Predicting protein stability changes upon mutations with dual-view ensemble learning from single sequence Article Swipe
 YOU?
·
· 2025
· Open Access
·
· DOI: https://doi.org/10.1093/bib/bbaf319
YOU?
·
· 2025
· Open Access
·
· DOI: https://doi.org/10.1093/bib/bbaf319
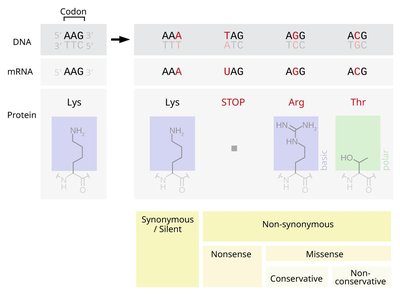
Predicting the protein stability changes upon mutations is one of the effective ways to improve the efficiency of protein engineering. Here, we propose a dual-view ensemble learning-based framework, DVE-stability, for mutation-induced protein stability change prediction from single sequence. DVE-stability integrates the global and local dependencies of mutations to capture the intramolecular interactions from two views through ensemble learning, in which a structural microenvironment simulation module is designed to indirectly introduce the information of structural microenvironment at the sequence level. DVE-stability achieved state-of-the-art prediction performance on seven single-point mutation benchmark datasets, and comprehensively surpassed other methods on five of them. Furthermore, DVE-stability outperformed other methods comprehensively through zero-shot inference on multiple-point mutation prediction task, demonstrating superior model generalizability to capture the epistasis of multiple-point mutations. More importantly, DVE-stability exhibited superior generalization performance in predicting rare beneficial mutations that are crucial for practical protein directed evolution scenarios. In addition, DVE-stability identified important intramolecular interactions via attention scores, demonstrating interpretable. Overall, DVE-stability provides a flexible and efficient tool for mutation-induced protein stability change prediction in an interpretable ensemble learning manner.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.1093/bib/bbaf319
- https://academic.oup.com/bib/article-pdf/26/4/bbaf319/63725431/bbaf319.pdf
- OA Status
- gold
- References
- 65
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4412184079
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4412184079Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1093/bib/bbaf319Digital Object Identifier
- Title
-
Predicting protein stability changes upon mutations with dual-view ensemble learning from single sequenceWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2025Year of publication
- Publication date
-
2025-06-17Full publication date if available
- Authors
-
Zhiwei Nie, Yiming Ma, Yutian Liu, Xiansong Huang, Zhihong Liu, Yang Peng, Fan Xu, Feng Yin, Zigang Li, J. Fu, Zhixiang Ren, Wenbin Zhang, Jie ChenList of authors in order
- Landing page
-
https://doi.org/10.1093/bib/bbaf319Publisher landing page
- PDF URL
-
https://academic.oup.com/bib/article-pdf/26/4/bbaf319/63725431/bbaf319.pdfDirect link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
goldOpen access status per OpenAlex
- OA URL
-
https://academic.oup.com/bib/article-pdf/26/4/bbaf319/63725431/bbaf319.pdfDirect OA link when available
- Concepts
-
Mutation, Stability (learning theory), Benchmark (surveying), Computer science, Epistasis, Ensemble learning, Ensemble forecasting, Artificial intelligence, Sequence (biology), Protein sequencing, Point mutation, Machine learning, Biology, Genetics, Peptide sequence, Geography, Gene, GeodesyTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
0Total citation count in OpenAlex
- References (count)
-
65Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4412184079 |
|---|---|
| doi | https://doi.org/10.1093/bib/bbaf319 |
| ids.doi | https://doi.org/10.1093/bib/bbaf319 |
| ids.pmid | https://pubmed.ncbi.nlm.nih.gov/40641045 |
| ids.openalex | https://openalex.org/W4412184079 |
| fwci | 0.0 |
| mesh[0].qualifier_ui | |
| mesh[0].descriptor_ui | D055550 |
| mesh[0].is_major_topic | False |
| mesh[0].qualifier_name | |
| mesh[0].descriptor_name | Protein Stability |
| mesh[1].qualifier_ui | Q000235 |
| mesh[1].descriptor_ui | D011506 |
| mesh[1].is_major_topic | True |
| mesh[1].qualifier_name | genetics |
| mesh[1].descriptor_name | Proteins |
| mesh[2].qualifier_ui | Q000737 |
| mesh[2].descriptor_ui | D011506 |
| mesh[2].is_major_topic | True |
| mesh[2].qualifier_name | chemistry |
| mesh[2].descriptor_name | Proteins |
| mesh[3].qualifier_ui | |
| mesh[3].descriptor_ui | D009154 |
| mesh[3].is_major_topic | True |
| mesh[3].qualifier_name | |
| mesh[3].descriptor_name | Mutation |
| mesh[4].qualifier_ui | |
| mesh[4].descriptor_ui | D000069550 |
| mesh[4].is_major_topic | True |
| mesh[4].qualifier_name | |
| mesh[4].descriptor_name | Machine Learning |
| mesh[5].qualifier_ui | Q000379 |
| mesh[5].descriptor_ui | D019295 |
| mesh[5].is_major_topic | True |
| mesh[5].qualifier_name | methods |
| mesh[5].descriptor_name | Computational Biology |
| mesh[6].qualifier_ui | |
| mesh[6].descriptor_ui | D000098406 |
| mesh[6].is_major_topic | False |
| mesh[6].qualifier_name | |
| mesh[6].descriptor_name | Ensemble Learning |
| mesh[7].qualifier_ui | |
| mesh[7].descriptor_ui | D055550 |
| mesh[7].is_major_topic | False |
| mesh[7].qualifier_name | |
| mesh[7].descriptor_name | Protein Stability |
| mesh[8].qualifier_ui | Q000235 |
| mesh[8].descriptor_ui | D011506 |
| mesh[8].is_major_topic | True |
| mesh[8].qualifier_name | genetics |
| mesh[8].descriptor_name | Proteins |
| mesh[9].qualifier_ui | Q000737 |
| mesh[9].descriptor_ui | D011506 |
| mesh[9].is_major_topic | True |
| mesh[9].qualifier_name | chemistry |
| mesh[9].descriptor_name | Proteins |
| mesh[10].qualifier_ui | |
| mesh[10].descriptor_ui | D009154 |
| mesh[10].is_major_topic | True |
| mesh[10].qualifier_name | |
| mesh[10].descriptor_name | Mutation |
| mesh[11].qualifier_ui | |
| mesh[11].descriptor_ui | D000069550 |
| mesh[11].is_major_topic | True |
| mesh[11].qualifier_name | |
| mesh[11].descriptor_name | Machine Learning |
| mesh[12].qualifier_ui | Q000379 |
| mesh[12].descriptor_ui | D019295 |
| mesh[12].is_major_topic | True |
| mesh[12].qualifier_name | methods |
| mesh[12].descriptor_name | Computational Biology |
| mesh[13].qualifier_ui | |
| mesh[13].descriptor_ui | D000098406 |
| mesh[13].is_major_topic | False |
| mesh[13].qualifier_name | |
| mesh[13].descriptor_name | Ensemble Learning |
| type | article |
| title | Predicting protein stability changes upon mutations with dual-view ensemble learning from single sequence |
| awards[0].id | https://openalex.org/G3229855046 |
| awards[0].funder_id | https://openalex.org/F4320321001 |
| awards[0].display_name | |
| awards[0].funder_award_id | 61972217 |
| awards[0].funder_display_name | National Natural Science Foundation of China |
| awards[1].id | https://openalex.org/G8282592590 |
| awards[1].funder_id | https://openalex.org/F4320335795 |
| awards[1].display_name | |
| awards[1].funder_award_id | 2024B0101010003 |
| awards[1].funder_display_name | Science and Technology Planning Project of Guangdong Province |
| awards[2].id | https://openalex.org/G3262489766 |
| awards[2].funder_id | https://openalex.org/F4320321001 |
| awards[2].display_name | |
| awards[2].funder_award_id | 62176249 |
| awards[2].funder_display_name | National Natural Science Foundation of China |
| awards[3].id | https://openalex.org/G5598344535 |
| awards[3].funder_id | https://openalex.org/F4320321001 |
| awards[3].display_name | |
| awards[3].funder_award_id | 62006133 |
| awards[3].funder_display_name | National Natural Science Foundation of China |
| awards[4].id | https://openalex.org/G7571408944 |
| awards[4].funder_id | https://openalex.org/F4320321001 |
| awards[4].display_name | |
| awards[4].funder_award_id | 62271465 |
| awards[4].funder_display_name | National Natural Science Foundation of China |
| awards[5].id | https://openalex.org/G4768617711 |
| awards[5].funder_id | https://openalex.org/F4320321001 |
| awards[5].display_name | |
| awards[5].funder_award_id | 32071459 |
| awards[5].funder_display_name | National Natural Science Foundation of China |
| awards[6].id | https://openalex.org/G136855964 |
| awards[6].funder_id | https://openalex.org/F4320321001 |
| awards[6].display_name | |
| awards[6].funder_award_id | 22331003 |
| awards[6].funder_display_name | National Natural Science Foundation of China |
| biblio.issue | 4 |
| biblio.volume | 26 |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T10044 |
| topics[0].field.id | https://openalex.org/fields/13 |
| topics[0].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[0].score | 0.9958000183105469 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1312 |
| topics[0].subfield.display_name | Molecular Biology |
| topics[0].display_name | Protein Structure and Dynamics |
| topics[1].id | https://openalex.org/T10015 |
| topics[1].field.id | https://openalex.org/fields/13 |
| topics[1].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[1].score | 0.9923999905586243 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1312 |
| topics[1].subfield.display_name | Molecular Biology |
| topics[1].display_name | Genomics and Phylogenetic Studies |
| topics[2].id | https://openalex.org/T10521 |
| topics[2].field.id | https://openalex.org/fields/13 |
| topics[2].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[2].score | 0.9865999817848206 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1312 |
| topics[2].subfield.display_name | Molecular Biology |
| topics[2].display_name | RNA and protein synthesis mechanisms |
| funders[0].id | https://openalex.org/F4320321001 |
| funders[0].ror | https://ror.org/01h0zpd94 |
| funders[0].display_name | National Natural Science Foundation of China |
| funders[1].id | https://openalex.org/F4320335795 |
| funders[1].ror | |
| funders[1].display_name | Science and Technology Planning Project of Guangdong Province |
| is_xpac | False |
| apc_list.value | 4011 |
| apc_list.currency | USD |
| apc_list.value_usd | 4011 |
| apc_paid.value | 4011 |
| apc_paid.currency | USD |
| apc_paid.value_usd | 4011 |
| concepts[0].id | https://openalex.org/C501734568 |
| concepts[0].level | 3 |
| concepts[0].score | 0.6931123733520508 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q42918 |
| concepts[0].display_name | Mutation |
| concepts[1].id | https://openalex.org/C112972136 |
| concepts[1].level | 2 |
| concepts[1].score | 0.6805262565612793 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q7595718 |
| concepts[1].display_name | Stability (learning theory) |
| concepts[2].id | https://openalex.org/C185798385 |
| concepts[2].level | 2 |
| concepts[2].score | 0.6721440553665161 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q1161707 |
| concepts[2].display_name | Benchmark (surveying) |
| concepts[3].id | https://openalex.org/C41008148 |
| concepts[3].level | 0 |
| concepts[3].score | 0.6634979844093323 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q21198 |
| concepts[3].display_name | Computer science |
| concepts[4].id | https://openalex.org/C61727976 |
| concepts[4].level | 3 |
| concepts[4].score | 0.5394566655158997 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q751748 |
| concepts[4].display_name | Epistasis |
| concepts[5].id | https://openalex.org/C45942800 |
| concepts[5].level | 2 |
| concepts[5].score | 0.5330385565757751 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q245652 |
| concepts[5].display_name | Ensemble learning |
| concepts[6].id | https://openalex.org/C119898033 |
| concepts[6].level | 2 |
| concepts[6].score | 0.5058536529541016 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q3433888 |
| concepts[6].display_name | Ensemble forecasting |
| concepts[7].id | https://openalex.org/C154945302 |
| concepts[7].level | 1 |
| concepts[7].score | 0.495776504278183 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q11660 |
| concepts[7].display_name | Artificial intelligence |
| concepts[8].id | https://openalex.org/C2778112365 |
| concepts[8].level | 2 |
| concepts[8].score | 0.4901467263698578 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q3511065 |
| concepts[8].display_name | Sequence (biology) |
| concepts[9].id | https://openalex.org/C10010492 |
| concepts[9].level | 4 |
| concepts[9].score | 0.46650972962379456 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q3142557 |
| concepts[9].display_name | Protein sequencing |
| concepts[10].id | https://openalex.org/C176944494 |
| concepts[10].level | 4 |
| concepts[10].score | 0.4487055838108063 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q1415380 |
| concepts[10].display_name | Point mutation |
| concepts[11].id | https://openalex.org/C119857082 |
| concepts[11].level | 1 |
| concepts[11].score | 0.367371141910553 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q2539 |
| concepts[11].display_name | Machine learning |
| concepts[12].id | https://openalex.org/C86803240 |
| concepts[12].level | 0 |
| concepts[12].score | 0.19391080737113953 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[12].display_name | Biology |
| concepts[13].id | https://openalex.org/C54355233 |
| concepts[13].level | 1 |
| concepts[13].score | 0.1634749174118042 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[13].display_name | Genetics |
| concepts[14].id | https://openalex.org/C167625842 |
| concepts[14].level | 3 |
| concepts[14].score | 0.14224332571029663 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q899763 |
| concepts[14].display_name | Peptide sequence |
| concepts[15].id | https://openalex.org/C205649164 |
| concepts[15].level | 0 |
| concepts[15].score | 0.0 |
| concepts[15].wikidata | https://www.wikidata.org/wiki/Q1071 |
| concepts[15].display_name | Geography |
| concepts[16].id | https://openalex.org/C104317684 |
| concepts[16].level | 2 |
| concepts[16].score | 0.0 |
| concepts[16].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[16].display_name | Gene |
| concepts[17].id | https://openalex.org/C13280743 |
| concepts[17].level | 1 |
| concepts[17].score | 0.0 |
| concepts[17].wikidata | https://www.wikidata.org/wiki/Q131089 |
| concepts[17].display_name | Geodesy |
| keywords[0].id | https://openalex.org/keywords/mutation |
| keywords[0].score | 0.6931123733520508 |
| keywords[0].display_name | Mutation |
| keywords[1].id | https://openalex.org/keywords/stability |
| keywords[1].score | 0.6805262565612793 |
| keywords[1].display_name | Stability (learning theory) |
| keywords[2].id | https://openalex.org/keywords/benchmark |
| keywords[2].score | 0.6721440553665161 |
| keywords[2].display_name | Benchmark (surveying) |
| keywords[3].id | https://openalex.org/keywords/computer-science |
| keywords[3].score | 0.6634979844093323 |
| keywords[3].display_name | Computer science |
| keywords[4].id | https://openalex.org/keywords/epistasis |
| keywords[4].score | 0.5394566655158997 |
| keywords[4].display_name | Epistasis |
| keywords[5].id | https://openalex.org/keywords/ensemble-learning |
| keywords[5].score | 0.5330385565757751 |
| keywords[5].display_name | Ensemble learning |
| keywords[6].id | https://openalex.org/keywords/ensemble-forecasting |
| keywords[6].score | 0.5058536529541016 |
| keywords[6].display_name | Ensemble forecasting |
| keywords[7].id | https://openalex.org/keywords/artificial-intelligence |
| keywords[7].score | 0.495776504278183 |
| keywords[7].display_name | Artificial intelligence |
| keywords[8].id | https://openalex.org/keywords/sequence |
| keywords[8].score | 0.4901467263698578 |
| keywords[8].display_name | Sequence (biology) |
| keywords[9].id | https://openalex.org/keywords/protein-sequencing |
| keywords[9].score | 0.46650972962379456 |
| keywords[9].display_name | Protein sequencing |
| keywords[10].id | https://openalex.org/keywords/point-mutation |
| keywords[10].score | 0.4487055838108063 |
| keywords[10].display_name | Point mutation |
| keywords[11].id | https://openalex.org/keywords/machine-learning |
| keywords[11].score | 0.367371141910553 |
| keywords[11].display_name | Machine learning |
| keywords[12].id | https://openalex.org/keywords/biology |
| keywords[12].score | 0.19391080737113953 |
| keywords[12].display_name | Biology |
| keywords[13].id | https://openalex.org/keywords/genetics |
| keywords[13].score | 0.1634749174118042 |
| keywords[13].display_name | Genetics |
| keywords[14].id | https://openalex.org/keywords/peptide-sequence |
| keywords[14].score | 0.14224332571029663 |
| keywords[14].display_name | Peptide sequence |
| language | en |
| locations[0].id | doi:10.1093/bib/bbaf319 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S91767247 |
| locations[0].source.issn | 1467-5463, 1477-4054 |
| locations[0].source.type | journal |
| locations[0].source.is_oa | True |
| locations[0].source.issn_l | 1467-5463 |
| locations[0].source.is_core | True |
| locations[0].source.is_in_doaj | True |
| locations[0].source.display_name | Briefings in Bioinformatics |
| locations[0].source.host_organization | https://openalex.org/P4310311648 |
| locations[0].source.host_organization_name | Oxford University Press |
| locations[0].source.host_organization_lineage | https://openalex.org/P4310311648, https://openalex.org/P4310311647 |
| locations[0].source.host_organization_lineage_names | Oxford University Press, University of Oxford |
| locations[0].license | cc-by-nc |
| locations[0].pdf_url | https://academic.oup.com/bib/article-pdf/26/4/bbaf319/63725431/bbaf319.pdf |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | https://openalex.org/licenses/cc-by-nc |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | Briefings in Bioinformatics |
| locations[0].landing_page_url | https://doi.org/10.1093/bib/bbaf319 |
| locations[1].id | pmid:40641045 |
| locations[1].is_oa | False |
| locations[1].source.id | https://openalex.org/S4306525036 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | PubMed |
| locations[1].source.host_organization | https://openalex.org/I1299303238 |
| locations[1].source.host_organization_name | National Institutes of Health |
| locations[1].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[1].license | |
| locations[1].pdf_url | |
| locations[1].version | publishedVersion |
| locations[1].raw_type | |
| locations[1].license_id | |
| locations[1].is_accepted | True |
| locations[1].is_published | True |
| locations[1].raw_source_name | Briefings in bioinformatics |
| locations[1].landing_page_url | https://pubmed.ncbi.nlm.nih.gov/40641045 |
| indexed_in | crossref, doaj, pubmed |
| authorships[0].author.id | https://openalex.org/A5066870142 |
| authorships[0].author.orcid | https://orcid.org/0000-0001-5198-4212 |
| authorships[0].author.display_name | Zhiwei Nie |
| authorships[0].countries | CN |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I20231570 |
| authorships[0].affiliations[0].raw_affiliation_string | School of Electronic and Computer Engineering , Peking University, Shenzhen, |
| authorships[0].affiliations[1].institution_ids | https://openalex.org/I4210136793 |
| authorships[0].affiliations[1].raw_affiliation_string | Pengcheng Laboratory , Shenzhen, |
| authorships[0].affiliations[2].institution_ids | https://openalex.org/I20231570 |
| authorships[0].affiliations[2].raw_affiliation_string | AI for Science (AI4S)-Preferred Program , Peking University Shenzhen Graduate School, Shenzhen, |
| authorships[0].institutions[0].id | https://openalex.org/I20231570 |
| authorships[0].institutions[0].ror | https://ror.org/02v51f717 |
| authorships[0].institutions[0].type | education |
| authorships[0].institutions[0].lineage | https://openalex.org/I20231570 |
| authorships[0].institutions[0].country_code | CN |
| authorships[0].institutions[0].display_name | Peking University |
| authorships[0].institutions[1].id | https://openalex.org/I4210136793 |
| authorships[0].institutions[1].ror | https://ror.org/03qdqbt06 |
| authorships[0].institutions[1].type | facility |
| authorships[0].institutions[1].lineage | https://openalex.org/I4210136793 |
| authorships[0].institutions[1].country_code | CN |
| authorships[0].institutions[1].display_name | Peng Cheng Laboratory |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Zhiwei Nie |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | AI for Science (AI4S)-Preferred Program , Peking University Shenzhen Graduate School, Shenzhen,, Pengcheng Laboratory , Shenzhen,, School of Electronic and Computer Engineering , Peking University, Shenzhen, |
| authorships[1].author.id | https://openalex.org/A5101569453 |
| authorships[1].author.orcid | https://orcid.org/0009-0007-2780-426X |
| authorships[1].author.display_name | Yiming Ma |
| authorships[1].countries | CN |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I20231570 |
| authorships[1].affiliations[0].raw_affiliation_string | School of Electronic and Computer Engineering , Peking University, Shenzhen, |
| authorships[1].affiliations[1].institution_ids | https://openalex.org/I20231570 |
| authorships[1].affiliations[1].raw_affiliation_string | AI for Science (AI4S)-Preferred Program , Peking University Shenzhen Graduate School, Shenzhen, |
| authorships[1].institutions[0].id | https://openalex.org/I20231570 |
| authorships[1].institutions[0].ror | https://ror.org/02v51f717 |
| authorships[1].institutions[0].type | education |
| authorships[1].institutions[0].lineage | https://openalex.org/I20231570 |
| authorships[1].institutions[0].country_code | CN |
| authorships[1].institutions[0].display_name | Peking University |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Yiming Ma |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | AI for Science (AI4S)-Preferred Program , Peking University Shenzhen Graduate School, Shenzhen,, School of Electronic and Computer Engineering , Peking University, Shenzhen, |
| authorships[2].author.id | https://openalex.org/A5058363207 |
| authorships[2].author.orcid | https://orcid.org/0000-0001-6932-5207 |
| authorships[2].author.display_name | Yutian Liu |
| authorships[2].countries | CN |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I20231570 |
| authorships[2].affiliations[0].raw_affiliation_string | School of Computer Science , Peking University, Beijing, |
| authorships[2].institutions[0].id | https://openalex.org/I20231570 |
| authorships[2].institutions[0].ror | https://ror.org/02v51f717 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I20231570 |
| authorships[2].institutions[0].country_code | CN |
| authorships[2].institutions[0].display_name | Peking University |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Yutian Liu |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | School of Computer Science , Peking University, Beijing, |
| authorships[3].author.id | https://openalex.org/A5049111913 |
| authorships[3].author.orcid | |
| authorships[3].author.display_name | Xiansong Huang |
| authorships[3].countries | CN |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I4210136793 |
| authorships[3].affiliations[0].raw_affiliation_string | Pengcheng Laboratory , Shenzhen, |
| authorships[3].institutions[0].id | https://openalex.org/I4210136793 |
| authorships[3].institutions[0].ror | https://ror.org/03qdqbt06 |
| authorships[3].institutions[0].type | facility |
| authorships[3].institutions[0].lineage | https://openalex.org/I4210136793 |
| authorships[3].institutions[0].country_code | CN |
| authorships[3].institutions[0].display_name | Peng Cheng Laboratory |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Xiansong Huang |
| authorships[3].is_corresponding | False |
| authorships[3].raw_affiliation_strings | Pengcheng Laboratory , Shenzhen, |
| authorships[4].author.id | https://openalex.org/A5102724893 |
| authorships[4].author.orcid | https://orcid.org/0000-0003-3314-4943 |
| authorships[4].author.display_name | Zhihong Liu |
| authorships[4].countries | CN |
| authorships[4].affiliations[0].institution_ids | https://openalex.org/I4210102541 |
| authorships[4].affiliations[0].raw_affiliation_string | Pingshan Translational Medicine Center , Shenzhen Bay Laboratory, Shenzhen, |
| authorships[4].institutions[0].id | https://openalex.org/I4210102541 |
| authorships[4].institutions[0].ror | https://ror.org/00sdcjz77 |
| authorships[4].institutions[0].type | facility |
| authorships[4].institutions[0].lineage | https://openalex.org/I4210102541 |
| authorships[4].institutions[0].country_code | CN |
| authorships[4].institutions[0].display_name | Shenzhen Bay Laboratory |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Zhihong Liu |
| authorships[4].is_corresponding | False |
| authorships[4].raw_affiliation_strings | Pingshan Translational Medicine Center , Shenzhen Bay Laboratory, Shenzhen, |
| authorships[5].author.id | https://openalex.org/A5101698101 |
| authorships[5].author.orcid | https://orcid.org/0009-0003-2824-5843 |
| authorships[5].author.display_name | Yang Peng |
| authorships[5].countries | CN, IR |
| authorships[5].affiliations[0].institution_ids | https://openalex.org/I20231570, https://openalex.org/I4210116048 |
| authorships[5].affiliations[0].raw_affiliation_string | Beijing National Laboratory for Molecular Sciences , Key Laboratory of Polymer Chemistry & Physics of Ministry of Education, Center for Soft Matter Science and Engineering, College of Chemistry and Molecular Engineering, Peking University, Beijing, |
| authorships[5].affiliations[1].institution_ids | https://openalex.org/I4210108141 |
| authorships[5].affiliations[1].raw_affiliation_string | Key Laboratory of Polymer Chemistry & Physics of Ministry of Education , Center for Soft Matter Science and Engineering, College of Chemistry and Molecular Engineering, Peking University, |
| authorships[5].institutions[0].id | https://openalex.org/I4210116048 |
| authorships[5].institutions[0].ror | https://ror.org/02601yx74 |
| authorships[5].institutions[0].type | facility |
| authorships[5].institutions[0].lineage | https://openalex.org/I4210116048 |
| authorships[5].institutions[0].country_code | CN |
| authorships[5].institutions[0].display_name | Beijing National Laboratory for Molecular Sciences |
| authorships[5].institutions[1].id | https://openalex.org/I20231570 |
| authorships[5].institutions[1].ror | https://ror.org/02v51f717 |
| authorships[5].institutions[1].type | education |
| authorships[5].institutions[1].lineage | https://openalex.org/I20231570 |
| authorships[5].institutions[1].country_code | CN |
| authorships[5].institutions[1].display_name | Peking University |
| authorships[5].institutions[2].id | https://openalex.org/I4210108141 |
| authorships[5].institutions[2].ror | https://ror.org/01hw7wf62 |
| authorships[5].institutions[2].type | government |
| authorships[5].institutions[2].lineage | https://openalex.org/I4210108141 |
| authorships[5].institutions[2].country_code | IR |
| authorships[5].institutions[2].display_name | Ministry of Education |
| authorships[5].author_position | middle |
| authorships[5].raw_author_name | Peng Yang |
| authorships[5].is_corresponding | False |
| authorships[5].raw_affiliation_strings | Beijing National Laboratory for Molecular Sciences , Key Laboratory of Polymer Chemistry & Physics of Ministry of Education, Center for Soft Matter Science and Engineering, College of Chemistry and Molecular Engineering, Peking University, Beijing,, Key Laboratory of Polymer Chemistry & Physics of Ministry of Education , Center for Soft Matter Science and Engineering, College of Chemistry and Molecular Engineering, Peking University, |
| authorships[6].author.id | https://openalex.org/A5100578497 |
| authorships[6].author.orcid | https://orcid.org/0009-0000-5479-3067 |
| authorships[6].author.display_name | Fan Xu |
| authorships[6].countries | CN |
| authorships[6].affiliations[0].institution_ids | https://openalex.org/I4210136793 |
| authorships[6].affiliations[0].raw_affiliation_string | Pengcheng Laboratory , Shenzhen, |
| authorships[6].institutions[0].id | https://openalex.org/I4210136793 |
| authorships[6].institutions[0].ror | https://ror.org/03qdqbt06 |
| authorships[6].institutions[0].type | facility |
| authorships[6].institutions[0].lineage | https://openalex.org/I4210136793 |
| authorships[6].institutions[0].country_code | CN |
| authorships[6].institutions[0].display_name | Peng Cheng Laboratory |
| authorships[6].author_position | middle |
| authorships[6].raw_author_name | Fan Xu |
| authorships[6].is_corresponding | False |
| authorships[6].raw_affiliation_strings | Pengcheng Laboratory , Shenzhen, |
| authorships[7].author.id | https://openalex.org/A5101648965 |
| authorships[7].author.orcid | https://orcid.org/0000-0001-7407-9047 |
| authorships[7].author.display_name | Feng Yin |
| authorships[7].countries | CN |
| authorships[7].affiliations[0].institution_ids | https://openalex.org/I4210102541 |
| authorships[7].affiliations[0].raw_affiliation_string | Pingshan Translational Medicine Center , Shenzhen Bay Laboratory, Shenzhen, |
| authorships[7].institutions[0].id | https://openalex.org/I4210102541 |
| authorships[7].institutions[0].ror | https://ror.org/00sdcjz77 |
| authorships[7].institutions[0].type | facility |
| authorships[7].institutions[0].lineage | https://openalex.org/I4210102541 |
| authorships[7].institutions[0].country_code | CN |
| authorships[7].institutions[0].display_name | Shenzhen Bay Laboratory |
| authorships[7].author_position | middle |
| authorships[7].raw_author_name | Feng Yin |
| authorships[7].is_corresponding | False |
| authorships[7].raw_affiliation_strings | Pingshan Translational Medicine Center , Shenzhen Bay Laboratory, Shenzhen, |
| authorships[8].author.id | https://openalex.org/A5100784551 |
| authorships[8].author.orcid | https://orcid.org/0000-0002-4649-1592 |
| authorships[8].author.display_name | Zigang Li |
| authorships[8].countries | CN |
| authorships[8].affiliations[0].institution_ids | https://openalex.org/I20231570 |
| authorships[8].affiliations[0].raw_affiliation_string | State Key Laboratory of Chemical Oncogenomics , School of Chemical Biology and Biotechnology, Peking University Shenzhen Graduate School, Shenzhen, |
| authorships[8].affiliations[1].institution_ids | https://openalex.org/I4210102541 |
| authorships[8].affiliations[1].raw_affiliation_string | Pingshan Translational Medicine Center , Shenzhen Bay Laboratory, Shenzhen, |
| authorships[8].institutions[0].id | https://openalex.org/I20231570 |
| authorships[8].institutions[0].ror | https://ror.org/02v51f717 |
| authorships[8].institutions[0].type | education |
| authorships[8].institutions[0].lineage | https://openalex.org/I20231570 |
| authorships[8].institutions[0].country_code | CN |
| authorships[8].institutions[0].display_name | Peking University |
| authorships[8].institutions[1].id | https://openalex.org/I4210102541 |
| authorships[8].institutions[1].ror | https://ror.org/00sdcjz77 |
| authorships[8].institutions[1].type | facility |
| authorships[8].institutions[1].lineage | https://openalex.org/I4210102541 |
| authorships[8].institutions[1].country_code | CN |
| authorships[8].institutions[1].display_name | Shenzhen Bay Laboratory |
| authorships[8].author_position | middle |
| authorships[8].raw_author_name | Zigang Li |
| authorships[8].is_corresponding | False |
| authorships[8].raw_affiliation_strings | Pingshan Translational Medicine Center , Shenzhen Bay Laboratory, Shenzhen,, State Key Laboratory of Chemical Oncogenomics , School of Chemical Biology and Biotechnology, Peking University Shenzhen Graduate School, Shenzhen, |
| authorships[9].author.id | https://openalex.org/A5114376171 |
| authorships[9].author.orcid | https://orcid.org/0000-0003-3177-2700 |
| authorships[9].author.display_name | J. Fu |
| authorships[9].affiliations[0].raw_affiliation_string | Shanghai AI Laboratory , Shanghai, |
| authorships[9].author_position | middle |
| authorships[9].raw_author_name | Jie Fu |
| authorships[9].is_corresponding | False |
| authorships[9].raw_affiliation_strings | Shanghai AI Laboratory , Shanghai, |
| authorships[10].author.id | https://openalex.org/A5070028902 |
| authorships[10].author.orcid | https://orcid.org/0000-0002-4104-3790 |
| authorships[10].author.display_name | Zhixiang Ren |
| authorships[10].countries | CN |
| authorships[10].affiliations[0].institution_ids | https://openalex.org/I4210136793 |
| authorships[10].affiliations[0].raw_affiliation_string | Pengcheng Laboratory , Shenzhen, |
| authorships[10].institutions[0].id | https://openalex.org/I4210136793 |
| authorships[10].institutions[0].ror | https://ror.org/03qdqbt06 |
| authorships[10].institutions[0].type | facility |
| authorships[10].institutions[0].lineage | https://openalex.org/I4210136793 |
| authorships[10].institutions[0].country_code | CN |
| authorships[10].institutions[0].display_name | Peng Cheng Laboratory |
| authorships[10].author_position | middle |
| authorships[10].raw_author_name | Zhixiang Ren |
| authorships[10].is_corresponding | False |
| authorships[10].raw_affiliation_strings | Pengcheng Laboratory , Shenzhen, |
| authorships[11].author.id | https://openalex.org/A5100710814 |
| authorships[11].author.orcid | https://orcid.org/0000-0003-3024-5415 |
| authorships[11].author.display_name | Wenbin Zhang |
| authorships[11].countries | CN, IR |
| authorships[11].affiliations[0].institution_ids | https://openalex.org/I20231570, https://openalex.org/I4210116048 |
| authorships[11].affiliations[0].raw_affiliation_string | Beijing National Laboratory for Molecular Sciences , Key Laboratory of Polymer Chemistry & Physics of Ministry of Education, Center for Soft Matter Science and Engineering, College of Chemistry and Molecular Engineering, Peking University, Beijing, |
| authorships[11].affiliations[1].institution_ids | https://openalex.org/I4210108141 |
| authorships[11].affiliations[1].raw_affiliation_string | Key Laboratory of Polymer Chemistry & Physics of Ministry of Education , Center for Soft Matter Science and Engineering, College of Chemistry and Molecular Engineering, Peking University, |
| authorships[11].affiliations[2].institution_ids | https://openalex.org/I20231570 |
| authorships[11].affiliations[2].raw_affiliation_string | AI for Science (AI4S)-Preferred Program , Peking University Shenzhen Graduate School, Shenzhen, |
| authorships[11].institutions[0].id | https://openalex.org/I4210116048 |
| authorships[11].institutions[0].ror | https://ror.org/02601yx74 |
| authorships[11].institutions[0].type | facility |
| authorships[11].institutions[0].lineage | https://openalex.org/I4210116048 |
| authorships[11].institutions[0].country_code | CN |
| authorships[11].institutions[0].display_name | Beijing National Laboratory for Molecular Sciences |
| authorships[11].institutions[1].id | https://openalex.org/I20231570 |
| authorships[11].institutions[1].ror | https://ror.org/02v51f717 |
| authorships[11].institutions[1].type | education |
| authorships[11].institutions[1].lineage | https://openalex.org/I20231570 |
| authorships[11].institutions[1].country_code | CN |
| authorships[11].institutions[1].display_name | Peking University |
| authorships[11].institutions[2].id | https://openalex.org/I4210108141 |
| authorships[11].institutions[2].ror | https://ror.org/01hw7wf62 |
| authorships[11].institutions[2].type | government |
| authorships[11].institutions[2].lineage | https://openalex.org/I4210108141 |
| authorships[11].institutions[2].country_code | IR |
| authorships[11].institutions[2].display_name | Ministry of Education |
| authorships[11].author_position | middle |
| authorships[11].raw_author_name | Wen-Bin Zhang |
| authorships[11].is_corresponding | False |
| authorships[11].raw_affiliation_strings | AI for Science (AI4S)-Preferred Program , Peking University Shenzhen Graduate School, Shenzhen,, Beijing National Laboratory for Molecular Sciences , Key Laboratory of Polymer Chemistry & Physics of Ministry of Education, Center for Soft Matter Science and Engineering, College of Chemistry and Molecular Engineering, Peking University, Beijing,, Key Laboratory of Polymer Chemistry & Physics of Ministry of Education , Center for Soft Matter Science and Engineering, College of Chemistry and Molecular Engineering, Peking University, |
| authorships[12].author.id | https://openalex.org/A5100332846 |
| authorships[12].author.orcid | https://orcid.org/0000-0001-7925-3729 |
| authorships[12].author.display_name | Jie Chen |
| authorships[12].countries | CN |
| authorships[12].affiliations[0].institution_ids | https://openalex.org/I20231570 |
| authorships[12].affiliations[0].raw_affiliation_string | School of Electronic and Computer Engineering , Peking University, Shenzhen, |
| authorships[12].affiliations[1].institution_ids | https://openalex.org/I4210136793 |
| authorships[12].affiliations[1].raw_affiliation_string | Pengcheng Laboratory , Shenzhen, |
| authorships[12].affiliations[2].institution_ids | https://openalex.org/I20231570 |
| authorships[12].affiliations[2].raw_affiliation_string | AI for Science (AI4S)-Preferred Program , Peking University Shenzhen Graduate School, Shenzhen, |
| authorships[12].institutions[0].id | https://openalex.org/I20231570 |
| authorships[12].institutions[0].ror | https://ror.org/02v51f717 |
| authorships[12].institutions[0].type | education |
| authorships[12].institutions[0].lineage | https://openalex.org/I20231570 |
| authorships[12].institutions[0].country_code | CN |
| authorships[12].institutions[0].display_name | Peking University |
| authorships[12].institutions[1].id | https://openalex.org/I4210136793 |
| authorships[12].institutions[1].ror | https://ror.org/03qdqbt06 |
| authorships[12].institutions[1].type | facility |
| authorships[12].institutions[1].lineage | https://openalex.org/I4210136793 |
| authorships[12].institutions[1].country_code | CN |
| authorships[12].institutions[1].display_name | Peng Cheng Laboratory |
| authorships[12].author_position | last |
| authorships[12].raw_author_name | Jie Chen |
| authorships[12].is_corresponding | False |
| authorships[12].raw_affiliation_strings | AI for Science (AI4S)-Preferred Program , Peking University Shenzhen Graduate School, Shenzhen,, Pengcheng Laboratory , Shenzhen,, School of Electronic and Computer Engineering , Peking University, Shenzhen, |
| has_content.pdf | True |
| has_content.grobid_xml | True |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://academic.oup.com/bib/article-pdf/26/4/bbaf319/63725431/bbaf319.pdf |
| open_access.oa_status | gold |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Predicting protein stability changes upon mutations with dual-view ensemble learning from single sequence |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-23T05:10:03.516525 |
| primary_topic.id | https://openalex.org/T10044 |
| primary_topic.field.id | https://openalex.org/fields/13 |
| primary_topic.field.display_name | Biochemistry, Genetics and Molecular Biology |
| primary_topic.score | 0.9958000183105469 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1312 |
| primary_topic.subfield.display_name | Molecular Biology |
| primary_topic.display_name | Protein Structure and Dynamics |
| related_works | https://openalex.org/W2891633941, https://openalex.org/W3202800081, https://openalex.org/W3101614107, https://openalex.org/W1909207154, https://openalex.org/W4390971112, https://openalex.org/W3036530763, https://openalex.org/W3124390867, https://openalex.org/W1514365828, https://openalex.org/W3204228978, https://openalex.org/W2155806188 |
| cited_by_count | 0 |
| locations_count | 2 |
| best_oa_location.id | doi:10.1093/bib/bbaf319 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S91767247 |
| best_oa_location.source.issn | 1467-5463, 1477-4054 |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | True |
| best_oa_location.source.issn_l | 1467-5463 |
| best_oa_location.source.is_core | True |
| best_oa_location.source.is_in_doaj | True |
| best_oa_location.source.display_name | Briefings in Bioinformatics |
| best_oa_location.source.host_organization | https://openalex.org/P4310311648 |
| best_oa_location.source.host_organization_name | Oxford University Press |
| best_oa_location.source.host_organization_lineage | https://openalex.org/P4310311648, https://openalex.org/P4310311647 |
| best_oa_location.source.host_organization_lineage_names | Oxford University Press, University of Oxford |
| best_oa_location.license | cc-by-nc |
| best_oa_location.pdf_url | https://academic.oup.com/bib/article-pdf/26/4/bbaf319/63725431/bbaf319.pdf |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | journal-article |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by-nc |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | Briefings in Bioinformatics |
| best_oa_location.landing_page_url | https://doi.org/10.1093/bib/bbaf319 |
| primary_location.id | doi:10.1093/bib/bbaf319 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S91767247 |
| primary_location.source.issn | 1467-5463, 1477-4054 |
| primary_location.source.type | journal |
| primary_location.source.is_oa | True |
| primary_location.source.issn_l | 1467-5463 |
| primary_location.source.is_core | True |
| primary_location.source.is_in_doaj | True |
| primary_location.source.display_name | Briefings in Bioinformatics |
| primary_location.source.host_organization | https://openalex.org/P4310311648 |
| primary_location.source.host_organization_name | Oxford University Press |
| primary_location.source.host_organization_lineage | https://openalex.org/P4310311648, https://openalex.org/P4310311647 |
| primary_location.source.host_organization_lineage_names | Oxford University Press, University of Oxford |
| primary_location.license | cc-by-nc |
| primary_location.pdf_url | https://academic.oup.com/bib/article-pdf/26/4/bbaf319/63725431/bbaf319.pdf |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | https://openalex.org/licenses/cc-by-nc |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | Briefings in Bioinformatics |
| primary_location.landing_page_url | https://doi.org/10.1093/bib/bbaf319 |
| publication_date | 2025-06-17 |
| publication_year | 2025 |
| referenced_works | https://openalex.org/W2111326065, https://openalex.org/W2147650094, https://openalex.org/W2145317354, https://openalex.org/W2794503008, https://openalex.org/W3035847987, https://openalex.org/W2100538849, https://openalex.org/W3014856454, https://openalex.org/W4206007286, https://openalex.org/W4200321201, https://openalex.org/W3048947632, https://openalex.org/W4394983192, https://openalex.org/W2023490488, https://openalex.org/W2151693654, https://openalex.org/W2125677968, https://openalex.org/W2103459989, https://openalex.org/W3109312535, https://openalex.org/W2324374958, https://openalex.org/W4391308485, https://openalex.org/W4400921915, https://openalex.org/W4403770916, https://openalex.org/W4296032638, https://openalex.org/W3171481040, https://openalex.org/W2262896633, https://openalex.org/W2953906682, https://openalex.org/W2121519777, https://openalex.org/W4388486442, https://openalex.org/W6858130012, https://openalex.org/W4401900858, https://openalex.org/W4327550249, https://openalex.org/W3177828909, https://openalex.org/W2980708516, https://openalex.org/W3116968030, https://openalex.org/W3176047188, https://openalex.org/W3093098684, https://openalex.org/W4224988655, https://openalex.org/W4392549818, https://openalex.org/W6780110802, https://openalex.org/W3146944767, https://openalex.org/W3083008290, https://openalex.org/W4379600237, https://openalex.org/W2767044445, https://openalex.org/W4366823175, https://openalex.org/W3121331573, https://openalex.org/W2003890287, https://openalex.org/W1554288134, https://openalex.org/W2139236870, https://openalex.org/W4233269677, https://openalex.org/W2801256450, https://openalex.org/W2922444498, https://openalex.org/W3205324928, https://openalex.org/W3119518863, https://openalex.org/W4384820649, https://openalex.org/W2950954328, https://openalex.org/W2172034373, https://openalex.org/W3105751341, https://openalex.org/W3093556707, https://openalex.org/W3104081477, https://openalex.org/W4315620405, https://openalex.org/W3117622225, https://openalex.org/W4292002633, https://openalex.org/W2030351692, https://openalex.org/W4386154677, https://openalex.org/W3012929814, https://openalex.org/W4390521376, https://openalex.org/W4387839235 |
| referenced_works_count | 65 |
| abstract_inverted_index.a | 24, 61, 161 |
| abstract_inverted_index.In | 146 |
| abstract_inverted_index.an | 173 |
| abstract_inverted_index.at | 76 |
| abstract_inverted_index.in | 59, 132, 172 |
| abstract_inverted_index.is | 8, 66 |
| abstract_inverted_index.of | 10, 18, 46, 73, 98, 122 |
| abstract_inverted_index.on | 85, 96, 109 |
| abstract_inverted_index.to | 14, 48, 68, 118 |
| abstract_inverted_index.we | 22 |
| abstract_inverted_index.and | 43, 91, 163 |
| abstract_inverted_index.are | 138 |
| abstract_inverted_index.for | 30, 140, 166 |
| abstract_inverted_index.one | 9 |
| abstract_inverted_index.the | 2, 11, 16, 41, 50, 71, 77, 120 |
| abstract_inverted_index.two | 54 |
| abstract_inverted_index.via | 153 |
| abstract_inverted_index.More | 125 |
| abstract_inverted_index.five | 97 |
| abstract_inverted_index.from | 36, 53 |
| abstract_inverted_index.rare | 134 |
| abstract_inverted_index.that | 137 |
| abstract_inverted_index.tool | 165 |
| abstract_inverted_index.upon | 6 |
| abstract_inverted_index.ways | 13 |
| abstract_inverted_index.Here, | 21 |
| abstract_inverted_index.local | 44 |
| abstract_inverted_index.model | 116 |
| abstract_inverted_index.other | 94, 103 |
| abstract_inverted_index.seven | 86 |
| abstract_inverted_index.task, | 113 |
| abstract_inverted_index.them. | 99 |
| abstract_inverted_index.views | 55 |
| abstract_inverted_index.which | 60 |
| abstract_inverted_index.change | 34, 170 |
| abstract_inverted_index.global | 42 |
| abstract_inverted_index.level. | 79 |
| abstract_inverted_index.module | 65 |
| abstract_inverted_index.single | 37 |
| abstract_inverted_index.capture | 49, 119 |
| abstract_inverted_index.changes | 5 |
| abstract_inverted_index.crucial | 139 |
| abstract_inverted_index.improve | 15 |
| abstract_inverted_index.manner. | 177 |
| abstract_inverted_index.methods | 95, 104 |
| abstract_inverted_index.propose | 23 |
| abstract_inverted_index.protein | 3, 19, 32, 142, 168 |
| abstract_inverted_index.scores, | 155 |
| abstract_inverted_index.through | 56, 106 |
| abstract_inverted_index.Abstract | 0 |
| abstract_inverted_index.Overall, | 158 |
| abstract_inverted_index.achieved | 81 |
| abstract_inverted_index.designed | 67 |
| abstract_inverted_index.directed | 143 |
| abstract_inverted_index.ensemble | 26, 57, 175 |
| abstract_inverted_index.flexible | 162 |
| abstract_inverted_index.learning | 176 |
| abstract_inverted_index.mutation | 88, 111 |
| abstract_inverted_index.provides | 160 |
| abstract_inverted_index.sequence | 78 |
| abstract_inverted_index.superior | 115, 129 |
| abstract_inverted_index.addition, | 147 |
| abstract_inverted_index.attention | 154 |
| abstract_inverted_index.benchmark | 89 |
| abstract_inverted_index.datasets, | 90 |
| abstract_inverted_index.dual-view | 25 |
| abstract_inverted_index.effective | 12 |
| abstract_inverted_index.efficient | 164 |
| abstract_inverted_index.epistasis | 121 |
| abstract_inverted_index.evolution | 144 |
| abstract_inverted_index.exhibited | 128 |
| abstract_inverted_index.important | 150 |
| abstract_inverted_index.inference | 108 |
| abstract_inverted_index.introduce | 70 |
| abstract_inverted_index.learning, | 58 |
| abstract_inverted_index.mutations | 7, 47, 136 |
| abstract_inverted_index.practical | 141 |
| abstract_inverted_index.sequence. | 38 |
| abstract_inverted_index.stability | 4, 33, 169 |
| abstract_inverted_index.surpassed | 93 |
| abstract_inverted_index.zero-shot | 107 |
| abstract_inverted_index.Predicting | 1 |
| abstract_inverted_index.beneficial | 135 |
| abstract_inverted_index.efficiency | 17 |
| abstract_inverted_index.framework, | 28 |
| abstract_inverted_index.identified | 149 |
| abstract_inverted_index.indirectly | 69 |
| abstract_inverted_index.integrates | 40 |
| abstract_inverted_index.mutations. | 124 |
| abstract_inverted_index.predicting | 133 |
| abstract_inverted_index.prediction | 35, 83, 112, 171 |
| abstract_inverted_index.scenarios. | 145 |
| abstract_inverted_index.simulation | 64 |
| abstract_inverted_index.structural | 62, 74 |
| abstract_inverted_index.information | 72 |
| abstract_inverted_index.performance | 84, 131 |
| abstract_inverted_index.Furthermore, | 100 |
| abstract_inverted_index.dependencies | 45 |
| abstract_inverted_index.engineering. | 20 |
| abstract_inverted_index.importantly, | 126 |
| abstract_inverted_index.interactions | 52, 152 |
| abstract_inverted_index.outperformed | 102 |
| abstract_inverted_index.single-point | 87 |
| abstract_inverted_index.DVE-stability | 39, 80, 101, 127, 148, 159 |
| abstract_inverted_index.demonstrating | 114, 156 |
| abstract_inverted_index.interpretable | 174 |
| abstract_inverted_index.DVE-stability, | 29 |
| abstract_inverted_index.generalization | 130 |
| abstract_inverted_index.interpretable. | 157 |
| abstract_inverted_index.intramolecular | 51, 151 |
| abstract_inverted_index.learning-based | 27 |
| abstract_inverted_index.multiple-point | 110, 123 |
| abstract_inverted_index.comprehensively | 92, 105 |
| abstract_inverted_index.generalizability | 117 |
| abstract_inverted_index.microenvironment | 63, 75 |
| abstract_inverted_index.mutation-induced | 31, 167 |
| abstract_inverted_index.state-of-the-art | 82 |
| cited_by_percentile_year | |
| countries_distinct_count | 2 |
| institutions_distinct_count | 13 |
| citation_normalized_percentile.value | 0.25938404 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |