Phylogenetic and Protein Domain Architecture Analysis of Toll- like Receptor 4-10 Genes in Camel (Camelus dromedarius) Article Swipe
 YOU?
·
· 2021
· Open Access
·
· DOI: https://doi.org/10.20546/ijcmas.2021.1001.335
YOU?
·
· 2021
· Open Access
·
· DOI: https://doi.org/10.20546/ijcmas.2021.1001.335
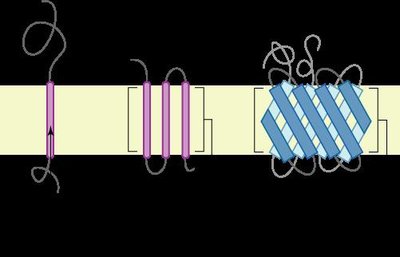
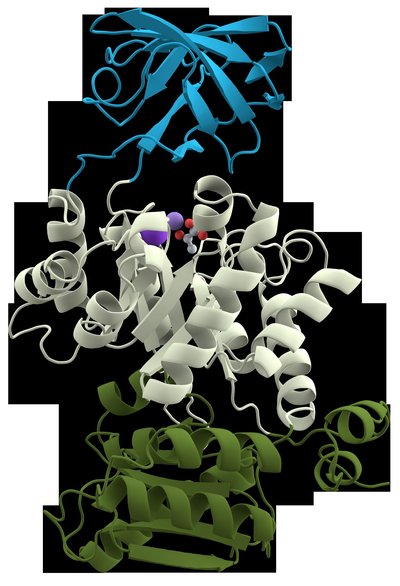
Toll-like receptors (TLRs) are evolutionary conserved protein and most extensively studied pathogen recognition receptors (PRRs) which provoke innate immune responses after recognition of invading pathogens.The present research is the first report of determination of domain architecture of camel (camelus dromedarius) TLRs4-10 proteins by Simple Modular Domain Architecture (SMART) tool.The study revealed that all the TLR proteins showed the typical structure containing Leucine Reach Repeats (ectodomain), transmembrane domain (TM), Toll/interleukin-1 receptor (TIR) domains except for TLR9 which was devoid of TM domain.The TLR 7,8,9 possessed more Leucine Rich Repeats (LRRs) motifsin their ectodomains than the other TLRs (4, 5, 6, 10).Further, the TM domain in TLRs (4-10) was exactly of similar length.However, the TIR domain was constituted of almost equal number of amino acids (140-144aa).Based on a phylogenetic analysis of the camel TLRs (1-10), the intracellularly expressed genes TLR7, TLR8 and TLR9 were clustered together whereas extracellularly expressed TLR1, 2, 6, and 10 branched out close to each other. K e y w o r d sCamel, domain analysis, Leucine rich repeats (LRRs), phylogenetic analysis, Toll like receptors (TLRs)
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.20546/ijcmas.2021.1001.335
- https://www.ijcmas.com/10-1-2021/Jyoti Choudhary, et al.pdf
- OA Status
- diamond
- Cited By
- 2
- References
- 23
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W3148566801
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W3148566801Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.20546/ijcmas.2021.1001.335Digital Object Identifier
- Title
-
Phylogenetic and Protein Domain Architecture Analysis of Toll- like Receptor 4-10 Genes in Camel (Camelus dromedarius)Work title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2021Year of publication
- Publication date
-
2021-01-10Full publication date if available
- Authors
-
Jyoti Choudhary, Mukul Purva, Mahendra Milind, Kritika Gahlot, Taruna Bhati, Sunil Maherchandani, Amit PandeyList of authors in order
- Landing page
-
https://doi.org/10.20546/ijcmas.2021.1001.335Publisher landing page
- PDF URL
-
https://www.ijcmas.com/10-1-2021/Jyoti Choudhary, et al.pdfDirect link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
diamondOpen access status per OpenAlex
- OA URL
-
https://www.ijcmas.com/10-1-2021/Jyoti Choudhary, et al.pdfDirect OA link when available
- Concepts
-
Ectodomain, Leucine-rich repeat, Architecture domain, Biology, Innate immune system, Transmembrane domain, TLR2, TLR9, Phylogenetic tree, Gene, Transmembrane protein, Receptor, Protein domain, Toll-like receptor, Toll-Like Receptor 9, Genetics, Computational biology, Gene expression, Architecture, Visual arts, DNA methylation, Art, Enterprise architecture management, Enterprise architectureTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
2Total citation count in OpenAlex
- Citations by year (recent)
-
2025: 1, 2023: 1Per-year citation counts (last 5 years)
- References (count)
-
23Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W3148566801 |
|---|---|
| doi | https://doi.org/10.20546/ijcmas.2021.1001.335 |
| ids.doi | https://doi.org/10.20546/ijcmas.2021.1001.335 |
| ids.mag | 3148566801 |
| ids.openalex | https://openalex.org/W3148566801 |
| fwci | 0.18178396 |
| type | article |
| title | Phylogenetic and Protein Domain Architecture Analysis of Toll- like Receptor 4-10 Genes in Camel (Camelus dromedarius) |
| biblio.issue | 01 |
| biblio.volume | 10 |
| biblio.last_page | 2895 |
| biblio.first_page | 2887 |
| topics[0].id | https://openalex.org/T13540 |
| topics[0].field.id | https://openalex.org/fields/11 |
| topics[0].field.display_name | Agricultural and Biological Sciences |
| topics[0].score | 0.9962999820709229 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1106 |
| topics[0].subfield.display_name | Food Science |
| topics[0].display_name | Animal Diversity and Health Studies |
| topics[1].id | https://openalex.org/T10371 |
| topics[1].field.id | https://openalex.org/fields/24 |
| topics[1].field.display_name | Immunology and Microbiology |
| topics[1].score | 0.9732000231742859 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/2403 |
| topics[1].subfield.display_name | Immunology |
| topics[1].display_name | Immune Response and Inflammation |
| topics[2].id | https://openalex.org/T11103 |
| topics[2].field.id | https://openalex.org/fields/24 |
| topics[2].field.display_name | Immunology and Microbiology |
| topics[2].score | 0.9480000138282776 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/2404 |
| topics[2].subfield.display_name | Microbiology |
| topics[2].display_name | Antimicrobial Peptides and Activities |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C1009742 |
| concepts[0].level | 3 |
| concepts[0].score | 0.9727557897567749 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q5333998 |
| concepts[0].display_name | Ectodomain |
| concepts[1].id | https://openalex.org/C99545290 |
| concepts[1].level | 3 |
| concepts[1].score | 0.8359972238540649 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q935326 |
| concepts[1].display_name | Leucine-rich repeat |
| concepts[2].id | https://openalex.org/C194167682 |
| concepts[2].level | 5 |
| concepts[2].score | 0.7070634365081787 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q4787088 |
| concepts[2].display_name | Architecture domain |
| concepts[3].id | https://openalex.org/C86803240 |
| concepts[3].level | 0 |
| concepts[3].score | 0.6447202563285828 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[3].display_name | Biology |
| concepts[4].id | https://openalex.org/C136449434 |
| concepts[4].level | 3 |
| concepts[4].score | 0.6445555090904236 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q428253 |
| concepts[4].display_name | Innate immune system |
| concepts[5].id | https://openalex.org/C118892022 |
| concepts[5].level | 3 |
| concepts[5].score | 0.6338085532188416 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q7834587 |
| concepts[5].display_name | Transmembrane domain |
| concepts[6].id | https://openalex.org/C2781236682 |
| concepts[6].level | 4 |
| concepts[6].score | 0.6254563927650452 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q410015 |
| concepts[6].display_name | TLR2 |
| concepts[7].id | https://openalex.org/C2779138994 |
| concepts[7].level | 5 |
| concepts[7].score | 0.6137874126434326 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q15332550 |
| concepts[7].display_name | TLR9 |
| concepts[8].id | https://openalex.org/C193252679 |
| concepts[8].level | 3 |
| concepts[8].score | 0.5995297431945801 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q242125 |
| concepts[8].display_name | Phylogenetic tree |
| concepts[9].id | https://openalex.org/C104317684 |
| concepts[9].level | 2 |
| concepts[9].score | 0.5712236166000366 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[9].display_name | Gene |
| concepts[10].id | https://openalex.org/C24530287 |
| concepts[10].level | 3 |
| concepts[10].score | 0.5135488510131836 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q424204 |
| concepts[10].display_name | Transmembrane protein |
| concepts[11].id | https://openalex.org/C170493617 |
| concepts[11].level | 2 |
| concepts[11].score | 0.4774487614631653 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q208467 |
| concepts[11].display_name | Receptor |
| concepts[12].id | https://openalex.org/C144292202 |
| concepts[12].level | 3 |
| concepts[12].score | 0.4537544250488281 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q898273 |
| concepts[12].display_name | Protein domain |
| concepts[13].id | https://openalex.org/C2776709828 |
| concepts[13].level | 4 |
| concepts[13].score | 0.43499892950057983 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q408004 |
| concepts[13].display_name | Toll-like receptor |
| concepts[14].id | https://openalex.org/C2908999202 |
| concepts[14].level | 5 |
| concepts[14].score | 0.4142468273639679 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q15332550 |
| concepts[14].display_name | Toll-Like Receptor 9 |
| concepts[15].id | https://openalex.org/C54355233 |
| concepts[15].level | 1 |
| concepts[15].score | 0.4006405770778656 |
| concepts[15].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[15].display_name | Genetics |
| concepts[16].id | https://openalex.org/C70721500 |
| concepts[16].level | 1 |
| concepts[16].score | 0.34770238399505615 |
| concepts[16].wikidata | https://www.wikidata.org/wiki/Q177005 |
| concepts[16].display_name | Computational biology |
| concepts[17].id | https://openalex.org/C150194340 |
| concepts[17].level | 3 |
| concepts[17].score | 0.1595432162284851 |
| concepts[17].wikidata | https://www.wikidata.org/wiki/Q26972 |
| concepts[17].display_name | Gene expression |
| concepts[18].id | https://openalex.org/C123657996 |
| concepts[18].level | 2 |
| concepts[18].score | 0.10485416650772095 |
| concepts[18].wikidata | https://www.wikidata.org/wiki/Q12271 |
| concepts[18].display_name | Architecture |
| concepts[19].id | https://openalex.org/C153349607 |
| concepts[19].level | 1 |
| concepts[19].score | 0.0 |
| concepts[19].wikidata | https://www.wikidata.org/wiki/Q36649 |
| concepts[19].display_name | Visual arts |
| concepts[20].id | https://openalex.org/C190727270 |
| concepts[20].level | 4 |
| concepts[20].score | 0.0 |
| concepts[20].wikidata | https://www.wikidata.org/wiki/Q874745 |
| concepts[20].display_name | DNA methylation |
| concepts[21].id | https://openalex.org/C142362112 |
| concepts[21].level | 0 |
| concepts[21].score | 0.0 |
| concepts[21].wikidata | https://www.wikidata.org/wiki/Q735 |
| concepts[21].display_name | Art |
| concepts[22].id | https://openalex.org/C163352659 |
| concepts[22].level | 4 |
| concepts[22].score | 0.0 |
| concepts[22].wikidata | https://www.wikidata.org/wiki/Q5380367 |
| concepts[22].display_name | Enterprise architecture management |
| concepts[23].id | https://openalex.org/C10590034 |
| concepts[23].level | 3 |
| concepts[23].score | 0.0 |
| concepts[23].wikidata | https://www.wikidata.org/wiki/Q1048431 |
| concepts[23].display_name | Enterprise architecture |
| keywords[0].id | https://openalex.org/keywords/ectodomain |
| keywords[0].score | 0.9727557897567749 |
| keywords[0].display_name | Ectodomain |
| keywords[1].id | https://openalex.org/keywords/leucine-rich-repeat |
| keywords[1].score | 0.8359972238540649 |
| keywords[1].display_name | Leucine-rich repeat |
| keywords[2].id | https://openalex.org/keywords/architecture-domain |
| keywords[2].score | 0.7070634365081787 |
| keywords[2].display_name | Architecture domain |
| keywords[3].id | https://openalex.org/keywords/biology |
| keywords[3].score | 0.6447202563285828 |
| keywords[3].display_name | Biology |
| keywords[4].id | https://openalex.org/keywords/innate-immune-system |
| keywords[4].score | 0.6445555090904236 |
| keywords[4].display_name | Innate immune system |
| keywords[5].id | https://openalex.org/keywords/transmembrane-domain |
| keywords[5].score | 0.6338085532188416 |
| keywords[5].display_name | Transmembrane domain |
| keywords[6].id | https://openalex.org/keywords/tlr2 |
| keywords[6].score | 0.6254563927650452 |
| keywords[6].display_name | TLR2 |
| keywords[7].id | https://openalex.org/keywords/tlr9 |
| keywords[7].score | 0.6137874126434326 |
| keywords[7].display_name | TLR9 |
| keywords[8].id | https://openalex.org/keywords/phylogenetic-tree |
| keywords[8].score | 0.5995297431945801 |
| keywords[8].display_name | Phylogenetic tree |
| keywords[9].id | https://openalex.org/keywords/gene |
| keywords[9].score | 0.5712236166000366 |
| keywords[9].display_name | Gene |
| keywords[10].id | https://openalex.org/keywords/transmembrane-protein |
| keywords[10].score | 0.5135488510131836 |
| keywords[10].display_name | Transmembrane protein |
| keywords[11].id | https://openalex.org/keywords/receptor |
| keywords[11].score | 0.4774487614631653 |
| keywords[11].display_name | Receptor |
| keywords[12].id | https://openalex.org/keywords/protein-domain |
| keywords[12].score | 0.4537544250488281 |
| keywords[12].display_name | Protein domain |
| keywords[13].id | https://openalex.org/keywords/toll-like-receptor |
| keywords[13].score | 0.43499892950057983 |
| keywords[13].display_name | Toll-like receptor |
| keywords[14].id | https://openalex.org/keywords/toll-like-receptor-9 |
| keywords[14].score | 0.4142468273639679 |
| keywords[14].display_name | Toll-Like Receptor 9 |
| keywords[15].id | https://openalex.org/keywords/genetics |
| keywords[15].score | 0.4006405770778656 |
| keywords[15].display_name | Genetics |
| keywords[16].id | https://openalex.org/keywords/computational-biology |
| keywords[16].score | 0.34770238399505615 |
| keywords[16].display_name | Computational biology |
| keywords[17].id | https://openalex.org/keywords/gene-expression |
| keywords[17].score | 0.1595432162284851 |
| keywords[17].display_name | Gene expression |
| keywords[18].id | https://openalex.org/keywords/architecture |
| keywords[18].score | 0.10485416650772095 |
| keywords[18].display_name | Architecture |
| language | en |
| locations[0].id | doi:10.20546/ijcmas.2021.1001.335 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S2765017008 |
| locations[0].source.issn | 2319-7692, 2319-7706 |
| locations[0].source.type | journal |
| locations[0].source.is_oa | True |
| locations[0].source.issn_l | 2319-7692 |
| locations[0].source.is_core | False |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | International Journal of Current Microbiology and Applied Sciences |
| locations[0].source.host_organization | https://openalex.org/P4322614448 |
| locations[0].source.host_organization_name | Excellent Publishers |
| locations[0].source.host_organization_lineage | https://openalex.org/P4322614448 |
| locations[0].license | |
| locations[0].pdf_url | https://www.ijcmas.com/10-1-2021/Jyoti Choudhary, et al.pdf |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | International Journal of Current Microbiology and Applied Sciences |
| locations[0].landing_page_url | https://doi.org/10.20546/ijcmas.2021.1001.335 |
| indexed_in | crossref |
| authorships[0].author.id | https://openalex.org/A5108041395 |
| authorships[0].author.orcid | https://orcid.org/0000-0001-8309-5954 |
| authorships[0].author.display_name | Jyoti Choudhary |
| authorships[0].affiliations[0].raw_affiliation_string | Department of Veterinary Microbiology and Biotechnology, |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Jyoti Choudhary |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Department of Veterinary Microbiology and Biotechnology, |
| authorships[1].author.id | https://openalex.org/A5018790131 |
| authorships[1].author.orcid | https://orcid.org/0000-0003-4863-5358 |
| authorships[1].author.display_name | Mukul Purva |
| authorships[1].affiliations[0].raw_affiliation_string | Department of Veterinary Microbiology and Biotechnology, |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Mukul Purva |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | Department of Veterinary Microbiology and Biotechnology, |
| authorships[2].author.id | https://openalex.org/A5067153754 |
| authorships[2].author.orcid | |
| authorships[2].author.display_name | Mahendra Milind |
| authorships[2].affiliations[0].raw_affiliation_string | Department of Veterinary Microbiology and Biotechnology, |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Mahendra Milind |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | Department of Veterinary Microbiology and Biotechnology, |
| authorships[3].author.id | https://openalex.org/A5070264063 |
| authorships[3].author.orcid | |
| authorships[3].author.display_name | Kritika Gahlot |
| authorships[3].affiliations[0].raw_affiliation_string | Department of Veterinary Microbiology and Biotechnology, |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Kritika Gahlot |
| authorships[3].is_corresponding | False |
| authorships[3].raw_affiliation_strings | Department of Veterinary Microbiology and Biotechnology, |
| authorships[4].author.id | https://openalex.org/A5047595797 |
| authorships[4].author.orcid | https://orcid.org/0000-0001-8460-5733 |
| authorships[4].author.display_name | Taruna Bhati |
| authorships[4].affiliations[0].raw_affiliation_string | Department of Veterinary Microbiology and Biotechnology, |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Taruna Bhati |
| authorships[4].is_corresponding | False |
| authorships[4].raw_affiliation_strings | Department of Veterinary Microbiology and Biotechnology, |
| authorships[5].author.id | https://openalex.org/A5000207188 |
| authorships[5].author.orcid | |
| authorships[5].author.display_name | Sunil Maherchandani |
| authorships[5].affiliations[0].raw_affiliation_string | Department of Veterinary Microbiology and Biotechnology, |
| authorships[5].author_position | middle |
| authorships[5].raw_author_name | Sunil Maherchandani |
| authorships[5].is_corresponding | False |
| authorships[5].raw_affiliation_strings | Department of Veterinary Microbiology and Biotechnology, |
| authorships[6].author.id | https://openalex.org/A5103784174 |
| authorships[6].author.orcid | https://orcid.org/0009-0000-8788-8791 |
| authorships[6].author.display_name | Amit Pandey |
| authorships[6].countries | IN |
| authorships[6].affiliations[0].institution_ids | https://openalex.org/I4210128267 |
| authorships[6].affiliations[0].raw_affiliation_string | College of Veterinary and Animal Science, RAJUVAS, Bikaner, Rajasthan, India |
| authorships[6].affiliations[1].raw_affiliation_string | Department of Biochemistry, |
| authorships[6].institutions[0].id | https://openalex.org/I4210128267 |
| authorships[6].institutions[0].ror | https://ror.org/03jjxkw75 |
| authorships[6].institutions[0].type | education |
| authorships[6].institutions[0].lineage | https://openalex.org/I4210128267 |
| authorships[6].institutions[0].country_code | IN |
| authorships[6].institutions[0].display_name | Rajasthan University of Veterinary and Animal Sciences |
| authorships[6].author_position | last |
| authorships[6].raw_author_name | Amit Pandey |
| authorships[6].is_corresponding | False |
| authorships[6].raw_affiliation_strings | College of Veterinary and Animal Science, RAJUVAS, Bikaner, Rajasthan, India, Department of Biochemistry, |
| has_content.pdf | True |
| has_content.grobid_xml | True |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://www.ijcmas.com/10-1-2021/Jyoti Choudhary, et al.pdf |
| open_access.oa_status | diamond |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Phylogenetic and Protein Domain Architecture Analysis of Toll- like Receptor 4-10 Genes in Camel (Camelus dromedarius) |
| has_fulltext | True |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T13540 |
| primary_topic.field.id | https://openalex.org/fields/11 |
| primary_topic.field.display_name | Agricultural and Biological Sciences |
| primary_topic.score | 0.9962999820709229 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1106 |
| primary_topic.subfield.display_name | Food Science |
| primary_topic.display_name | Animal Diversity and Health Studies |
| related_works | https://openalex.org/W580083713, https://openalex.org/W2314264549, https://openalex.org/W2129961630, https://openalex.org/W2148845369, https://openalex.org/W2374198738, https://openalex.org/W2795535550, https://openalex.org/W2032783993, https://openalex.org/W2056254683, https://openalex.org/W2042939195, https://openalex.org/W1924119614 |
| cited_by_count | 2 |
| counts_by_year[0].year | 2025 |
| counts_by_year[0].cited_by_count | 1 |
| counts_by_year[1].year | 2023 |
| counts_by_year[1].cited_by_count | 1 |
| locations_count | 1 |
| best_oa_location.id | doi:10.20546/ijcmas.2021.1001.335 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S2765017008 |
| best_oa_location.source.issn | 2319-7692, 2319-7706 |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | True |
| best_oa_location.source.issn_l | 2319-7692 |
| best_oa_location.source.is_core | False |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | International Journal of Current Microbiology and Applied Sciences |
| best_oa_location.source.host_organization | https://openalex.org/P4322614448 |
| best_oa_location.source.host_organization_name | Excellent Publishers |
| best_oa_location.source.host_organization_lineage | https://openalex.org/P4322614448 |
| best_oa_location.license | |
| best_oa_location.pdf_url | https://www.ijcmas.com/10-1-2021/Jyoti Choudhary, et al.pdf |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | journal-article |
| best_oa_location.license_id | |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | International Journal of Current Microbiology and Applied Sciences |
| best_oa_location.landing_page_url | https://doi.org/10.20546/ijcmas.2021.1001.335 |
| primary_location.id | doi:10.20546/ijcmas.2021.1001.335 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S2765017008 |
| primary_location.source.issn | 2319-7692, 2319-7706 |
| primary_location.source.type | journal |
| primary_location.source.is_oa | True |
| primary_location.source.issn_l | 2319-7692 |
| primary_location.source.is_core | False |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | International Journal of Current Microbiology and Applied Sciences |
| primary_location.source.host_organization | https://openalex.org/P4322614448 |
| primary_location.source.host_organization_name | Excellent Publishers |
| primary_location.source.host_organization_lineage | https://openalex.org/P4322614448 |
| primary_location.license | |
| primary_location.pdf_url | https://www.ijcmas.com/10-1-2021/Jyoti Choudhary, et al.pdf |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | International Journal of Current Microbiology and Applied Sciences |
| primary_location.landing_page_url | https://doi.org/10.20546/ijcmas.2021.1001.335 |
| publication_date | 2021-01-10 |
| publication_year | 2021 |
| referenced_works | https://openalex.org/W2033869585, https://openalex.org/W1969354039, https://openalex.org/W6659884305, https://openalex.org/W2138811813, https://openalex.org/W2063042300, https://openalex.org/W6675890380, https://openalex.org/W1503037824, https://openalex.org/W2256564896, https://openalex.org/W2149024057, https://openalex.org/W1995586746, https://openalex.org/W1987027200, https://openalex.org/W2796050959, https://openalex.org/W1968693613, https://openalex.org/W1994942139, https://openalex.org/W2432929256, https://openalex.org/W2046231927, https://openalex.org/W2105174344, https://openalex.org/W2004465286, https://openalex.org/W2794551838, https://openalex.org/W2036693151, https://openalex.org/W2111260918, https://openalex.org/W2120587073, https://openalex.org/W2025495699 |
| referenced_works_count | 23 |
| abstract_inverted_index.K | 158 |
| abstract_inverted_index.a | 125 |
| abstract_inverted_index.d | 164 |
| abstract_inverted_index.e | 159 |
| abstract_inverted_index.o | 162 |
| abstract_inverted_index.r | 163 |
| abstract_inverted_index.w | 161 |
| abstract_inverted_index.y | 160 |
| abstract_inverted_index.10 | 151 |
| abstract_inverted_index.2, | 148 |
| abstract_inverted_index.5, | 97 |
| abstract_inverted_index.6, | 98, 149 |
| abstract_inverted_index.TM | 79, 101 |
| abstract_inverted_index.by | 42 |
| abstract_inverted_index.in | 103 |
| abstract_inverted_index.is | 27 |
| abstract_inverted_index.of | 22, 31, 33, 36, 78, 108, 116, 120, 128 |
| abstract_inverted_index.on | 124 |
| abstract_inverted_index.to | 155 |
| abstract_inverted_index.(4, | 96 |
| abstract_inverted_index.TIR | 112 |
| abstract_inverted_index.TLR | 54, 81 |
| abstract_inverted_index.all | 52 |
| abstract_inverted_index.and | 7, 139, 150 |
| abstract_inverted_index.are | 3 |
| abstract_inverted_index.for | 73 |
| abstract_inverted_index.out | 153 |
| abstract_inverted_index.the | 28, 53, 57, 93, 100, 111, 129, 133 |
| abstract_inverted_index.was | 76, 106, 114 |
| abstract_inverted_index.Rich | 86 |
| abstract_inverted_index.TLR8 | 138 |
| abstract_inverted_index.TLR9 | 74, 140 |
| abstract_inverted_index.TLRs | 95, 104, 131 |
| abstract_inverted_index.Toll | 174 |
| abstract_inverted_index.each | 156 |
| abstract_inverted_index.like | 175 |
| abstract_inverted_index.more | 84 |
| abstract_inverted_index.most | 8 |
| abstract_inverted_index.rich | 169 |
| abstract_inverted_index.than | 92 |
| abstract_inverted_index.that | 51 |
| abstract_inverted_index.were | 141 |
| abstract_inverted_index.(TIR) | 70 |
| abstract_inverted_index.(TM), | 67 |
| abstract_inverted_index.7,8,9 | 82 |
| abstract_inverted_index.Reach | 62 |
| abstract_inverted_index.TLR1, | 147 |
| abstract_inverted_index.TLR7, | 137 |
| abstract_inverted_index.acids | 122 |
| abstract_inverted_index.after | 20 |
| abstract_inverted_index.amino | 121 |
| abstract_inverted_index.camel | 37, 130 |
| abstract_inverted_index.close | 154 |
| abstract_inverted_index.equal | 118 |
| abstract_inverted_index.first | 29 |
| abstract_inverted_index.genes | 136 |
| abstract_inverted_index.other | 94 |
| abstract_inverted_index.study | 49 |
| abstract_inverted_index.their | 90 |
| abstract_inverted_index.which | 15, 75 |
| abstract_inverted_index.(4-10) | 105 |
| abstract_inverted_index.(LRRs) | 88 |
| abstract_inverted_index.(PRRs) | 14 |
| abstract_inverted_index.(TLRs) | 2, 177 |
| abstract_inverted_index.Domain | 45 |
| abstract_inverted_index.Simple | 43 |
| abstract_inverted_index.almost | 117 |
| abstract_inverted_index.devoid | 77 |
| abstract_inverted_index.domain | 34, 66, 102, 113, 166 |
| abstract_inverted_index.except | 72 |
| abstract_inverted_index.immune | 18 |
| abstract_inverted_index.innate | 17 |
| abstract_inverted_index.number | 119 |
| abstract_inverted_index.other. | 157 |
| abstract_inverted_index.report | 30 |
| abstract_inverted_index.showed | 56 |
| abstract_inverted_index.(1-10), | 132 |
| abstract_inverted_index.(LRRs), | 171 |
| abstract_inverted_index.(SMART) | 47 |
| abstract_inverted_index.Leucine | 61, 85, 168 |
| abstract_inverted_index.Modular | 44 |
| abstract_inverted_index.Repeats | 63, 87 |
| abstract_inverted_index.domains | 71 |
| abstract_inverted_index.exactly | 107 |
| abstract_inverted_index.present | 25 |
| abstract_inverted_index.protein | 6 |
| abstract_inverted_index.provoke | 16 |
| abstract_inverted_index.repeats | 170 |
| abstract_inverted_index.sCamel, | 165 |
| abstract_inverted_index.similar | 109 |
| abstract_inverted_index.studied | 10 |
| abstract_inverted_index.typical | 58 |
| abstract_inverted_index.whereas | 144 |
| abstract_inverted_index.(camelus | 38 |
| abstract_inverted_index.TLRs4-10 | 40 |
| abstract_inverted_index.analysis | 127 |
| abstract_inverted_index.branched | 152 |
| abstract_inverted_index.invading | 23 |
| abstract_inverted_index.motifsin | 89 |
| abstract_inverted_index.pathogen | 11 |
| abstract_inverted_index.proteins | 41, 55 |
| abstract_inverted_index.receptor | 69 |
| abstract_inverted_index.research | 26 |
| abstract_inverted_index.revealed | 50 |
| abstract_inverted_index.together | 143 |
| abstract_inverted_index.tool.The | 48 |
| abstract_inverted_index.Toll-like | 0 |
| abstract_inverted_index.analysis, | 167, 173 |
| abstract_inverted_index.clustered | 142 |
| abstract_inverted_index.conserved | 5 |
| abstract_inverted_index.expressed | 135, 146 |
| abstract_inverted_index.possessed | 83 |
| abstract_inverted_index.receptors | 1, 13, 176 |
| abstract_inverted_index.responses | 19 |
| abstract_inverted_index.structure | 59 |
| abstract_inverted_index.containing | 60 |
| abstract_inverted_index.domain.The | 80 |
| abstract_inverted_index.constituted | 115 |
| abstract_inverted_index.ectodomains | 91 |
| abstract_inverted_index.extensively | 9 |
| abstract_inverted_index.recognition | 12, 21 |
| abstract_inverted_index.10).Further, | 99 |
| abstract_inverted_index.Architecture | 46 |
| abstract_inverted_index.architecture | 35 |
| abstract_inverted_index.dromedarius) | 39 |
| abstract_inverted_index.evolutionary | 4 |
| abstract_inverted_index.phylogenetic | 126, 172 |
| abstract_inverted_index.(ectodomain), | 64 |
| abstract_inverted_index.determination | 32 |
| abstract_inverted_index.pathogens.The | 24 |
| abstract_inverted_index.transmembrane | 65 |
| abstract_inverted_index.extracellularly | 145 |
| abstract_inverted_index.intracellularly | 134 |
| abstract_inverted_index.length.However, | 110 |
| abstract_inverted_index.(140-144aa).Based | 123 |
| abstract_inverted_index.Toll/interleukin-1 | 68 |
| cited_by_percentile_year.max | 95 |
| cited_by_percentile_year.min | 89 |
| countries_distinct_count | 1 |
| institutions_distinct_count | 7 |
| citation_normalized_percentile.value | 0.48879812 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |