-system |
| keywords[5].score | 0.4985694885253906 |
| keywords[5].display_name | Innate immune system |
| keywords[6].id | https://openalex.org/keywords/immune-system |
| keywords[6].score | 0.4535747766494751 |
| keywords[6].display_name | Immune system |
| keywords[7].id | https://openalex.org/keywords/alternative-complement-pathway |
| keywords[7].score | 0.45074185729026794 |
| keywords[7].display_name | Alternative complement pathway |
| keywords[8].id | https://openalex.org/keywords/biological-pathway |
| keywords[8].score | 0.42500805854797363 |
| keywords[8].display_name | Biological pathway |
| keywords[9].id | https://openalex.org/keywords/bioinformatics |
| keywords[9].score | 0.41033878922462463 |
| keywords[9].display_name | Bioinformatics |
| keywords[10].id | https://openalex.org/keywords/complement-system |
| keywords[10].score | 0.3918585777282715 |
| keywords[10].display_name | Complement system |
| keywords[11].id | https://openalex.org/keywords/medicine |
| keywords[11].score | 0.33196157217025757 |
| keywords[11].display_name | Medicine |
| keywords[12].id | https://openalex.org/keywords/immunology |
| keywords[12].score | 0.29818195104599 |
| keywords[12].display_name | Immunology |
| keywords[13].id | https://openalex.org/keywords/gene |
| keywords[13].score | 0.2548319101333618 |
| keywords[13].display_name | Gene |
| keywords[14].id | https://openalex.org/keywords/cell-biology |
| keywords[14].score | 0.19973719120025635 |
| keywords[14].display_name | Cell biology |
| keywords[15].id | https://openalex.org/keywords/genetics |
| keywords[15].score | 0.18328434228897095 |
| keywords[15].display_name | Genetics |
| keywords[16].id | https://openalex.org/keywords/gene-expression |
| keywords[16].score | 0.1492024064064026 |
| keywords[16].display_name | Gene expression |
| language | en |
| locations[0].id | doi:10.3389/fimmu.2023.1103392 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S2595292759 |
| locations[0].source.issn | 1664-3224 |
| locations[0].source.type | journal |
| locations[0].source.is_oa | True |
| locations[0].source.issn_l | 1664-3224 |
| locations[0].source.is_core | True |
| locations[0].source.is_in_doaj | True |
| locations[0].source.display_name | Frontiers in Immunology |
| locations[0].source.host_organization | https://openalex.org/P4310320527 |
| locations[0].source.host_organization_name | Frontiers Media |
| locations[0].source.host_organization_lineage | https://openalex.org/P4310320527 |
| locations[0].source.host_organization_lineage_names | Frontiers Media |
| locations[0].license | cc-by |
| locations[0].pdf_url | |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | Frontiers in Immunology |
| locations[0].landing_page_url | https://doi.org/10.3389/fimmu.2023.1103392 |
| locations[1].id | pmid:37033947 |
| locations[1].is_oa | False |
| locations[1].source.id | https://openalex.org/S4306525036 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | PubMed |
| locations[1].source.host_organization | https://openalex.org/I1299303238 |
| locations[1].source.host_organization_name | National Institutes of Health |
| locations[1].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[1].license | |
| locations[1].pdf_url | |
| locations[1].version | publishedVersion |
| locations[1].raw_type | |
| locations[1].license_id | |
| locations[1].is_accepted | True |
| locations[1].is_published | True |
| locations[1].raw_source_name | Frontiers in immunology |
| locations[1].landing_page_url | https://pubmed.ncbi.nlm.nih.gov/37033947 |
| locations[2].id | pmh:oai:pubmedcentral.nih.gov:10080220 |
| locations[2].is_oa | True |
| locations[2].source.id | https://openalex.org/S2764455111 |
| locations[2].source.issn | |
| locations[2].source.type | repository |
| locations[2].source.is_oa | False |
| locations[2].source.issn_l | |
| locations[2].source.is_core | False |
| locations[2].source.is_in_doaj | False |
| locations[2].source.display_name | PubMed Central |
| locations[2].source.host_organization | https://openalex.org/I1299303238 |
| locations[2].source.host_organization_name | National Institutes of Health |
| locations[2].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[2].license | cc-by |
| locations[2].pdf_url | https://pmc.ncbi.nlm.nih.gov/articles/PMC10080220/pdf/fimmu-14-1103392.pdf |
| locations[2].version | submittedVersion |
| locations[2].raw_type | Text |
| locations[2].license_id | https://openalex.org/licenses/cc-by |
| locations[2].is_accepted | False |
| locations[2].is_published | False |
| locations[2].raw_source_name | Front Immunol |
| locations[2].landing_page_url | https://www.ncbi.nlm.nih.gov/pmc/articles/10080220 |
| locations[3].id | pmh:oai:doaj.org/article:2e87193b4bde48d29f17598bc29e266c |
| locations[3].is_oa | False |
| locations[3].source.id | https://openalex.org/S4306401280 |
| locations[3].source.issn | |
| locations[3].source.type | repository |
| locations[3].source.is_oa | False |
| locations[3].source.issn_l | |
| locations[3].source.is_core | False |
| locations[3].source.is_in_doaj | False |
| locations[3].source.display_name | DOAJ (DOAJ: Directory of Open Access Journals) |
| locations[3].source.host_organization | |
| locations[3].source.host_organization_name | |
| locations[3].license | |
| locations[3].pdf_url | |
| locations[3].version | submittedVersion |
| locations[3].raw_type | article |
| locations[3].license_id | |
| locations[3].is_accepted | False |
| locations[3].is_published | False |
| locations[3].raw_source_name | Frontiers in Immunology, Vol 14 (2023) |
| locations[3].landing_page_url | https://doaj.org/article/2e87193b4bde48d29f17598bc29e266c |
| indexed_in | crossref, doaj, pubmed |
| authorships[0].author.id | https://openalex.org/A5058871894 |
| authorships[0].author.orcid | https://orcid.org/0000-0002-1394-150X |
| authorships[0].author.display_name | Hannan Ai |
| authorships[0].countries | CN, US |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I157773358 |
| authorships[0].affiliations[0].raw_affiliation_string | State Key Laboratory for Biocontrol, School of Life Sciences, Sun Yat-sen University, Guangzhou, China |
| authorships[0].affiliations[1].institution_ids | https://openalex.org/I4210137956 |
| authorships[0].affiliations[1].raw_affiliation_string | National Center for Quality Supervision and Inspection of Automatic Equipment, National Center for Testing and Evaluation of Robots (Guangzhou), CRAT, SINOMACH-IT, Guangzhou, China |
| authorships[0].affiliations[2].institution_ids | https://openalex.org/I157725225 |
| authorships[0].affiliations[2].raw_affiliation_string | Department of Electrical and Computer Engineering, The Grainger College of Engineering, University of Illinois at Urbana-Champaign, Urbana, IL, United States |
| authorships[0].institutions[0].id | https://openalex.org/I4210137956 |
| authorships[0].institutions[0].ror | https://ror.org/03wdfwr71 |
| authorships[0].institutions[0].type | education |
| authorships[0].institutions[0].lineage | https://openalex.org/I4210137956 |
| authorships[0].institutions[0].country_code | CN |
| authorships[0].institutions[0].display_name | Guangdong Testing Institute for Product Quality Supervision |
| authorships[0].institutions[1].id | https://openalex.org/I157773358 |
| authorships[0].institutions[1].ror | https://ror.org/0064kty71 |
| authorships[0].institutions[1].type | education |
| authorships[0].institutions[1].lineage | https://openalex.org/I157773358 |
| authorships[0].institutions[1].country_code | CN |
| authorships[0].institutions[1].display_name | Sun Yat-sen University |
| authorships[0].institutions[2].id | https://openalex.org/I157725225 |
| authorships[0].institutions[2].ror | https://ror.org/047426m28 |
| authorships[0].institutions[2].type | education |
| authorships[0].institutions[2].lineage | https://openalex.org/I157725225 |
| authorships[0].institutions[2].country_code | US |
| authorships[0].institutions[2].display_name | University of Illinois Urbana-Champaign |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Hannan Ai |
| authorships[0].is_corresponding | True |
| authorships[0].raw_affiliation_strings | Department of Electrical and Computer Engineering, The Grainger College of Engineering, University of Illinois at Urbana-Champaign, Urbana, IL, United States, National Center for Quality Supervision and Inspection of Automatic Equipment, National Center for Testing and Evaluation of Robots (Guangzhou), CRAT, SINOMACH-IT, Guangzhou, China, State Key Laboratory for Biocontrol, School of Life Sciences, Sun Yat-sen University, Guangzhou, China |
| authorships[1].author.id | https://openalex.org/A5102947476 |
| authorships[1].author.orcid | https://orcid.org/0000-0002-4287-2240 |
| authorships[1].author.display_name | Fanmei Meng |
| authorships[1].countries | CN |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I157773358 |
| authorships[1].affiliations[0].raw_affiliation_string | State Key Laboratory for Biocontrol, School of Life Sciences, Sun Yat-sen University, Guangzhou, China |
| authorships[1].institutions[0].id | https://openalex.org/I157773358 |
| authorships[1].institutions[0].ror | https://ror.org/0064kty71 |
| authorships[1].institutions[0].type | education |
| authorships[1].institutions[0].lineage | https://openalex.org/I157773358 |
| authorships[1].institutions[0].country_code | CN |
| authorships[1].institutions[0].display_name | Sun Yat-sen University |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Fanmei Meng |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | State Key Laboratory for Biocontrol, School of Life Sciences, Sun Yat-sen University, Guangzhou, China |
| authorships[2].author.id | https://openalex.org/A5091789944 |
| authorships[2].author.orcid | https://orcid.org/0000-0003-4151-6313 |
| authorships[2].author.display_name | Yuncan Ai |
| authorships[2].countries | CN |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I157773358 |
| authorships[2].affiliations[0].raw_affiliation_string | State Key Laboratory for Biocontrol, School of Life Sciences, Sun Yat-sen University, Guangzhou, China |
| authorships[2].affiliations[1].institution_ids | https://openalex.org/I4210090868, https://openalex.org/I92039509 |
| authorships[2].affiliations[1].raw_affiliation_string | The Second Affiliated Hospital, Guangdong Provincial Key Laboratory of Allergy & Clinical Immunology, Center for Inflammation, Immunity & Immune-mediated Disease, Sino-French Hoffmann Institute, Guangzhou Medical University, Guangzhou, Guangdong, China |
| authorships[2].institutions[0].id | https://openalex.org/I92039509 |
| authorships[2].institutions[0].ror | https://ror.org/00zat6v61 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I92039509 |
| authorships[2].institutions[0].country_code | CN |
| authorships[2].institutions[0].display_name | Guangzhou Medical University |
| authorships[2].institutions[1].id | https://openalex.org/I4210090868 |
| authorships[2].institutions[1].ror | https://ror.org/00a98yf63 |
| authorships[2].institutions[1].type | healthcare |
| authorships[2].institutions[1].lineage | https://openalex.org/I4210090868 |
| authorships[2].institutions[1].country_code | CN |
| authorships[2].institutions[1].display_name | Second Affiliated Hospital of Guangzhou Medical University |
| authorships[2].institutions[2].id | https://openalex.org/I157773358 |
| authorships[2].institutions[2].ror | https://ror.org/0064kty71 |
| authorships[2].institutions[2].type | education |
| authorships[2].institutions[2].lineage | https://openalex.org/I157773358 |
| authorships[2].institutions[2].country_code | CN |
| authorships[2].institutions[2].display_name | Sun Yat-sen University |
| authorships[2].author_position | last |
| authorships[2].raw_author_name | Yuncan Ai |
| authorships[2].is_corresponding | True |
| authorships[2].raw_affiliation_strings | State Key Laboratory for Biocontrol, School of Life Sciences, Sun Yat-sen University, Guangzhou, China, The Second Affiliated Hospital, Guangdong Provincial Key Laboratory of Allergy & Clinical Immunology, Center for Inflammation, Immunity & Immune-mediated Disease, Sino-French Hoffmann Institute, Guangzhou Medical University, Guangzhou, Guangdong, China |
| has_content.pdf | False |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://doi.org/10.3389/fimmu.2023.1103392 |
| open_access.oa_status | gold |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | PathwayKO: An integrated platform for deciphering the systems-level signaling pathways |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T10211 |
| primary_topic.field.id | https://openalex.org/fields/17 |
| primary_topic.field.display_name | Computer Science |
| primary_topic.score | 0.9918000102043152 |
| primary_topic.domain.id | https://openalex.org/domains/3 |
| primary_topic.domain.display_name | Physical Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1703 |
| primary_topic.subfield.display_name | Computational Theory and Mathematics |
| primary_topic.display_name | Computational Drug Discovery Methods |
| related_works | https://openalex.org/W2161149782, https://openalex.org/W2029482690, https://openalex.org/W1985841818, https://openalex.org/W1588529067, https://openalex.org/W2117028746, https://openalex.org/W4214774859, https://openalex.org/W1990395889, https://openalex.org/W4206912673, https://openalex.org/W2162173224, https://openalex.org/W2469420430 |
| cited_by_count | 0 |
| locations_count | 4 |
| best_oa_location.id | doi:10.3389/fimmu.2023.1103392 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S2595292759 |
| best_oa_location.source.issn | 1664-3224 |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | True |
| best_oa_location.source.issn_l | 1664-3224 |
| best_oa_location.source.is_core | True |
| best_oa_location.source.is_in_doaj | True |
| best_oa_location.source.display_name | Frontiers in Immunology |
| best_oa_location.source.host_organization | https://openalex.org/P4310320527 |
| best_oa_location.source.host_organization_name | Frontiers Media |
| best_oa_location.source.host_organization_lineage | https://openalex.org/P4310320527 |
| best_oa_location.source.host_organization_lineage_names | Frontiers Media |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | journal-article |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | Frontiers in Immunology |
| best_oa_location.landing_page_url | https://doi.org/10.3389/fimmu.2023.1103392 |
| primary_location.id | doi:10.3389/fimmu.2023.1103392 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S2595292759 |
| primary_location.source.issn | 1664-3224 |
| primary_location.source.type | journal |
| primary_location.source.is_oa | True |
| primary_location.source.issn_l | 1664-3224 |
| primary_location.source.is_core | True |
| primary_location.source.is_in_doaj | True |
| primary_location.source.display_name | Frontiers in Immunology |
| primary_location.source.host_organization | https://openalex.org/P4310320527 |
| primary_location.source.host_organization_name | Frontiers Media |
| primary_location.source.host_organization_lineage | https://openalex.org/P4310320527 |
| primary_location.source.host_organization_lineage_names | Frontiers Media |
| primary_location.license | cc-by |
| primary_location.pdf_url | |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | Frontiers in Immunology |
| primary_location.landing_page_url | https://doi.org/10.3389/fimmu.2023.1103392 |
| publication_date | 2023-03-23 |
| publication_year | 2023 |
| referenced_works | https://openalex.org/W4282822205, https://openalex.org/W2135203259, https://openalex.org/W2029421341, https://openalex.org/W2976957583, https://openalex.org/W2979832776, https://openalex.org/W2110256992, https://openalex.org/W2796586233, https://openalex.org/W2330304652, https://openalex.org/W2139036189, https://openalex.org/W2130410032, https://openalex.org/W2020859582, https://openalex.org/W2129099812, https://openalex.org/W2121824548, https://openalex.org/W2134736660, https://openalex.org/W2110687686, https://openalex.org/W2154972152, https://openalex.org/W2613052384, https://openalex.org/W1793984240, https://openalex.org/W2006617902, https://openalex.org/W2141224543, https://openalex.org/W2148708580, https://openalex.org/W2081098333, https://openalex.org/W2146512944, https://openalex.org/W2100625056, https://openalex.org/W2078622638, https://openalex.org/W4298878824, https://openalex.org/W6676591658, https://openalex.org/W1596515083, https://openalex.org/W2120842870, https://openalex.org/W1566535612, https://openalex.org/W2108375396, https://openalex.org/W1963912918, https://openalex.org/W2127620487, https://openalex.org/W2166342119, https://openalex.org/W3047433286, https://openalex.org/W3138310216, https://openalex.org/W3165655050, https://openalex.org/W2941894376, https://openalex.org/W4212920968, https://openalex.org/W3096351980, https://openalex.org/W2896517057, https://openalex.org/W3009862485, https://openalex.org/W3041304817, https://openalex.org/W3172158917, https://openalex.org/W2919197532, https://openalex.org/W3039150496, https://openalex.org/W2045729458, https://openalex.org/W2923098009, https://openalex.org/W2152575748 |
| referenced_works_count | 49 |
| abstract_inverted_index.a | 13, 44, 76, 281 |
| abstract_inverted_index.(2 | 134 |
| abstract_inverted_index.21 | 163 |
| abstract_inverted_index.KO | 30, 37, 82, 115, 164 |
| abstract_inverted_index.We | 88 |
| abstract_inverted_index.as | 226 |
| abstract_inverted_index.at | 39, 85, 295 |
| abstract_inverted_index.be | 108 |
| abstract_inverted_index.by | 36, 130 |
| abstract_inverted_index.in | 5, 15, 75, 190, 222, 248, 289 |
| abstract_inverted_index.is | 12, 43 |
| abstract_inverted_index.of | 2, 60, 98, 162, 195, 246, 274, 283, 302 |
| abstract_inverted_index.on | 90 |
| abstract_inverted_index.to | 69, 110, 240 |
| abstract_inverted_index.we | 101, 260 |
| abstract_inverted_index.(12 | 145 |
| abstract_inverted_index.85% | 137 |
| abstract_inverted_index.and | 8, 66, 79, 95, 118, 141, 182, 186, 198, 211, 230, 237, 306 |
| abstract_inverted_index.are | 25, 34 |
| abstract_inverted_index.can | 107, 268 |
| abstract_inverted_index.for | 27, 46, 228, 291 |
| abstract_inverted_index.has | 56 |
| abstract_inverted_index.hit | 157 |
| abstract_inverted_index.may | 224, 298 |
| abstract_inverted_index.our | 151 |
| abstract_inverted_index.set | 161 |
| abstract_inverted_index.the | 40, 52, 86, 92, 104, 125, 158, 169, 174, 178, 183, 193, 234, 242, 263, 271, 296, 300 |
| abstract_inverted_index.who | 252 |
| abstract_inverted_index.(KO) | 23 |
| abstract_inverted_index.MAPK | 179 |
| abstract_inverted_index.PD-1 | 187 |
| abstract_inverted_index.Such | 220 |
| abstract_inverted_index.This | 49 |
| abstract_inverted_index.both | 155 |
| abstract_inverted_index.care | 250 |
| abstract_inverted_index.core | 160 |
| abstract_inverted_index.cues | 214 |
| abstract_inverted_index.from | 21 |
| abstract_inverted_index.have | 253 |
| abstract_inverted_index.high | 243 |
| abstract_inverted_index.mice | 223 |
| abstract_inverted_index.same | 159 |
| abstract_inverted_index.such | 287 |
| abstract_inverted_index.that | 33, 103, 154, 215, 262 |
| abstract_inverted_index.when | 278 |
| abstract_inverted_index.with | 280 |
| abstract_inverted_index.NFκB | 175 |
| abstract_inverted_index.PD-L1 | 184 |
| abstract_inverted_index.There | 42 |
| abstract_inverted_index.These | 201 |
| abstract_inverted_index.after | 136, 147 |
| abstract_inverted_index.cases | 11, 156 |
| abstract_inverted_index.focus | 89 |
| abstract_inverted_index.genes | 38 |
| abstract_inverted_index.guide | 232 |
| abstract_inverted_index.hours | 135, 146 |
| abstract_inverted_index.liver | 138 |
| abstract_inverted_index.mouse | 114 |
| abstract_inverted_index.offer | 212 |
| abstract_inverted_index.rates | 245 |
| abstract_inverted_index.serve | 225 |
| abstract_inverted_index.these | 208 |
| abstract_inverted_index.units | 251 |
| abstract_inverted_index.viral | 197 |
| abstract_inverted_index.which | 55, 120 |
| abstract_inverted_index.First, | 100 |
| abstract_inverted_index.common | 204 |
| abstract_inverted_index.demand | 45 |
| abstract_inverted_index.fields | 301 |
| abstract_inverted_index.future | 217 |
| abstract_inverted_index.humans | 229 |
| abstract_inverted_index.immune | 3, 127, 209 |
| abstract_inverted_index.innate | 126 |
| abstract_inverted_index.models | 227 |
| abstract_inverted_index.modern | 16 |
| abstract_inverted_index.proper | 284 |
| abstract_inverted_index.reduce | 241 |
| abstract_inverted_index.reveal | 121 |
| abstract_inverted_index.sepsis | 144 |
| abstract_inverted_index.target | 29, 81 |
| abstract_inverted_index.Second, | 259 |
| abstract_inverted_index.Systems | 0 |
| abstract_inverted_index.analyze | 112 |
| abstract_inverted_index.article | 50 |
| abstract_inverted_index.benefit | 299 |
| abstract_inverted_index.between | 207 |
| abstract_inverted_index.beyond. | 307 |
| abstract_inverted_index.cancer, | 191 |
| abstract_inverted_index.conduct | 70 |
| abstract_inverted_index.crucial | 26 |
| abstract_inverted_index.disease | 7 |
| abstract_inverted_index.fashion | 78 |
| abstract_inverted_index.health, | 6 |
| abstract_inverted_index.methods | 59, 277, 290 |
| abstract_inverted_index.pathway | 61, 73, 189, 275 |
| abstract_inverted_index.results | 152 |
| abstract_inverted_index.suggest | 203 |
| abstract_inverted_index.systems | 304 |
| abstract_inverted_index.warrant | 216 |
| abstract_inverted_index.advances | 288 |
| abstract_inverted_index.analysis | 68, 74, 276 |
| abstract_inverted_index.clinical | 9 |
| abstract_inverted_index.decipher | 80 |
| abstract_inverted_index.evaluate | 270 |
| abstract_inverted_index.features | 97 |
| abstract_inverted_index.findings | 202 |
| abstract_inverted_index.impaired | 35 |
| abstract_inverted_index.indicate | 153 |
| abstract_inverted_index.insights | 294 |
| abstract_inverted_index.pathway, | 173, 177, 181 |
| abstract_inverted_index.pathways | 32, 84, 194 |
| abstract_inverted_index.patients | 247 |
| abstract_inverted_index.pipeline | 77 |
| abstract_inverted_index.platform | 106, 265 |
| abstract_inverted_index.priority | 14 |
| abstract_inverted_index.receptor | 171 |
| abstract_inverted_index.research | 235 |
| abstract_inverted_index.surgery) | 140 |
| abstract_inverted_index.surgical | 257 |
| abstract_inverted_index.systemic | 122 |
| abstract_inverted_index.utilized | 109 |
| abstract_inverted_index.(GSE22873 | 117 |
| abstract_inverted_index.PathwayKO | 53, 105, 264 |
| abstract_inverted_index.Together, | 286 |
| abstract_inverted_index.Toll-like | 170 |
| abstract_inverted_index.alongside | 192 |
| abstract_inverted_index.analysis, | 63, 65 |
| abstract_inverted_index.composite | 238 |
| abstract_inverted_index.describes | 51 |
| abstract_inverted_index.extensive | 132 |
| abstract_inverted_index.including | 168 |
| abstract_inverted_index.intensive | 249 |
| abstract_inverted_index.medicine. | 17 |
| abstract_inverted_index.mortality | 244 |
| abstract_inverted_index.paradigms | 236 |
| abstract_inverted_index.parasitic | 199 |
| abstract_inverted_index.pathways, | 167 |
| abstract_inverted_index.platform, | 54 |
| abstract_inverted_index.resection | 139 |
| abstract_inverted_index.responses | 128, 210 |
| abstract_inverted_index.signaling | 31, 83, 166, 172, 176, 180 |
| abstract_inverted_index.surgery). | 149 |
| abstract_inverted_index.traumatic | 256 |
| abstract_inverted_index.triggered | 129 |
| abstract_inverted_index.undergone | 254 |
| abstract_inverted_index.CASP-model | 143, 148 |
| abstract_inverted_index.GSE24327), | 119 |
| abstract_inverted_index.PathwayKO. | 99 |
| abstract_inverted_index.bacterial, | 196 |
| abstract_inverted_index.biological | 293 |
| abstract_inverted_index.checkpoint | 188 |
| abstract_inverted_index.collection | 282 |
| abstract_inverted_index.describing | 91 |
| abstract_inverted_index.difference | 273 |
| abstract_inverted_index.enrichment | 62 |
| abstract_inverted_index.expression | 185 |
| abstract_inverted_index.immunology | 305 |
| abstract_inverted_index.infectious | 142 |
| abstract_inverted_index.integrated | 57 |
| abstract_inverted_index.landscapes | 4 |
| abstract_inverted_index.mechanisms | 123, 206, 221 |
| abstract_inverted_index.platforms. | 48 |
| abstract_inverted_index.principles | 94 |
| abstract_inverted_index.real-world | 113 |
| abstract_inverted_index.statistics | 64 |
| abstract_inverted_index.strategies | 239 |
| abstract_inverted_index.successful | 255 |
| abstract_inverted_index.ultimately | 231 |
| abstract_inverted_index.underlying | 124 |
| abstract_inverted_index.Strikingly, | 150 |
| abstract_inverted_index.accumulated | 20 |
| abstract_inverted_index.application | 96 |
| abstract_inverted_index.assessments | 267 |
| abstract_inverted_index.benchmarked | 279 |
| abstract_inverted_index.deciphering | 28, 292 |
| abstract_inverted_index.demonstrate | 102, 261 |
| abstract_inverted_index.effectively | 269 |
| abstract_inverted_index.experiments | 24 |
| abstract_inverted_index.formulating | 233 |
| abstract_inverted_index.fundamental | 205 |
| abstract_inverted_index.hepatectomy | 133 |
| abstract_inverted_index.infections. | 200 |
| abstract_inverted_index.informative | 213 |
| abstract_inverted_index.integrative | 47, 72 |
| abstract_inverted_index.model-based | 266 |
| abstract_inverted_index.performance | 272 |
| abstract_inverted_index.treatments. | 258 |
| abstract_inverted_index.validation. | 219 |
| abstract_inverted_index.visualizing | 67 |
| abstract_inverted_index.cutting-edge | 71 |
| abstract_inverted_index.experimental | 218 |
| abstract_inverted_index.intervention | 10 |
| abstract_inverted_index.methodology, | 93 |
| abstract_inverted_index.gene-knockout | 22 |
| abstract_inverted_index.systems-level | 297 |
| abstract_inverted_index.non-infectious | 131 |
| abstract_inverted_index.systems-level. | 41, 87 |
| abstract_inverted_index.transcriptomes | 19, 116 |
| abstract_inverted_index.High-throughput | 18 |
| abstract_inverted_index.bioinformatics, | 303 |
| abstract_inverted_index.comprehensively | 111 |
| abstract_inverted_index.transcriptomes. | 285 |
| abstract_inverted_index.MyD88-associated | 165 |
| abstract_inverted_index.characterization | 1 |
| abstract_inverted_index.state-of-the-art | 58 |
| cited_by_percentile_year | |
| corresponding_author_ids | https://openalex.org/A5058871894, https://openalex.org/A5091789944 |
| countries_distinct_count | 2 |
| institutions_distinct_count | 3 |
| corresponding_institution_ids | https://openalex.org/I157725225, https://openalex.org/I157773358, https://openalex.org/I4210090868, https://openalex.org/I4210137956, https://openalex.org/I92039509 |
| sustainable_development_goals[0].id | https://metadata.un.org/sdg/3 |
| sustainable_development_goals[0].score | 0.8799999952316284 |
| sustainable_development_goals[0].display_name | Good health and well-being |
| citation_normalized_percentile.value | 0.05368811 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |