PGS Browser: a public platform for personalized polygenic score interpretation Article Swipe
 YOU?
·
· 2025
· Open Access
·
· DOI: https://doi.org/10.1101/2025.07.03.25330851
YOU?
·
· 2025
· Open Access
·
· DOI: https://doi.org/10.1101/2025.07.03.25330851
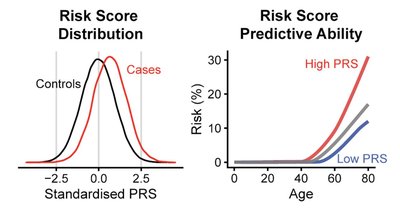
Identifying individuals at elevated risk before disease onset is a cornerstone of personalized prevention, and polygenic scores (PGSs) have proven to be nstrumental in this regard. In this study, we systematically benchmarked 3,168 PGS models from the PGS Catalog in 473,681 FinnGen participants, revealing top-performing scores across major ancestry groups and highlighting traits where non-target PGSs significantly improve prediction. We conducted 10,531 phenome-wide association studies, uncovering 439,070 significant associations between PGSs and 4,739 clinical endpoints. To support individual score interpretation, we provide public access to ancestry-adjusted reference PGS distributions derived from the large-scale FinnGen cohort, alongside elastic-net Cox models for predicting time-to-event for 22 major disorders All data and predictive tools are accessible via the PGS Browser ( pgs.nchigm.org ), an interactive web-based platform for PGS analysis and interpretation. We envision that this esource will greatly advance both research and translational applications of polygenic scores.
Related Topics
- Type
- preprint
- Language
- en
- Landing Page
- https://doi.org/10.1101/2025.07.03.25330851
- https://www.medrxiv.org/content/medrxiv/early/2025/07/05/2025.07.03.25330851.full.pdf
- OA Status
- green
- References
- 44
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4412053001
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4412053001Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1101/2025.07.03.25330851Digital Object Identifier
- Title
-
PGS Browser: a public platform for personalized polygenic score interpretationWork title
- Type
-
preprintOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2025Year of publication
- Publication date
-
2025-07-05Full publication date if available
- Authors
-
Nikita Kolosov, Mary Pat Reeve, Pietro Della Briotta Parolo, Mitja I. Kurki, Vincent Llorens, Timo P. Sipilä, Adam Herman, Ivan Molotkov, Mervi Aavikko, Samuli Ripatti, Aarno Palotie, Mark J. Daly, Mykyta ArtomovList of authors in order
- Landing page
-
https://doi.org/10.1101/2025.07.03.25330851Publisher landing page
- PDF URL
-
https://www.medrxiv.org/content/medrxiv/early/2025/07/05/2025.07.03.25330851.full.pdfDirect link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
greenOpen access status per OpenAlex
- OA URL
-
https://www.medrxiv.org/content/medrxiv/early/2025/07/05/2025.07.03.25330851.full.pdfDirect OA link when available
- Concepts
-
Interpretation (philosophy), Polygenic risk score, Computer science, Personalized medicine, F1 score, Natural language processing, World Wide Web, Artificial intelligence, Psychology, Bioinformatics, Biology, Genetics, Gene, Single-nucleotide polymorphism, Programming language, GenotypeTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
0Total citation count in OpenAlex
- References (count)
-
44Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4412053001 |
|---|---|
| doi | https://doi.org/10.1101/2025.07.03.25330851 |
| ids.doi | https://doi.org/10.1101/2025.07.03.25330851 |
| ids.openalex | https://openalex.org/W4412053001 |
| fwci | |
| type | preprint |
| title | PGS Browser: a public platform for personalized polygenic score interpretation |
| biblio.issue | |
| biblio.volume | |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T11309 |
| topics[0].field.id | https://openalex.org/fields/17 |
| topics[0].field.display_name | Computer Science |
| topics[0].score | 0.9894000291824341 |
| topics[0].domain.id | https://openalex.org/domains/3 |
| topics[0].domain.display_name | Physical Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1711 |
| topics[0].subfield.display_name | Signal Processing |
| topics[0].display_name | Music and Audio Processing |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C527412718 |
| concepts[0].level | 2 |
| concepts[0].score | 0.7480637431144714 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q855395 |
| concepts[0].display_name | Interpretation (philosophy) |
| concepts[1].id | https://openalex.org/C2993137441 |
| concepts[1].level | 5 |
| concepts[1].score | 0.583706796169281 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q28135712 |
| concepts[1].display_name | Polygenic risk score |
| concepts[2].id | https://openalex.org/C41008148 |
| concepts[2].level | 0 |
| concepts[2].score | 0.4829265773296356 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q21198 |
| concepts[2].display_name | Computer science |
| concepts[3].id | https://openalex.org/C32220436 |
| concepts[3].level | 2 |
| concepts[3].score | 0.452958881855011 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q2072214 |
| concepts[3].display_name | Personalized medicine |
| concepts[4].id | https://openalex.org/C148524875 |
| concepts[4].level | 2 |
| concepts[4].score | 0.4116840362548828 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q6975395 |
| concepts[4].display_name | F1 score |
| concepts[5].id | https://openalex.org/C204321447 |
| concepts[5].level | 1 |
| concepts[5].score | 0.3996903598308563 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q30642 |
| concepts[5].display_name | Natural language processing |
| concepts[6].id | https://openalex.org/C136764020 |
| concepts[6].level | 1 |
| concepts[6].score | 0.3654334545135498 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q466 |
| concepts[6].display_name | World Wide Web |
| concepts[7].id | https://openalex.org/C154945302 |
| concepts[7].level | 1 |
| concepts[7].score | 0.35322970151901245 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q11660 |
| concepts[7].display_name | Artificial intelligence |
| concepts[8].id | https://openalex.org/C15744967 |
| concepts[8].level | 0 |
| concepts[8].score | 0.3426014184951782 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q9418 |
| concepts[8].display_name | Psychology |
| concepts[9].id | https://openalex.org/C60644358 |
| concepts[9].level | 1 |
| concepts[9].score | 0.24158722162246704 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q128570 |
| concepts[9].display_name | Bioinformatics |
| concepts[10].id | https://openalex.org/C86803240 |
| concepts[10].level | 0 |
| concepts[10].score | 0.23530688881874084 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[10].display_name | Biology |
| concepts[11].id | https://openalex.org/C54355233 |
| concepts[11].level | 1 |
| concepts[11].score | 0.17888963222503662 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[11].display_name | Genetics |
| concepts[12].id | https://openalex.org/C104317684 |
| concepts[12].level | 2 |
| concepts[12].score | 0.08249059319496155 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[12].display_name | Gene |
| concepts[13].id | https://openalex.org/C153209595 |
| concepts[13].level | 4 |
| concepts[13].score | 0.0 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q501128 |
| concepts[13].display_name | Single-nucleotide polymorphism |
| concepts[14].id | https://openalex.org/C199360897 |
| concepts[14].level | 1 |
| concepts[14].score | 0.0 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q9143 |
| concepts[14].display_name | Programming language |
| concepts[15].id | https://openalex.org/C135763542 |
| concepts[15].level | 3 |
| concepts[15].score | 0.0 |
| concepts[15].wikidata | https://www.wikidata.org/wiki/Q106016 |
| concepts[15].display_name | Genotype |
| keywords[0].id | https://openalex.org/keywords/interpretation |
| keywords[0].score | 0.7480637431144714 |
| keywords[0].display_name | Interpretation (philosophy) |
| keywords[1].id | https://openalex.org/keywords/polygenic-risk-score |
| keywords[1].score | 0.583706796169281 |
| keywords[1].display_name | Polygenic risk score |
| keywords[2].id | https://openalex.org/keywords/computer-science |
| keywords[2].score | 0.4829265773296356 |
| keywords[2].display_name | Computer science |
| keywords[3].id | https://openalex.org/keywords/personalized-medicine |
| keywords[3].score | 0.452958881855011 |
| keywords[3].display_name | Personalized medicine |
| keywords[4].id | https://openalex.org/keywords/f1-score |
| keywords[4].score | 0.4116840362548828 |
| keywords[4].display_name | F1 score |
| keywords[5].id | https://openalex.org/keywords/natural-language-processing |
| keywords[5].score | 0.3996903598308563 |
| keywords[5].display_name | Natural language processing |
| keywords[6].id | https://openalex.org/keywords/world-wide-web |
| keywords[6].score | 0.3654334545135498 |
| keywords[6].display_name | World Wide Web |
| keywords[7].id | https://openalex.org/keywords/artificial-intelligence |
| keywords[7].score | 0.35322970151901245 |
| keywords[7].display_name | Artificial intelligence |
| keywords[8].id | https://openalex.org/keywords/psychology |
| keywords[8].score | 0.3426014184951782 |
| keywords[8].display_name | Psychology |
| keywords[9].id | https://openalex.org/keywords/bioinformatics |
| keywords[9].score | 0.24158722162246704 |
| keywords[9].display_name | Bioinformatics |
| keywords[10].id | https://openalex.org/keywords/biology |
| keywords[10].score | 0.23530688881874084 |
| keywords[10].display_name | Biology |
| keywords[11].id | https://openalex.org/keywords/genetics |
| keywords[11].score | 0.17888963222503662 |
| keywords[11].display_name | Genetics |
| keywords[12].id | https://openalex.org/keywords/gene |
| keywords[12].score | 0.08249059319496155 |
| keywords[12].display_name | Gene |
| language | en |
| locations[0].id | doi:10.1101/2025.07.03.25330851 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4306402567 |
| locations[0].source.issn | |
| locations[0].source.type | repository |
| locations[0].source.is_oa | False |
| locations[0].source.issn_l | |
| locations[0].source.is_core | False |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| locations[0].source.host_organization | https://openalex.org/I2750212522 |
| locations[0].source.host_organization_name | Cold Spring Harbor Laboratory |
| locations[0].source.host_organization_lineage | https://openalex.org/I2750212522 |
| locations[0].license | cc-by-nc-nd |
| locations[0].pdf_url | https://www.medrxiv.org/content/medrxiv/early/2025/07/05/2025.07.03.25330851.full.pdf |
| locations[0].version | acceptedVersion |
| locations[0].raw_type | posted-content |
| locations[0].license_id | https://openalex.org/licenses/cc-by-nc-nd |
| locations[0].is_accepted | True |
| locations[0].is_published | False |
| locations[0].raw_source_name | |
| locations[0].landing_page_url | https://doi.org/10.1101/2025.07.03.25330851 |
| indexed_in | crossref |
| authorships[0].author.id | https://openalex.org/A5047488994 |
| authorships[0].author.orcid | https://orcid.org/0000-0002-2139-6775 |
| authorships[0].author.display_name | Nikita Kolosov |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Nikita Kolosov |
| authorships[0].is_corresponding | False |
| authorships[1].author.id | https://openalex.org/A5043982944 |
| authorships[1].author.orcid | https://orcid.org/0000-0003-1788-2321 |
| authorships[1].author.display_name | Mary Pat Reeve |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Mary P. Reeve |
| authorships[1].is_corresponding | False |
| authorships[2].author.id | https://openalex.org/A5104127292 |
| authorships[2].author.orcid | |
| authorships[2].author.display_name | Pietro Della Briotta Parolo |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Pietro Della Briotta Parolo |
| authorships[2].is_corresponding | False |
| authorships[3].author.id | https://openalex.org/A5108114461 |
| authorships[3].author.orcid | |
| authorships[3].author.display_name | Mitja I. Kurki |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Mitja I. Kurki |
| authorships[3].is_corresponding | False |
| authorships[4].author.id | https://openalex.org/A5015118948 |
| authorships[4].author.orcid | |
| authorships[4].author.display_name | Vincent Llorens |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Vincent Llorens |
| authorships[4].is_corresponding | False |
| authorships[5].author.id | https://openalex.org/A5087434511 |
| authorships[5].author.orcid | https://orcid.org/0000-0001-6739-1606 |
| authorships[5].author.display_name | Timo P. Sipilä |
| authorships[5].author_position | middle |
| authorships[5].raw_author_name | Timo Petteri Sipila |
| authorships[5].is_corresponding | False |
| authorships[6].author.id | https://openalex.org/A5028458306 |
| authorships[6].author.orcid | https://orcid.org/0000-0001-7463-9462 |
| authorships[6].author.display_name | Adam Herman |
| authorships[6].author_position | middle |
| authorships[6].raw_author_name | Adam Herman |
| authorships[6].is_corresponding | False |
| authorships[7].author.id | https://openalex.org/A5092832874 |
| authorships[7].author.orcid | https://orcid.org/0009-0008-3566-0160 |
| authorships[7].author.display_name | Ivan Molotkov |
| authorships[7].author_position | middle |
| authorships[7].raw_author_name | Ivan Molotkov |
| authorships[7].is_corresponding | False |
| authorships[8].author.id | https://openalex.org/A5063496761 |
| authorships[8].author.orcid | https://orcid.org/0000-0003-2583-0150 |
| authorships[8].author.display_name | Mervi Aavikko |
| authorships[8].author_position | middle |
| authorships[8].raw_author_name | Mervi Aavikko |
| authorships[8].is_corresponding | False |
| authorships[9].author.id | https://openalex.org/A5111111902 |
| authorships[9].author.orcid | |
| authorships[9].author.display_name | Samuli Ripatti |
| authorships[9].author_position | middle |
| authorships[9].raw_author_name | Samuli Ripatti |
| authorships[9].is_corresponding | False |
| authorships[10].author.id | https://openalex.org/A5108117156 |
| authorships[10].author.orcid | |
| authorships[10].author.display_name | Aarno Palotie |
| authorships[10].author_position | middle |
| authorships[10].raw_author_name | Aarno Palotie |
| authorships[10].is_corresponding | False |
| authorships[11].author.id | https://openalex.org/A5107596541 |
| authorships[11].author.orcid | https://orcid.org/0000-0002-0949-8752 |
| authorships[11].author.display_name | Mark J. Daly |
| authorships[11].author_position | middle |
| authorships[11].raw_author_name | Mark J. Daly |
| authorships[11].is_corresponding | False |
| authorships[12].author.id | https://openalex.org/A5082319199 |
| authorships[12].author.orcid | https://orcid.org/0000-0001-5282-8764 |
| authorships[12].author.display_name | Mykyta Artomov |
| authorships[12].author_position | last |
| authorships[12].raw_author_name | Mykyta Artomov |
| authorships[12].is_corresponding | False |
| has_content.pdf | True |
| has_content.grobid_xml | True |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://www.medrxiv.org/content/medrxiv/early/2025/07/05/2025.07.03.25330851.full.pdf |
| open_access.oa_status | green |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | PGS Browser: a public platform for personalized polygenic score interpretation |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T11309 |
| primary_topic.field.id | https://openalex.org/fields/17 |
| primary_topic.field.display_name | Computer Science |
| primary_topic.score | 0.9894000291824341 |
| primary_topic.domain.id | https://openalex.org/domains/3 |
| primary_topic.domain.display_name | Physical Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1711 |
| primary_topic.subfield.display_name | Signal Processing |
| primary_topic.display_name | Music and Audio Processing |
| related_works | https://openalex.org/W4313320911, https://openalex.org/W4327743144, https://openalex.org/W4245077728, https://openalex.org/W2607424049, https://openalex.org/W4390922876, https://openalex.org/W3183204001, https://openalex.org/W4206302830, https://openalex.org/W2185941092, https://openalex.org/W4386782890, https://openalex.org/W3210948575 |
| cited_by_count | 0 |
| locations_count | 1 |
| best_oa_location.id | doi:10.1101/2025.07.03.25330851 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4306402567 |
| best_oa_location.source.issn | |
| best_oa_location.source.type | repository |
| best_oa_location.source.is_oa | False |
| best_oa_location.source.issn_l | |
| best_oa_location.source.is_core | False |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| best_oa_location.source.host_organization | https://openalex.org/I2750212522 |
| best_oa_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| best_oa_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| best_oa_location.license | cc-by-nc-nd |
| best_oa_location.pdf_url | https://www.medrxiv.org/content/medrxiv/early/2025/07/05/2025.07.03.25330851.full.pdf |
| best_oa_location.version | acceptedVersion |
| best_oa_location.raw_type | posted-content |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by-nc-nd |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | False |
| best_oa_location.raw_source_name | |
| best_oa_location.landing_page_url | https://doi.org/10.1101/2025.07.03.25330851 |
| primary_location.id | doi:10.1101/2025.07.03.25330851 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4306402567 |
| primary_location.source.issn | |
| primary_location.source.type | repository |
| primary_location.source.is_oa | False |
| primary_location.source.issn_l | |
| primary_location.source.is_core | False |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| primary_location.source.host_organization | https://openalex.org/I2750212522 |
| primary_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| primary_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| primary_location.license | cc-by-nc-nd |
| primary_location.pdf_url | https://www.medrxiv.org/content/medrxiv/early/2025/07/05/2025.07.03.25330851.full.pdf |
| primary_location.version | acceptedVersion |
| primary_location.raw_type | posted-content |
| primary_location.license_id | https://openalex.org/licenses/cc-by-nc-nd |
| primary_location.is_accepted | True |
| primary_location.is_published | False |
| primary_location.raw_source_name | |
| primary_location.landing_page_url | https://doi.org/10.1101/2025.07.03.25330851 |
| publication_date | 2025-07-05 |
| publication_year | 2025 |
| referenced_works | https://openalex.org/W3023515526, https://openalex.org/W3015631361, https://openalex.org/W4383371561, https://openalex.org/W2899836172, https://openalex.org/W3088258981, https://openalex.org/W4296887932, https://openalex.org/W3134721632, https://openalex.org/W3134184401, https://openalex.org/W4390507853, https://openalex.org/W4409735485, https://openalex.org/W2104549677, https://openalex.org/W3011413344, https://openalex.org/W4292241266, https://openalex.org/W3106839128, https://openalex.org/W4221121740, https://openalex.org/W3146985094, https://openalex.org/W4319067573, https://openalex.org/W4223474278, https://openalex.org/W4404439917, https://openalex.org/W4200559672, https://openalex.org/W1608887304, https://openalex.org/W3213756041, https://openalex.org/W4402076117, https://openalex.org/W2012109041, https://openalex.org/W2335623839, https://openalex.org/W4317779009, https://openalex.org/W4380154191, https://openalex.org/W4401794450, https://openalex.org/W4223924706, https://openalex.org/W4317478560, https://openalex.org/W4308435067, https://openalex.org/W2135665336, https://openalex.org/W2902482795, https://openalex.org/W2951553270, https://openalex.org/W3111018742, https://openalex.org/W4402874940, https://openalex.org/W4407405480, https://openalex.org/W4392950969, https://openalex.org/W4401339546, https://openalex.org/W2302501749, https://openalex.org/W2161633633, https://openalex.org/W2461051203, https://openalex.org/W2794694102, https://openalex.org/W4398167347 |
| referenced_works_count | 44 |
| abstract_inverted_index.( | 118 |
| abstract_inverted_index.a | 10 |
| abstract_inverted_index.), | 120 |
| abstract_inverted_index.22 | 104 |
| abstract_inverted_index.In | 27 |
| abstract_inverted_index.To | 76 |
| abstract_inverted_index.We | 60, 130 |
| abstract_inverted_index.an | 121 |
| abstract_inverted_index.at | 3 |
| abstract_inverted_index.be | 22 |
| abstract_inverted_index.in | 24, 40 |
| abstract_inverted_index.is | 9 |
| abstract_inverted_index.of | 12, 143 |
| abstract_inverted_index.to | 21, 85 |
| abstract_inverted_index.we | 30, 81 |
| abstract_inverted_index.All | 107 |
| abstract_inverted_index.Cox | 98 |
| abstract_inverted_index.PGS | 34, 38, 88, 116, 126 |
| abstract_inverted_index.and | 15, 51, 72, 109, 128, 140 |
| abstract_inverted_index.are | 112 |
| abstract_inverted_index.for | 100, 103, 125 |
| abstract_inverted_index.the | 37, 92, 115 |
| abstract_inverted_index.via | 114 |
| abstract_inverted_index.PGSs | 56, 71 |
| abstract_inverted_index.both | 138 |
| abstract_inverted_index.data | 108 |
| abstract_inverted_index.from | 36, 91 |
| abstract_inverted_index.have | 19 |
| abstract_inverted_index.risk | 5 |
| abstract_inverted_index.that | 132 |
| abstract_inverted_index.this | 25, 28, 133 |
| abstract_inverted_index.will | 135 |
| abstract_inverted_index.3,168 | 33 |
| abstract_inverted_index.4,739 | 73 |
| abstract_inverted_index.major | 48, 105 |
| abstract_inverted_index.onset | 8 |
| abstract_inverted_index.score | 79 |
| abstract_inverted_index.tools | 111 |
| abstract_inverted_index.where | 54 |
| abstract_inverted_index.(PGSs) | 18 |
| abstract_inverted_index.10,531 | 62 |
| abstract_inverted_index.access | 84 |
| abstract_inverted_index.across | 47 |
| abstract_inverted_index.before | 6 |
| abstract_inverted_index.groups | 50 |
| abstract_inverted_index.models | 35, 99 |
| abstract_inverted_index.proven | 20 |
| abstract_inverted_index.public | 83 |
| abstract_inverted_index.scores | 17, 46 |
| abstract_inverted_index.study, | 29 |
| abstract_inverted_index.traits | 53 |
| abstract_inverted_index.439,070 | 67 |
| abstract_inverted_index.473,681 | 41 |
| abstract_inverted_index.Browser | 117 |
| abstract_inverted_index.Catalog | 39 |
| abstract_inverted_index.FinnGen | 42, 94 |
| abstract_inverted_index.advance | 137 |
| abstract_inverted_index.between | 70 |
| abstract_inverted_index.cohort, | 95 |
| abstract_inverted_index.derived | 90 |
| abstract_inverted_index.disease | 7 |
| abstract_inverted_index.esource | 134 |
| abstract_inverted_index.greatly | 136 |
| abstract_inverted_index.improve | 58 |
| abstract_inverted_index.provide | 82 |
| abstract_inverted_index.regard. | 26 |
| abstract_inverted_index.scores. | 145 |
| abstract_inverted_index.support | 77 |
| abstract_inverted_index.Abstract | 0 |
| abstract_inverted_index.analysis | 127 |
| abstract_inverted_index.ancestry | 49 |
| abstract_inverted_index.clinical | 74 |
| abstract_inverted_index.elevated | 4 |
| abstract_inverted_index.envision | 131 |
| abstract_inverted_index.platform | 124 |
| abstract_inverted_index.research | 139 |
| abstract_inverted_index.studies, | 65 |
| abstract_inverted_index.alongside | 96 |
| abstract_inverted_index.conducted | 61 |
| abstract_inverted_index.disorders | 106 |
| abstract_inverted_index.polygenic | 16, 144 |
| abstract_inverted_index.reference | 87 |
| abstract_inverted_index.revealing | 44 |
| abstract_inverted_index.web-based | 123 |
| abstract_inverted_index.accessible | 113 |
| abstract_inverted_index.endpoints. | 75 |
| abstract_inverted_index.individual | 78 |
| abstract_inverted_index.non-target | 55 |
| abstract_inverted_index.predicting | 101 |
| abstract_inverted_index.predictive | 110 |
| abstract_inverted_index.uncovering | 66 |
| abstract_inverted_index.Identifying | 1 |
| abstract_inverted_index.association | 64 |
| abstract_inverted_index.benchmarked | 32 |
| abstract_inverted_index.cornerstone | 11 |
| abstract_inverted_index.elastic-net | 97 |
| abstract_inverted_index.individuals | 2 |
| abstract_inverted_index.interactive | 122 |
| abstract_inverted_index.large-scale | 93 |
| abstract_inverted_index.nstrumental | 23 |
| abstract_inverted_index.prediction. | 59 |
| abstract_inverted_index.prevention, | 14 |
| abstract_inverted_index.significant | 68 |
| abstract_inverted_index.applications | 142 |
| abstract_inverted_index.associations | 69 |
| abstract_inverted_index.highlighting | 52 |
| abstract_inverted_index.personalized | 13 |
| abstract_inverted_index.phenome-wide | 63 |
| abstract_inverted_index.distributions | 89 |
| abstract_inverted_index.participants, | 43 |
| abstract_inverted_index.significantly | 57 |
| abstract_inverted_index.time-to-event | 102 |
| abstract_inverted_index.translational | 141 |
| abstract_inverted_index.pgs.nchigm.org | 119 |
| abstract_inverted_index.systematically | 31 |
| abstract_inverted_index.top-performing | 45 |
| abstract_inverted_index.interpretation, | 80 |
| abstract_inverted_index.interpretation. | 129 |
| abstract_inverted_index.ancestry-adjusted | 86 |
| cited_by_percentile_year | |
| countries_distinct_count | 0 |
| institutions_distinct_count | 13 |
| citation_normalized_percentile.value | 0.33335491 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |