Predicting Neoadjuvant Treatment Response in Triple-Negative Breast Cancer Using Machine Learning Article Swipe
 YOU?
·
· 2023
· Open Access
·
· DOI: https://doi.org/10.3390/diagnostics14010074
YOU?
·
· 2023
· Open Access
·
· DOI: https://doi.org/10.3390/diagnostics14010074
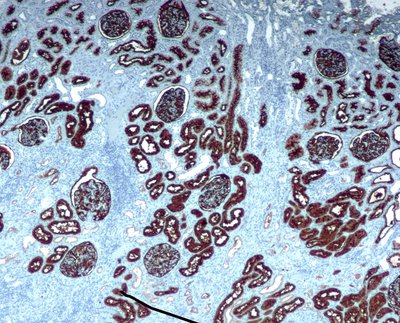
Background: Neoadjuvant chemotherapy (NAC) is the standard treatment for early-stage triple negative breast cancer (TNBC). The primary endpoint of NAC is a pathological complete response (pCR). NAC results in pCR in only 30–40% of TNBC patients. Tumor-infiltrating lymphocytes (TILs), Ki67 and phosphohistone H3 (pH3) are a few known biomarkers to predict NAC response. Currently, systematic evaluation of the combined value of these biomarkers in predicting NAC response is lacking. In this study, the predictive value of markers derived from H&E and IHC stained biopsy tissue was comprehensively evaluated using a supervised machine learning (ML)-based approach. Identifying predictive biomarkers could help guide therapeutic decisions by enabling precise stratification of TNBC patients into responders and partial or non-responders. Methods: Serial sections from core needle biopsies (n = 76) were stained with H&E and immunohistochemically for the Ki67 and pH3 markers, followed by whole-slide image (WSI) generation. The serial section stains in H&E stain, Ki67 and pH3 markers formed WSI triplets for each patient. The resulting WSI triplets were co-registered with H&E WSIs serving as the reference. Separate mask region-based CNN (MRCNN) models were trained with annotated H&E, Ki67 and pH3 images for detecting tumor cells, stromal and intratumoral TILs (sTILs and tTILs), Ki67+, and pH3+ cells. Top image patches with a high density of cells of interest were identified as hotspots. Best classifiers for NAC response prediction were identified by training multiple ML models and evaluating their performance by accuracy, area under curve, and confusion matrix analyses. Results: Highest prediction accuracy was achieved when hotspot regions were identified by tTIL counts and each hotspot was represented by measures of tTILs, sTILs, tumor cells, Ki67+, and pH3+ features. Regardless of the hotspot selection metric, a complementary use of multiple histological features (tTILs, sTILs) and molecular biomarkers (Ki67 and pH3) resulted in top ranked performance at the patient level. Conclusions: Overall, our results emphasize that prediction models for NAC response should be based on biomarkers in combination rather than in isolation. Our study provides compelling evidence to support the use of ML-based models to predict NAC response in patients with TNBC.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.3390/diagnostics14010074
- https://www.mdpi.com/2075-4418/14/1/74/pdf?version=1703759638
- OA Status
- gold
- Cited By
- 9
- References
- 54
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4390347463
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4390347463Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.3390/diagnostics14010074Digital Object Identifier
- Title
-
Predicting Neoadjuvant Treatment Response in Triple-Negative Breast Cancer Using Machine LearningWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2023Year of publication
- Publication date
-
2023-12-28Full publication date if available
- Authors
-
Shristi Bhattarai, Geetanjali Saini, Hongxiao Li, Gaurav Seth, Timothy B. Fisher, Emiel A. M. Janssen, Umay Kiraz, Jun Kong, Ritu AnejaList of authors in order
- Landing page
-
https://doi.org/10.3390/diagnostics14010074Publisher landing page
- PDF URL
-
https://www.mdpi.com/2075-4418/14/1/74/pdf?version=1703759638Direct link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
goldOpen access status per OpenAlex
- OA URL
-
https://www.mdpi.com/2075-4418/14/1/74/pdf?version=1703759638Direct OA link when available
- Concepts
-
Triple-negative breast cancer, Breast cancer, Medicine, Biopsy, Tumor-infiltrating lymphocytes, H&E stain, Digital pathology, Cancer, Pathology, Immunohistochemistry, Oncology, Internal medicine, ImmunotherapyTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
9Total citation count in OpenAlex
- Citations by year (recent)
-
2025: 7, 2024: 2Per-year citation counts (last 5 years)
- References (count)
-
54Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4390347463 |
|---|---|
| doi | https://doi.org/10.3390/diagnostics14010074 |
| ids.doi | https://doi.org/10.3390/diagnostics14010074 |
| ids.pmid | https://pubmed.ncbi.nlm.nih.gov/38201383 |
| ids.openalex | https://openalex.org/W4390347463 |
| fwci | 2.78149469 |
| type | article |
| title | Predicting Neoadjuvant Treatment Response in Triple-Negative Breast Cancer Using Machine Learning |
| biblio.issue | 1 |
| biblio.volume | 14 |
| biblio.last_page | 74 |
| biblio.first_page | 74 |
| topics[0].id | https://openalex.org/T12422 |
| topics[0].field.id | https://openalex.org/fields/27 |
| topics[0].field.display_name | Medicine |
| topics[0].score | 0.9998000264167786 |
| topics[0].domain.id | https://openalex.org/domains/4 |
| topics[0].domain.display_name | Health Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/2741 |
| topics[0].subfield.display_name | Radiology, Nuclear Medicine and Imaging |
| topics[0].display_name | Radiomics and Machine Learning in Medical Imaging |
| topics[1].id | https://openalex.org/T10183 |
| topics[1].field.id | https://openalex.org/fields/13 |
| topics[1].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[1].score | 0.9997000098228455 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1306 |
| topics[1].subfield.display_name | Cancer Research |
| topics[1].display_name | Breast Cancer Treatment Studies |
| topics[2].id | https://openalex.org/T10862 |
| topics[2].field.id | https://openalex.org/fields/17 |
| topics[2].field.display_name | Computer Science |
| topics[2].score | 0.9995999932289124 |
| topics[2].domain.id | https://openalex.org/domains/3 |
| topics[2].domain.display_name | Physical Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1702 |
| topics[2].subfield.display_name | Artificial Intelligence |
| topics[2].display_name | AI in cancer detection |
| is_xpac | False |
| apc_list.value | 2000 |
| apc_list.currency | CHF |
| apc_list.value_usd | 2165 |
| apc_paid.value | 2000 |
| apc_paid.currency | CHF |
| apc_paid.value_usd | 2165 |
| concepts[0].id | https://openalex.org/C2780110267 |
| concepts[0].level | 4 |
| concepts[0].score | 0.800706148147583 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q7843332 |
| concepts[0].display_name | Triple-negative breast cancer |
| concepts[1].id | https://openalex.org/C530470458 |
| concepts[1].level | 3 |
| concepts[1].score | 0.6316923499107361 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q128581 |
| concepts[1].display_name | Breast cancer |
| concepts[2].id | https://openalex.org/C71924100 |
| concepts[2].level | 0 |
| concepts[2].score | 0.6259180903434753 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q11190 |
| concepts[2].display_name | Medicine |
| concepts[3].id | https://openalex.org/C2775934546 |
| concepts[3].level | 2 |
| concepts[3].score | 0.5224137306213379 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q179991 |
| concepts[3].display_name | Biopsy |
| concepts[4].id | https://openalex.org/C2778326572 |
| concepts[4].level | 4 |
| concepts[4].score | 0.4701445400714874 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q7852668 |
| concepts[4].display_name | Tumor-infiltrating lymphocytes |
| concepts[5].id | https://openalex.org/C125473707 |
| concepts[5].level | 3 |
| concepts[5].score | 0.44690415263175964 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q9914 |
| concepts[5].display_name | H&E stain |
| concepts[6].id | https://openalex.org/C2777522853 |
| concepts[6].level | 2 |
| concepts[6].score | 0.43005573749542236 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q5276128 |
| concepts[6].display_name | Digital pathology |
| concepts[7].id | https://openalex.org/C121608353 |
| concepts[7].level | 2 |
| concepts[7].score | 0.3788311183452606 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q12078 |
| concepts[7].display_name | Cancer |
| concepts[8].id | https://openalex.org/C142724271 |
| concepts[8].level | 1 |
| concepts[8].score | 0.3759899437427521 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q7208 |
| concepts[8].display_name | Pathology |
| concepts[9].id | https://openalex.org/C204232928 |
| concepts[9].level | 2 |
| concepts[9].score | 0.3381648659706116 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q899285 |
| concepts[9].display_name | Immunohistochemistry |
| concepts[10].id | https://openalex.org/C143998085 |
| concepts[10].level | 1 |
| concepts[10].score | 0.3234425187110901 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q162555 |
| concepts[10].display_name | Oncology |
| concepts[11].id | https://openalex.org/C126322002 |
| concepts[11].level | 1 |
| concepts[11].score | 0.24163267016410828 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q11180 |
| concepts[11].display_name | Internal medicine |
| concepts[12].id | https://openalex.org/C2777701055 |
| concepts[12].level | 3 |
| concepts[12].score | 0.09107944369316101 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q1427096 |
| concepts[12].display_name | Immunotherapy |
| keywords[0].id | https://openalex.org/keywords/triple-negative-breast-cancer |
| keywords[0].score | 0.800706148147583 |
| keywords[0].display_name | Triple-negative breast cancer |
| keywords[1].id | https://openalex.org/keywords/breast-cancer |
| keywords[1].score | 0.6316923499107361 |
| keywords[1].display_name | Breast cancer |
| keywords[2].id | https://openalex.org/keywords/medicine |
| keywords[2].score | 0.6259180903434753 |
| keywords[2].display_name | Medicine |
| keywords[3].id | https://openalex.org/keywords/biopsy |
| keywords[3].score | 0.5224137306213379 |
| keywords[3].display_name | Biopsy |
| keywords[4].id | https://openalex.org/keywords/tumor-infiltrating-lymphocytes |
| keywords[4].score | 0.4701445400714874 |
| keywords[4].display_name | Tumor-infiltrating lymphocytes |
| keywords[5].id | https://openalex.org/keywords/h-e-stain |
| keywords[5].score | 0.44690415263175964 |
| keywords[5].display_name | H&E stain |
| keywords[6].id | https://openalex.org/keywords/digital-pathology |
| keywords[6].score | 0.43005573749542236 |
| keywords[6].display_name | Digital pathology |
| keywords[7].id | https://openalex.org/keywords/cancer |
| keywords[7].score | 0.3788311183452606 |
| keywords[7].display_name | Cancer |
| keywords[8].id | https://openalex.org/keywords/pathology |
| keywords[8].score | 0.3759899437427521 |
| keywords[8].display_name | Pathology |
| keywords[9].id | https://openalex.org/keywords/immunohistochemistry |
| keywords[9].score | 0.3381648659706116 |
| keywords[9].display_name | Immunohistochemistry |
| keywords[10].id | https://openalex.org/keywords/oncology |
| keywords[10].score | 0.3234425187110901 |
| keywords[10].display_name | Oncology |
| keywords[11].id | https://openalex.org/keywords/internal-medicine |
| keywords[11].score | 0.24163267016410828 |
| keywords[11].display_name | Internal medicine |
| keywords[12].id | https://openalex.org/keywords/immunotherapy |
| keywords[12].score | 0.09107944369316101 |
| keywords[12].display_name | Immunotherapy |
| language | en |
| locations[0].id | doi:10.3390/diagnostics14010074 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4210172076 |
| locations[0].source.issn | 2075-4418 |
| locations[0].source.type | journal |
| locations[0].source.is_oa | True |
| locations[0].source.issn_l | 2075-4418 |
| locations[0].source.is_core | True |
| locations[0].source.is_in_doaj | True |
| locations[0].source.display_name | Diagnostics |
| locations[0].source.host_organization | https://openalex.org/P4310310987 |
| locations[0].source.host_organization_name | Multidisciplinary Digital Publishing Institute |
| locations[0].source.host_organization_lineage | https://openalex.org/P4310310987 |
| locations[0].source.host_organization_lineage_names | Multidisciplinary Digital Publishing Institute |
| locations[0].license | cc-by |
| locations[0].pdf_url | https://www.mdpi.com/2075-4418/14/1/74/pdf?version=1703759638 |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | Diagnostics |
| locations[0].landing_page_url | https://doi.org/10.3390/diagnostics14010074 |
| locations[1].id | pmid:38201383 |
| locations[1].is_oa | False |
| locations[1].source.id | https://openalex.org/S4306525036 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | PubMed |
| locations[1].source.host_organization | https://openalex.org/I1299303238 |
| locations[1].source.host_organization_name | National Institutes of Health |
| locations[1].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[1].license | |
| locations[1].pdf_url | |
| locations[1].version | publishedVersion |
| locations[1].raw_type | |
| locations[1].license_id | |
| locations[1].is_accepted | True |
| locations[1].is_published | True |
| locations[1].raw_source_name | Diagnostics (Basel, Switzerland) |
| locations[1].landing_page_url | https://pubmed.ncbi.nlm.nih.gov/38201383 |
| locations[2].id | pmh:oai:pubmedcentral.nih.gov:10871101 |
| locations[2].is_oa | True |
| locations[2].source.id | https://openalex.org/S2764455111 |
| locations[2].source.issn | |
| locations[2].source.type | repository |
| locations[2].source.is_oa | False |
| locations[2].source.issn_l | |
| locations[2].source.is_core | False |
| locations[2].source.is_in_doaj | False |
| locations[2].source.display_name | PubMed Central |
| locations[2].source.host_organization | https://openalex.org/I1299303238 |
| locations[2].source.host_organization_name | National Institutes of Health |
| locations[2].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[2].license | cc-by |
| locations[2].pdf_url | https://pmc.ncbi.nlm.nih.gov/articles/PMC10871101/pdf/diagnostics-14-00074.pdf |
| locations[2].version | submittedVersion |
| locations[2].raw_type | Text |
| locations[2].license_id | https://openalex.org/licenses/cc-by |
| locations[2].is_accepted | False |
| locations[2].is_published | False |
| locations[2].raw_source_name | Diagnostics (Basel) |
| locations[2].landing_page_url | https://www.ncbi.nlm.nih.gov/pmc/articles/10871101 |
| locations[3].id | pmh:oai:doaj.org/article:76ffd498081d4fdea5f0b9520970ba1e |
| locations[3].is_oa | False |
| locations[3].source.id | https://openalex.org/S4306401280 |
| locations[3].source.issn | |
| locations[3].source.type | repository |
| locations[3].source.is_oa | False |
| locations[3].source.issn_l | |
| locations[3].source.is_core | False |
| locations[3].source.is_in_doaj | False |
| locations[3].source.display_name | DOAJ (DOAJ: Directory of Open Access Journals) |
| locations[3].source.host_organization | |
| locations[3].source.host_organization_name | |
| locations[3].license | |
| locations[3].pdf_url | |
| locations[3].version | submittedVersion |
| locations[3].raw_type | article |
| locations[3].license_id | |
| locations[3].is_accepted | False |
| locations[3].is_published | False |
| locations[3].raw_source_name | Diagnostics, Vol 14, Iss 1, p 74 (2023) |
| locations[3].landing_page_url | https://doaj.org/article/76ffd498081d4fdea5f0b9520970ba1e |
| indexed_in | crossref, doaj, pubmed |
| authorships[0].author.id | https://openalex.org/A5050041510 |
| authorships[0].author.orcid | |
| authorships[0].author.display_name | Shristi Bhattarai |
| authorships[0].countries | US |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I32389192 |
| authorships[0].affiliations[0].raw_affiliation_string | Department of Clinical and Diagnostic Sciences, School of Health Professions, University of Alabama at Birmingham, Birmingham, AL 35294, USA |
| authorships[0].institutions[0].id | https://openalex.org/I32389192 |
| authorships[0].institutions[0].ror | https://ror.org/008s83205 |
| authorships[0].institutions[0].type | education |
| authorships[0].institutions[0].lineage | https://openalex.org/I32389192 |
| authorships[0].institutions[0].country_code | US |
| authorships[0].institutions[0].display_name | University of Alabama at Birmingham |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Shristi Bhattarai |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Department of Clinical and Diagnostic Sciences, School of Health Professions, University of Alabama at Birmingham, Birmingham, AL 35294, USA |
| authorships[1].author.id | https://openalex.org/A5113083235 |
| authorships[1].author.orcid | |
| authorships[1].author.display_name | Geetanjali Saini |
| authorships[1].countries | US |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I32389192 |
| authorships[1].affiliations[0].raw_affiliation_string | Department of Clinical and Diagnostic Sciences, School of Health Professions, University of Alabama at Birmingham, Birmingham, AL 35294, USA |
| authorships[1].institutions[0].id | https://openalex.org/I32389192 |
| authorships[1].institutions[0].ror | https://ror.org/008s83205 |
| authorships[1].institutions[0].type | education |
| authorships[1].institutions[0].lineage | https://openalex.org/I32389192 |
| authorships[1].institutions[0].country_code | US |
| authorships[1].institutions[0].display_name | University of Alabama at Birmingham |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Geetanjali Saini |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | Department of Clinical and Diagnostic Sciences, School of Health Professions, University of Alabama at Birmingham, Birmingham, AL 35294, USA |
| authorships[2].author.id | https://openalex.org/A5101824486 |
| authorships[2].author.orcid | https://orcid.org/0000-0002-6114-8215 |
| authorships[2].author.display_name | Hongxiao Li |
| authorships[2].countries | US |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I181565077 |
| authorships[2].affiliations[0].raw_affiliation_string | Department of Mathematics and Statistics, Georgia State University, Atlanta, GA 30302, USA |
| authorships[2].institutions[0].id | https://openalex.org/I181565077 |
| authorships[2].institutions[0].ror | https://ror.org/03qt6ba18 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I181565077 |
| authorships[2].institutions[0].country_code | US |
| authorships[2].institutions[0].display_name | Georgia State University |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Hongxiao Li |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | Department of Mathematics and Statistics, Georgia State University, Atlanta, GA 30302, USA |
| authorships[3].author.id | https://openalex.org/A5027436323 |
| authorships[3].author.orcid | https://orcid.org/0009-0006-3466-9345 |
| authorships[3].author.display_name | Gaurav Seth |
| authorships[3].countries | US |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I32389192 |
| authorships[3].affiliations[0].raw_affiliation_string | Department of Clinical and Diagnostic Sciences, School of Health Professions, University of Alabama at Birmingham, Birmingham, AL 35294, USA |
| authorships[3].institutions[0].id | https://openalex.org/I32389192 |
| authorships[3].institutions[0].ror | https://ror.org/008s83205 |
| authorships[3].institutions[0].type | education |
| authorships[3].institutions[0].lineage | https://openalex.org/I32389192 |
| authorships[3].institutions[0].country_code | US |
| authorships[3].institutions[0].display_name | University of Alabama at Birmingham |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Gaurav Seth |
| authorships[3].is_corresponding | False |
| authorships[3].raw_affiliation_strings | Department of Clinical and Diagnostic Sciences, School of Health Professions, University of Alabama at Birmingham, Birmingham, AL 35294, USA |
| authorships[4].author.id | https://openalex.org/A5026106543 |
| authorships[4].author.orcid | |
| authorships[4].author.display_name | Timothy B. Fisher |
| authorships[4].countries | US |
| authorships[4].affiliations[0].institution_ids | https://openalex.org/I181565077 |
| authorships[4].affiliations[0].raw_affiliation_string | Department of Biology, Georgia State University, Atlanta, GA 30302, USA |
| authorships[4].institutions[0].id | https://openalex.org/I181565077 |
| authorships[4].institutions[0].ror | https://ror.org/03qt6ba18 |
| authorships[4].institutions[0].type | education |
| authorships[4].institutions[0].lineage | https://openalex.org/I181565077 |
| authorships[4].institutions[0].country_code | US |
| authorships[4].institutions[0].display_name | Georgia State University |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Timothy B. Fisher |
| authorships[4].is_corresponding | False |
| authorships[4].raw_affiliation_strings | Department of Biology, Georgia State University, Atlanta, GA 30302, USA |
| authorships[5].author.id | https://openalex.org/A5003110329 |
| authorships[5].author.orcid | https://orcid.org/0000-0002-7760-5396 |
| authorships[5].author.display_name | Emiel A. M. Janssen |
| authorships[5].countries | NO |
| authorships[5].affiliations[0].institution_ids | https://openalex.org/I92008406 |
| authorships[5].affiliations[0].raw_affiliation_string | Department of Chemistry, Bioscience and Environmental Engineering, Stavanger University, 4021 Stavanger, Norway |
| authorships[5].affiliations[1].institution_ids | https://openalex.org/I2800132241 |
| authorships[5].affiliations[1].raw_affiliation_string | Department of Pathology, Stavanger University Hospital, 4011 Stavanger, Norway |
| authorships[5].institutions[0].id | https://openalex.org/I2800132241 |
| authorships[5].institutions[0].ror | https://ror.org/04zn72g03 |
| authorships[5].institutions[0].type | healthcare |
| authorships[5].institutions[0].lineage | https://openalex.org/I2800132241 |
| authorships[5].institutions[0].country_code | NO |
| authorships[5].institutions[0].display_name | Stavanger University Hospital |
| authorships[5].institutions[1].id | https://openalex.org/I92008406 |
| authorships[5].institutions[1].ror | https://ror.org/02qte9q33 |
| authorships[5].institutions[1].type | education |
| authorships[5].institutions[1].lineage | https://openalex.org/I92008406 |
| authorships[5].institutions[1].country_code | NO |
| authorships[5].institutions[1].display_name | University of Stavanger |
| authorships[5].author_position | middle |
| authorships[5].raw_author_name | Emiel A. M. Janssen |
| authorships[5].is_corresponding | False |
| authorships[5].raw_affiliation_strings | Department of Chemistry, Bioscience and Environmental Engineering, Stavanger University, 4021 Stavanger, Norway, Department of Pathology, Stavanger University Hospital, 4011 Stavanger, Norway |
| authorships[6].author.id | https://openalex.org/A5025905653 |
| authorships[6].author.orcid | https://orcid.org/0000-0002-6721-4877 |
| authorships[6].author.display_name | Umay Kiraz |
| authorships[6].countries | NO |
| authorships[6].affiliations[0].institution_ids | https://openalex.org/I2800132241 |
| authorships[6].affiliations[0].raw_affiliation_string | Department of Pathology, Stavanger University Hospital, 4011 Stavanger, Norway |
| authorships[6].affiliations[1].institution_ids | https://openalex.org/I92008406 |
| authorships[6].affiliations[1].raw_affiliation_string | Department of Chemistry, Bioscience and Environmental Engineering, Stavanger University, 4021 Stavanger, Norway |
| authorships[6].institutions[0].id | https://openalex.org/I2800132241 |
| authorships[6].institutions[0].ror | https://ror.org/04zn72g03 |
| authorships[6].institutions[0].type | healthcare |
| authorships[6].institutions[0].lineage | https://openalex.org/I2800132241 |
| authorships[6].institutions[0].country_code | NO |
| authorships[6].institutions[0].display_name | Stavanger University Hospital |
| authorships[6].institutions[1].id | https://openalex.org/I92008406 |
| authorships[6].institutions[1].ror | https://ror.org/02qte9q33 |
| authorships[6].institutions[1].type | education |
| authorships[6].institutions[1].lineage | https://openalex.org/I92008406 |
| authorships[6].institutions[1].country_code | NO |
| authorships[6].institutions[1].display_name | University of Stavanger |
| authorships[6].author_position | middle |
| authorships[6].raw_author_name | Umay Kiraz |
| authorships[6].is_corresponding | False |
| authorships[6].raw_affiliation_strings | Department of Chemistry, Bioscience and Environmental Engineering, Stavanger University, 4021 Stavanger, Norway, Department of Pathology, Stavanger University Hospital, 4011 Stavanger, Norway |
| authorships[7].author.id | https://openalex.org/A5066949209 |
| authorships[7].author.orcid | https://orcid.org/0000-0001-8757-8339 |
| authorships[7].author.display_name | Jun Kong |
| authorships[7].countries | US |
| authorships[7].affiliations[0].institution_ids | https://openalex.org/I181565077 |
| authorships[7].affiliations[0].raw_affiliation_string | Department of Mathematics and Statistics, Georgia State University, Atlanta, GA 30302, USA |
| authorships[7].institutions[0].id | https://openalex.org/I181565077 |
| authorships[7].institutions[0].ror | https://ror.org/03qt6ba18 |
| authorships[7].institutions[0].type | education |
| authorships[7].institutions[0].lineage | https://openalex.org/I181565077 |
| authorships[7].institutions[0].country_code | US |
| authorships[7].institutions[0].display_name | Georgia State University |
| authorships[7].author_position | middle |
| authorships[7].raw_author_name | Jun Kong |
| authorships[7].is_corresponding | True |
| authorships[7].raw_affiliation_strings | Department of Mathematics and Statistics, Georgia State University, Atlanta, GA 30302, USA |
| authorships[8].author.id | https://openalex.org/A5019846100 |
| authorships[8].author.orcid | https://orcid.org/0000-0003-4489-5320 |
| authorships[8].author.display_name | Ritu Aneja |
| authorships[8].countries | US |
| authorships[8].affiliations[0].institution_ids | https://openalex.org/I32389192 |
| authorships[8].affiliations[0].raw_affiliation_string | Department of Clinical and Diagnostic Sciences, School of Health Professions, University of Alabama at Birmingham, Birmingham, AL 35294, USA |
| authorships[8].institutions[0].id | https://openalex.org/I32389192 |
| authorships[8].institutions[0].ror | https://ror.org/008s83205 |
| authorships[8].institutions[0].type | education |
| authorships[8].institutions[0].lineage | https://openalex.org/I32389192 |
| authorships[8].institutions[0].country_code | US |
| authorships[8].institutions[0].display_name | University of Alabama at Birmingham |
| authorships[8].author_position | last |
| authorships[8].raw_author_name | Ritu Aneja |
| authorships[8].is_corresponding | True |
| authorships[8].raw_affiliation_strings | Department of Clinical and Diagnostic Sciences, School of Health Professions, University of Alabama at Birmingham, Birmingham, AL 35294, USA |
| has_content.pdf | True |
| has_content.grobid_xml | True |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://www.mdpi.com/2075-4418/14/1/74/pdf?version=1703759638 |
| open_access.oa_status | gold |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Predicting Neoadjuvant Treatment Response in Triple-Negative Breast Cancer Using Machine Learning |
| has_fulltext | True |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T12422 |
| primary_topic.field.id | https://openalex.org/fields/27 |
| primary_topic.field.display_name | Medicine |
| primary_topic.score | 0.9998000264167786 |
| primary_topic.domain.id | https://openalex.org/domains/4 |
| primary_topic.domain.display_name | Health Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/2741 |
| primary_topic.subfield.display_name | Radiology, Nuclear Medicine and Imaging |
| primary_topic.display_name | Radiomics and Machine Learning in Medical Imaging |
| related_works | https://openalex.org/W2065857975, https://openalex.org/W1759030281, https://openalex.org/W4386103331, https://openalex.org/W3012179402, https://openalex.org/W4214718886, https://openalex.org/W3097689000, https://openalex.org/W2125942968, https://openalex.org/W4288034278, https://openalex.org/W4387618892, https://openalex.org/W4378952948 |
| cited_by_count | 9 |
| counts_by_year[0].year | 2025 |
| counts_by_year[0].cited_by_count | 7 |
| counts_by_year[1].year | 2024 |
| counts_by_year[1].cited_by_count | 2 |
| locations_count | 4 |
| best_oa_location.id | doi:10.3390/diagnostics14010074 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4210172076 |
| best_oa_location.source.issn | 2075-4418 |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | True |
| best_oa_location.source.issn_l | 2075-4418 |
| best_oa_location.source.is_core | True |
| best_oa_location.source.is_in_doaj | True |
| best_oa_location.source.display_name | Diagnostics |
| best_oa_location.source.host_organization | https://openalex.org/P4310310987 |
| best_oa_location.source.host_organization_name | Multidisciplinary Digital Publishing Institute |
| best_oa_location.source.host_organization_lineage | https://openalex.org/P4310310987 |
| best_oa_location.source.host_organization_lineage_names | Multidisciplinary Digital Publishing Institute |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | https://www.mdpi.com/2075-4418/14/1/74/pdf?version=1703759638 |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | journal-article |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | Diagnostics |
| best_oa_location.landing_page_url | https://doi.org/10.3390/diagnostics14010074 |
| primary_location.id | doi:10.3390/diagnostics14010074 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4210172076 |
| primary_location.source.issn | 2075-4418 |
| primary_location.source.type | journal |
| primary_location.source.is_oa | True |
| primary_location.source.issn_l | 2075-4418 |
| primary_location.source.is_core | True |
| primary_location.source.is_in_doaj | True |
| primary_location.source.display_name | Diagnostics |
| primary_location.source.host_organization | https://openalex.org/P4310310987 |
| primary_location.source.host_organization_name | Multidisciplinary Digital Publishing Institute |
| primary_location.source.host_organization_lineage | https://openalex.org/P4310310987 |
| primary_location.source.host_organization_lineage_names | Multidisciplinary Digital Publishing Institute |
| primary_location.license | cc-by |
| primary_location.pdf_url | https://www.mdpi.com/2075-4418/14/1/74/pdf?version=1703759638 |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | Diagnostics |
| primary_location.landing_page_url | https://doi.org/10.3390/diagnostics14010074 |
| publication_date | 2023-12-28 |
| publication_year | 2023 |
| referenced_works | https://openalex.org/W2738153741, https://openalex.org/W7074011258, https://openalex.org/W3031939267, https://openalex.org/W3006930853, https://openalex.org/W6719700345, https://openalex.org/W2886991068, https://openalex.org/W4313889797, https://openalex.org/W1990573783, https://openalex.org/W2999521665, https://openalex.org/W2612476887, https://openalex.org/W2968288061, https://openalex.org/W2082172177, https://openalex.org/W3110744452, https://openalex.org/W3087393791, https://openalex.org/W4280617720, https://openalex.org/W6754773263, https://openalex.org/W3211240094, https://openalex.org/W2628196379, https://openalex.org/W2120431466, https://openalex.org/W2806070179, https://openalex.org/W2150134401, https://openalex.org/W2168197820, https://openalex.org/W2161136893, https://openalex.org/W3006134372, https://openalex.org/W2937307539, https://openalex.org/W3204013916, https://openalex.org/W6803739019, https://openalex.org/W3176612188, https://openalex.org/W3200545581, https://openalex.org/W4281626918, https://openalex.org/W3018084241, https://openalex.org/W2420462095, https://openalex.org/W3093593152, https://openalex.org/W2132893003, https://openalex.org/W2466569086, https://openalex.org/W2572716935, https://openalex.org/W2048492216, https://openalex.org/W1977295091, https://openalex.org/W2561856923, https://openalex.org/W2126486896, https://openalex.org/W2154536753, https://openalex.org/W2789428140, https://openalex.org/W6774302454, https://openalex.org/W2886485146, https://openalex.org/W2037718815, https://openalex.org/W2131754688, https://openalex.org/W2150351703, https://openalex.org/W2086524378, https://openalex.org/W3006138035, https://openalex.org/W2892287597, https://openalex.org/W2112687290, https://openalex.org/W3008893950, https://openalex.org/W2044702943, https://openalex.org/W3210165781 |
| referenced_works_count | 54 |
| abstract_inverted_index.= | 124 |
| abstract_inverted_index.a | 21, 45, 89, 208, 281 |
| abstract_inverted_index.(n | 123 |
| abstract_inverted_index.H3 | 42 |
| abstract_inverted_index.In | 69 |
| abstract_inverted_index.ML | 230 |
| abstract_inverted_index.as | 171, 217 |
| abstract_inverted_index.at | 301 |
| abstract_inverted_index.be | 317 |
| abstract_inverted_index.by | 103, 139, 227, 236, 256, 264 |
| abstract_inverted_index.in | 28, 30, 63, 148, 297, 321, 325, 343 |
| abstract_inverted_index.is | 4, 20, 67 |
| abstract_inverted_index.of | 18, 33, 56, 60, 75, 107, 211, 213, 266, 276, 284, 336 |
| abstract_inverted_index.on | 319 |
| abstract_inverted_index.or | 114 |
| abstract_inverted_index.to | 49, 332, 339 |
| abstract_inverted_index.76) | 125 |
| abstract_inverted_index.CNN | 177 |
| abstract_inverted_index.IHC | 81 |
| abstract_inverted_index.NAC | 19, 26, 51, 65, 222, 314, 341 |
| abstract_inverted_index.Our | 327 |
| abstract_inverted_index.The | 15, 144, 161 |
| abstract_inverted_index.Top | 204 |
| abstract_inverted_index.WSI | 156, 163 |
| abstract_inverted_index.and | 40, 80, 112, 130, 135, 152, 186, 194, 198, 201, 232, 241, 259, 272, 290, 294 |
| abstract_inverted_index.are | 44 |
| abstract_inverted_index.few | 46 |
| abstract_inverted_index.for | 8, 132, 158, 189, 221, 313 |
| abstract_inverted_index.our | 307 |
| abstract_inverted_index.pCR | 29 |
| abstract_inverted_index.pH3 | 136, 153, 187 |
| abstract_inverted_index.the | 5, 57, 72, 133, 172, 277, 302, 334 |
| abstract_inverted_index.top | 298 |
| abstract_inverted_index.use | 283, 335 |
| abstract_inverted_index.was | 85, 249, 262 |
| abstract_inverted_index.Best | 219 |
| abstract_inverted_index.Ki67 | 39, 134, 151, 185 |
| abstract_inverted_index.TILs | 196 |
| abstract_inverted_index.TNBC | 34, 108 |
| abstract_inverted_index.WSIs | 169 |
| abstract_inverted_index.area | 238 |
| abstract_inverted_index.core | 120 |
| abstract_inverted_index.each | 159, 260 |
| abstract_inverted_index.from | 78, 119 |
| abstract_inverted_index.help | 99 |
| abstract_inverted_index.high | 209 |
| abstract_inverted_index.into | 110 |
| abstract_inverted_index.mask | 175 |
| abstract_inverted_index.only | 31 |
| abstract_inverted_index.pH3) | 295 |
| abstract_inverted_index.pH3+ | 202, 273 |
| abstract_inverted_index.tTIL | 257 |
| abstract_inverted_index.than | 324 |
| abstract_inverted_index.that | 310 |
| abstract_inverted_index.this | 70 |
| abstract_inverted_index.were | 126, 165, 180, 215, 225, 254 |
| abstract_inverted_index.when | 251 |
| abstract_inverted_index.with | 128, 167, 182, 207, 345 |
| abstract_inverted_index.(Ki67 | 293 |
| abstract_inverted_index.(NAC) | 3 |
| abstract_inverted_index.(WSI) | 142 |
| abstract_inverted_index.(pH3) | 43 |
| abstract_inverted_index.TNBC. | 346 |
| abstract_inverted_index.based | 318 |
| abstract_inverted_index.cells | 212 |
| abstract_inverted_index.could | 98 |
| abstract_inverted_index.guide | 100 |
| abstract_inverted_index.image | 141, 205 |
| abstract_inverted_index.known | 47 |
| abstract_inverted_index.study | 328 |
| abstract_inverted_index.their | 234 |
| abstract_inverted_index.these | 61 |
| abstract_inverted_index.tumor | 191, 269 |
| abstract_inverted_index.under | 239 |
| abstract_inverted_index.using | 88 |
| abstract_inverted_index.value | 59, 74 |
| abstract_inverted_index.(pCR). | 25 |
| abstract_inverted_index.(sTILs | 197 |
| abstract_inverted_index.Ki67+, | 200, 271 |
| abstract_inverted_index.Serial | 117 |
| abstract_inverted_index.biopsy | 83 |
| abstract_inverted_index.breast | 12 |
| abstract_inverted_index.cancer | 13 |
| abstract_inverted_index.cells, | 192, 270 |
| abstract_inverted_index.cells. | 203 |
| abstract_inverted_index.counts | 258 |
| abstract_inverted_index.curve, | 240 |
| abstract_inverted_index.formed | 155 |
| abstract_inverted_index.images | 188 |
| abstract_inverted_index.level. | 304 |
| abstract_inverted_index.matrix | 243 |
| abstract_inverted_index.models | 179, 231, 312, 338 |
| abstract_inverted_index.needle | 121 |
| abstract_inverted_index.ranked | 299 |
| abstract_inverted_index.rather | 323 |
| abstract_inverted_index.sTILs) | 289 |
| abstract_inverted_index.sTILs, | 268 |
| abstract_inverted_index.serial | 145 |
| abstract_inverted_index.should | 316 |
| abstract_inverted_index.stain, | 150 |
| abstract_inverted_index.stains | 147 |
| abstract_inverted_index.study, | 71 |
| abstract_inverted_index.tTILs, | 267 |
| abstract_inverted_index.tissue | 84 |
| abstract_inverted_index.triple | 10 |
| abstract_inverted_index.(MRCNN) | 178 |
| abstract_inverted_index.(TILs), | 38 |
| abstract_inverted_index.(TNBC). | 14 |
| abstract_inverted_index.(tTILs, | 288 |
| abstract_inverted_index.H&E | 79, 129, 149, 168 |
| abstract_inverted_index.Highest | 246 |
| abstract_inverted_index.density | 210 |
| abstract_inverted_index.derived | 77 |
| abstract_inverted_index.hotspot | 252, 261, 278 |
| abstract_inverted_index.machine | 91 |
| abstract_inverted_index.markers | 76, 154 |
| abstract_inverted_index.metric, | 280 |
| abstract_inverted_index.partial | 113 |
| abstract_inverted_index.patches | 206 |
| abstract_inverted_index.patient | 303 |
| abstract_inverted_index.precise | 105 |
| abstract_inverted_index.predict | 50, 340 |
| abstract_inverted_index.primary | 16 |
| abstract_inverted_index.regions | 253 |
| abstract_inverted_index.results | 27, 308 |
| abstract_inverted_index.section | 146 |
| abstract_inverted_index.serving | 170 |
| abstract_inverted_index.stained | 82, 127 |
| abstract_inverted_index.stromal | 193 |
| abstract_inverted_index.support | 333 |
| abstract_inverted_index.tTILs), | 199 |
| abstract_inverted_index.trained | 181 |
| abstract_inverted_index.30–40% | 32 |
| abstract_inverted_index.H&E, | 184 |
| abstract_inverted_index.ML-based | 337 |
| abstract_inverted_index.Methods: | 116 |
| abstract_inverted_index.Overall, | 306 |
| abstract_inverted_index.Results: | 245 |
| abstract_inverted_index.Separate | 174 |
| abstract_inverted_index.accuracy | 248 |
| abstract_inverted_index.achieved | 250 |
| abstract_inverted_index.biopsies | 122 |
| abstract_inverted_index.combined | 58 |
| abstract_inverted_index.complete | 23 |
| abstract_inverted_index.enabling | 104 |
| abstract_inverted_index.endpoint | 17 |
| abstract_inverted_index.evidence | 331 |
| abstract_inverted_index.features | 287 |
| abstract_inverted_index.followed | 138 |
| abstract_inverted_index.interest | 214 |
| abstract_inverted_index.lacking. | 68 |
| abstract_inverted_index.learning | 92 |
| abstract_inverted_index.markers, | 137 |
| abstract_inverted_index.measures | 265 |
| abstract_inverted_index.multiple | 229, 285 |
| abstract_inverted_index.negative | 11 |
| abstract_inverted_index.patient. | 160 |
| abstract_inverted_index.patients | 109, 344 |
| abstract_inverted_index.provides | 329 |
| abstract_inverted_index.response | 24, 66, 223, 315, 342 |
| abstract_inverted_index.resulted | 296 |
| abstract_inverted_index.sections | 118 |
| abstract_inverted_index.standard | 6 |
| abstract_inverted_index.training | 228 |
| abstract_inverted_index.triplets | 157, 164 |
| abstract_inverted_index.accuracy, | 237 |
| abstract_inverted_index.analyses. | 244 |
| abstract_inverted_index.annotated | 183 |
| abstract_inverted_index.approach. | 94 |
| abstract_inverted_index.confusion | 242 |
| abstract_inverted_index.decisions | 102 |
| abstract_inverted_index.detecting | 190 |
| abstract_inverted_index.emphasize | 309 |
| abstract_inverted_index.evaluated | 87 |
| abstract_inverted_index.features. | 274 |
| abstract_inverted_index.hotspots. | 218 |
| abstract_inverted_index.molecular | 291 |
| abstract_inverted_index.patients. | 35 |
| abstract_inverted_index.response. | 52 |
| abstract_inverted_index.resulting | 162 |
| abstract_inverted_index.selection | 279 |
| abstract_inverted_index.treatment | 7 |
| abstract_inverted_index.(ML)-based | 93 |
| abstract_inverted_index.Currently, | 53 |
| abstract_inverted_index.Regardless | 275 |
| abstract_inverted_index.biomarkers | 48, 62, 97, 292, 320 |
| abstract_inverted_index.compelling | 330 |
| abstract_inverted_index.evaluating | 233 |
| abstract_inverted_index.evaluation | 55 |
| abstract_inverted_index.identified | 216, 226, 255 |
| abstract_inverted_index.isolation. | 326 |
| abstract_inverted_index.predicting | 64 |
| abstract_inverted_index.prediction | 224, 247, 311 |
| abstract_inverted_index.predictive | 73, 96 |
| abstract_inverted_index.reference. | 173 |
| abstract_inverted_index.responders | 111 |
| abstract_inverted_index.supervised | 90 |
| abstract_inverted_index.systematic | 54 |
| abstract_inverted_index.Background: | 0 |
| abstract_inverted_index.Identifying | 95 |
| abstract_inverted_index.Neoadjuvant | 1 |
| abstract_inverted_index.classifiers | 220 |
| abstract_inverted_index.combination | 322 |
| abstract_inverted_index.early-stage | 9 |
| abstract_inverted_index.generation. | 143 |
| abstract_inverted_index.lymphocytes | 37 |
| abstract_inverted_index.performance | 235, 300 |
| abstract_inverted_index.represented | 263 |
| abstract_inverted_index.therapeutic | 101 |
| abstract_inverted_index.whole-slide | 140 |
| abstract_inverted_index.Conclusions: | 305 |
| abstract_inverted_index.chemotherapy | 2 |
| abstract_inverted_index.histological | 286 |
| abstract_inverted_index.intratumoral | 195 |
| abstract_inverted_index.pathological | 22 |
| abstract_inverted_index.region-based | 176 |
| abstract_inverted_index.co-registered | 166 |
| abstract_inverted_index.complementary | 282 |
| abstract_inverted_index.phosphohistone | 41 |
| abstract_inverted_index.stratification | 106 |
| abstract_inverted_index.comprehensively | 86 |
| abstract_inverted_index.non-responders. | 115 |
| abstract_inverted_index.Tumor-infiltrating | 36 |
| abstract_inverted_index.immunohistochemically | 131 |
| cited_by_percentile_year.max | 99 |
| cited_by_percentile_year.min | 94 |
| corresponding_author_ids | https://openalex.org/A5066949209, https://openalex.org/A5019846100 |
| countries_distinct_count | 2 |
| institutions_distinct_count | 9 |
| corresponding_institution_ids | https://openalex.org/I181565077, https://openalex.org/I32389192 |
| sustainable_development_goals[0].id | https://metadata.un.org/sdg/3 |
| sustainable_development_goals[0].score | 0.7900000214576721 |
| sustainable_development_goals[0].display_name | Good health and well-being |
| citation_normalized_percentile.value | 0.89554101 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | True |