Protein-protein interaction prediction via structure-based deep learning Article Swipe
 YOU?
·
· 2023
· Open Access
·
· DOI: https://doi.org/10.1101/2023.05.27.542552
YOU?
·
· 2023
· Open Access
·
· DOI: https://doi.org/10.1101/2023.05.27.542552
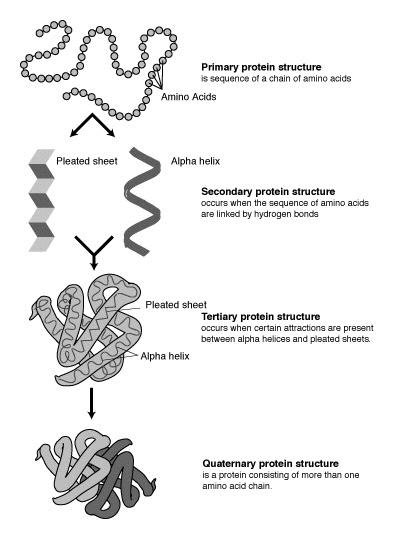
Protein-protein interactions (PPIs) play an essential role in life activities. Many machine learning algorithms based on protein sequence information have been developed to predict PPIs. However, these models have difficulty dealing with various sequence lengths and suffer from low generalization and prediction accuracy. In this study, we proposed a novel end-to-end deep learning framework, RSPPI, combining Residual Neural Network (ResNet) and Spatial Pyramid Pooling (SPP), to predict PPIs based on the protein sequence physicochemistry properties and spatial structural information. In the RSPPI model, ResNet was employed to extract the structural and physicochemical information from the protein 3D structure and primary sequence; the SPP layer was used to transform feature maps to a single vector and avoid the fixed-length requirement. The RSPPI model possessed excellent cross-species performance and outperformed several state-of-the-art methods based either on protein sequence or gene ontology in most evaluation metrics. The RSPPI model provides a novel strategic direction to develop an AI PPI prediction algorithm.
Related Topics
- Type
- preprint
- Language
- en
- Landing Page
- https://doi.org/10.1101/2023.05.27.542552
- https://www.biorxiv.org/content/biorxiv/early/2023/05/30/2023.05.27.542552.full.pdf
- OA Status
- green
- Cited By
- 3
- References
- 34
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4379106378
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4379106378Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1101/2023.05.27.542552Digital Object Identifier
- Title
-
Protein-protein interaction prediction via structure-based deep learningWork title
- Type
-
preprintOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2023Year of publication
- Publication date
-
2023-05-30Full publication date if available
- Authors
-
Yucong Liu, Zhenhai LiList of authors in order
- Landing page
-
https://doi.org/10.1101/2023.05.27.542552Publisher landing page
- PDF URL
-
https://www.biorxiv.org/content/biorxiv/early/2023/05/30/2023.05.27.542552.full.pdfDirect link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
greenOpen access status per OpenAlex
- OA URL
-
https://www.biorxiv.org/content/biorxiv/early/2023/05/30/2023.05.27.542552.full.pdfDirect OA link when available
- Concepts
-
Artificial intelligence, Computer science, Pooling, Sequence (biology), Deep learning, Protein structure prediction, Machine learning, Pyramid (geometry), Feature (linguistics), Generalization, Protein sequencing, Support vector machine, Artificial neural network, Pattern recognition (psychology), Residual, Data mining, Protein structure, Peptide sequence, Algorithm, Mathematics, Gene, Biology, Genetics, Mathematical analysis, Philosophy, Linguistics, Geometry, BiochemistryTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
3Total citation count in OpenAlex
- Citations by year (recent)
-
2025: 1, 2024: 2Per-year citation counts (last 5 years)
- References (count)
-
34Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4379106378 |
|---|---|
| doi | https://doi.org/10.1101/2023.05.27.542552 |
| ids.doi | https://doi.org/10.1101/2023.05.27.542552 |
| ids.openalex | https://openalex.org/W4379106378 |
| fwci | 0.92688361 |
| type | preprint |
| title | Protein-protein interaction prediction via structure-based deep learning |
| biblio.issue | |
| biblio.volume | |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T10211 |
| topics[0].field.id | https://openalex.org/fields/17 |
| topics[0].field.display_name | Computer Science |
| topics[0].score | 0.9990000128746033 |
| topics[0].domain.id | https://openalex.org/domains/3 |
| topics[0].domain.display_name | Physical Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1703 |
| topics[0].subfield.display_name | Computational Theory and Mathematics |
| topics[0].display_name | Computational Drug Discovery Methods |
| topics[1].id | https://openalex.org/T10044 |
| topics[1].field.id | https://openalex.org/fields/13 |
| topics[1].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[1].score | 0.9987999796867371 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1312 |
| topics[1].subfield.display_name | Molecular Biology |
| topics[1].display_name | Protein Structure and Dynamics |
| topics[2].id | https://openalex.org/T10887 |
| topics[2].field.id | https://openalex.org/fields/13 |
| topics[2].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[2].score | 0.9980000257492065 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1312 |
| topics[2].subfield.display_name | Molecular Biology |
| topics[2].display_name | Bioinformatics and Genomic Networks |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C154945302 |
| concepts[0].level | 1 |
| concepts[0].score | 0.6977975368499756 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q11660 |
| concepts[0].display_name | Artificial intelligence |
| concepts[1].id | https://openalex.org/C41008148 |
| concepts[1].level | 0 |
| concepts[1].score | 0.6569093465805054 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q21198 |
| concepts[1].display_name | Computer science |
| concepts[2].id | https://openalex.org/C70437156 |
| concepts[2].level | 2 |
| concepts[2].score | 0.6222468018531799 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q7228652 |
| concepts[2].display_name | Pooling |
| concepts[3].id | https://openalex.org/C2778112365 |
| concepts[3].level | 2 |
| concepts[3].score | 0.5970906019210815 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q3511065 |
| concepts[3].display_name | Sequence (biology) |
| concepts[4].id | https://openalex.org/C108583219 |
| concepts[4].level | 2 |
| concepts[4].score | 0.587956964969635 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q197536 |
| concepts[4].display_name | Deep learning |
| concepts[5].id | https://openalex.org/C18051474 |
| concepts[5].level | 3 |
| concepts[5].score | 0.5228598117828369 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q899656 |
| concepts[5].display_name | Protein structure prediction |
| concepts[6].id | https://openalex.org/C119857082 |
| concepts[6].level | 1 |
| concepts[6].score | 0.4793720543384552 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q2539 |
| concepts[6].display_name | Machine learning |
| concepts[7].id | https://openalex.org/C142575187 |
| concepts[7].level | 2 |
| concepts[7].score | 0.4610300660133362 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q3358290 |
| concepts[7].display_name | Pyramid (geometry) |
| concepts[8].id | https://openalex.org/C2776401178 |
| concepts[8].level | 2 |
| concepts[8].score | 0.46037158370018005 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q12050496 |
| concepts[8].display_name | Feature (linguistics) |
| concepts[9].id | https://openalex.org/C177148314 |
| concepts[9].level | 2 |
| concepts[9].score | 0.45510733127593994 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q170084 |
| concepts[9].display_name | Generalization |
| concepts[10].id | https://openalex.org/C10010492 |
| concepts[10].level | 4 |
| concepts[10].score | 0.45470860600471497 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q3142557 |
| concepts[10].display_name | Protein sequencing |
| concepts[11].id | https://openalex.org/C12267149 |
| concepts[11].level | 2 |
| concepts[11].score | 0.4456506371498108 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q282453 |
| concepts[11].display_name | Support vector machine |
| concepts[12].id | https://openalex.org/C50644808 |
| concepts[12].level | 2 |
| concepts[12].score | 0.4452388286590576 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q192776 |
| concepts[12].display_name | Artificial neural network |
| concepts[13].id | https://openalex.org/C153180895 |
| concepts[13].level | 2 |
| concepts[13].score | 0.4346785247325897 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q7148389 |
| concepts[13].display_name | Pattern recognition (psychology) |
| concepts[14].id | https://openalex.org/C155512373 |
| concepts[14].level | 2 |
| concepts[14].score | 0.43247461318969727 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q287450 |
| concepts[14].display_name | Residual |
| concepts[15].id | https://openalex.org/C124101348 |
| concepts[15].level | 1 |
| concepts[15].score | 0.321765661239624 |
| concepts[15].wikidata | https://www.wikidata.org/wiki/Q172491 |
| concepts[15].display_name | Data mining |
| concepts[16].id | https://openalex.org/C47701112 |
| concepts[16].level | 2 |
| concepts[16].score | 0.2902108132839203 |
| concepts[16].wikidata | https://www.wikidata.org/wiki/Q735188 |
| concepts[16].display_name | Protein structure |
| concepts[17].id | https://openalex.org/C167625842 |
| concepts[17].level | 3 |
| concepts[17].score | 0.23111951351165771 |
| concepts[17].wikidata | https://www.wikidata.org/wiki/Q899763 |
| concepts[17].display_name | Peptide sequence |
| concepts[18].id | https://openalex.org/C11413529 |
| concepts[18].level | 1 |
| concepts[18].score | 0.21311238408088684 |
| concepts[18].wikidata | https://www.wikidata.org/wiki/Q8366 |
| concepts[18].display_name | Algorithm |
| concepts[19].id | https://openalex.org/C33923547 |
| concepts[19].level | 0 |
| concepts[19].score | 0.1158246099948883 |
| concepts[19].wikidata | https://www.wikidata.org/wiki/Q395 |
| concepts[19].display_name | Mathematics |
| concepts[20].id | https://openalex.org/C104317684 |
| concepts[20].level | 2 |
| concepts[20].score | 0.09754407405853271 |
| concepts[20].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[20].display_name | Gene |
| concepts[21].id | https://openalex.org/C86803240 |
| concepts[21].level | 0 |
| concepts[21].score | 0.08736249804496765 |
| concepts[21].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[21].display_name | Biology |
| concepts[22].id | https://openalex.org/C54355233 |
| concepts[22].level | 1 |
| concepts[22].score | 0.0 |
| concepts[22].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[22].display_name | Genetics |
| concepts[23].id | https://openalex.org/C134306372 |
| concepts[23].level | 1 |
| concepts[23].score | 0.0 |
| concepts[23].wikidata | https://www.wikidata.org/wiki/Q7754 |
| concepts[23].display_name | Mathematical analysis |
| concepts[24].id | https://openalex.org/C138885662 |
| concepts[24].level | 0 |
| concepts[24].score | 0.0 |
| concepts[24].wikidata | https://www.wikidata.org/wiki/Q5891 |
| concepts[24].display_name | Philosophy |
| concepts[25].id | https://openalex.org/C41895202 |
| concepts[25].level | 1 |
| concepts[25].score | 0.0 |
| concepts[25].wikidata | https://www.wikidata.org/wiki/Q8162 |
| concepts[25].display_name | Linguistics |
| concepts[26].id | https://openalex.org/C2524010 |
| concepts[26].level | 1 |
| concepts[26].score | 0.0 |
| concepts[26].wikidata | https://www.wikidata.org/wiki/Q8087 |
| concepts[26].display_name | Geometry |
| concepts[27].id | https://openalex.org/C55493867 |
| concepts[27].level | 1 |
| concepts[27].score | 0.0 |
| concepts[27].wikidata | https://www.wikidata.org/wiki/Q7094 |
| concepts[27].display_name | Biochemistry |
| keywords[0].id | https://openalex.org/keywords/artificial-intelligence |
| keywords[0].score | 0.6977975368499756 |
| keywords[0].display_name | Artificial intelligence |
| keywords[1].id | https://openalex.org/keywords/computer-science |
| keywords[1].score | 0.6569093465805054 |
| keywords[1].display_name | Computer science |
| keywords[2].id | https://openalex.org/keywords/pooling |
| keywords[2].score | 0.6222468018531799 |
| keywords[2].display_name | Pooling |
| keywords[3].id | https://openalex.org/keywords/sequence |
| keywords[3].score | 0.5970906019210815 |
| keywords[3].display_name | Sequence (biology) |
| keywords[4].id | https://openalex.org/keywords/deep-learning |
| keywords[4].score | 0.587956964969635 |
| keywords[4].display_name | Deep learning |
| keywords[5].id | https://openalex.org/keywords/protein-structure-prediction |
| keywords[5].score | 0.5228598117828369 |
| keywords[5].display_name | Protein structure prediction |
| keywords[6].id | https://openalex.org/keywords/machine-learning |
| keywords[6].score | 0.4793720543384552 |
| keywords[6].display_name | Machine learning |
| keywords[7].id | https://openalex.org/keywords/pyramid |
| keywords[7].score | 0.4610300660133362 |
| keywords[7].display_name | Pyramid (geometry) |
| keywords[8].id | https://openalex.org/keywords/feature |
| keywords[8].score | 0.46037158370018005 |
| keywords[8].display_name | Feature (linguistics) |
| keywords[9].id | https://openalex.org/keywords/generalization |
| keywords[9].score | 0.45510733127593994 |
| keywords[9].display_name | Generalization |
| keywords[10].id | https://openalex.org/keywords/protein-sequencing |
| keywords[10].score | 0.45470860600471497 |
| keywords[10].display_name | Protein sequencing |
| keywords[11].id | https://openalex.org/keywords/support-vector-machine |
| keywords[11].score | 0.4456506371498108 |
| keywords[11].display_name | Support vector machine |
| keywords[12].id | https://openalex.org/keywords/artificial-neural-network |
| keywords[12].score | 0.4452388286590576 |
| keywords[12].display_name | Artificial neural network |
| keywords[13].id | https://openalex.org/keywords/pattern-recognition |
| keywords[13].score | 0.4346785247325897 |
| keywords[13].display_name | Pattern recognition (psychology) |
| keywords[14].id | https://openalex.org/keywords/residual |
| keywords[14].score | 0.43247461318969727 |
| keywords[14].display_name | Residual |
| keywords[15].id | https://openalex.org/keywords/data-mining |
| keywords[15].score | 0.321765661239624 |
| keywords[15].display_name | Data mining |
| keywords[16].id | https://openalex.org/keywords/protein-structure |
| keywords[16].score | 0.2902108132839203 |
| keywords[16].display_name | Protein structure |
| keywords[17].id | https://openalex.org/keywords/peptide-sequence |
| keywords[17].score | 0.23111951351165771 |
| keywords[17].display_name | Peptide sequence |
| keywords[18].id | https://openalex.org/keywords/algorithm |
| keywords[18].score | 0.21311238408088684 |
| keywords[18].display_name | Algorithm |
| keywords[19].id | https://openalex.org/keywords/mathematics |
| keywords[19].score | 0.1158246099948883 |
| keywords[19].display_name | Mathematics |
| keywords[20].id | https://openalex.org/keywords/gene |
| keywords[20].score | 0.09754407405853271 |
| keywords[20].display_name | Gene |
| keywords[21].id | https://openalex.org/keywords/biology |
| keywords[21].score | 0.08736249804496765 |
| keywords[21].display_name | Biology |
| language | en |
| locations[0].id | doi:10.1101/2023.05.27.542552 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4306402567 |
| locations[0].source.issn | |
| locations[0].source.type | repository |
| locations[0].source.is_oa | False |
| locations[0].source.issn_l | |
| locations[0].source.is_core | False |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| locations[0].source.host_organization | https://openalex.org/I2750212522 |
| locations[0].source.host_organization_name | Cold Spring Harbor Laboratory |
| locations[0].source.host_organization_lineage | https://openalex.org/I2750212522 |
| locations[0].license | cc-by-nc-nd |
| locations[0].pdf_url | https://www.biorxiv.org/content/biorxiv/early/2023/05/30/2023.05.27.542552.full.pdf |
| locations[0].version | acceptedVersion |
| locations[0].raw_type | posted-content |
| locations[0].license_id | https://openalex.org/licenses/cc-by-nc-nd |
| locations[0].is_accepted | True |
| locations[0].is_published | False |
| locations[0].raw_source_name | |
| locations[0].landing_page_url | https://doi.org/10.1101/2023.05.27.542552 |
| indexed_in | crossref |
| authorships[0].author.id | https://openalex.org/A5025152292 |
| authorships[0].author.orcid | https://orcid.org/0000-0002-3753-9670 |
| authorships[0].author.display_name | Yucong Liu |
| authorships[0].countries | CN |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I113940042 |
| authorships[0].affiliations[0].raw_affiliation_string | Shanghai Key Laboratory of Mechanics in Energy Engineering, Shanghai Institute of Applied Mathematics and Mechanics, School of Mechanics and Engineering Science, Shanghai University, Shanghai 200072, China |
| authorships[0].institutions[0].id | https://openalex.org/I113940042 |
| authorships[0].institutions[0].ror | https://ror.org/006teas31 |
| authorships[0].institutions[0].type | education |
| authorships[0].institutions[0].lineage | https://openalex.org/I113940042 |
| authorships[0].institutions[0].country_code | CN |
| authorships[0].institutions[0].display_name | Shanghai University |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Yucong Liu |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Shanghai Key Laboratory of Mechanics in Energy Engineering, Shanghai Institute of Applied Mathematics and Mechanics, School of Mechanics and Engineering Science, Shanghai University, Shanghai 200072, China |
| authorships[1].author.id | https://openalex.org/A5102814477 |
| authorships[1].author.orcid | https://orcid.org/0000-0002-0720-2807 |
| authorships[1].author.display_name | Zhenhai Li |
| authorships[1].countries | CN |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I113940042 |
| authorships[1].affiliations[0].raw_affiliation_string | Shanghai Key Laboratory of Mechanics in Energy Engineering, Shanghai Institute of Applied Mathematics and Mechanics, School of Mechanics and Engineering Science, Shanghai University, Shanghai 200072, China |
| authorships[1].institutions[0].id | https://openalex.org/I113940042 |
| authorships[1].institutions[0].ror | https://ror.org/006teas31 |
| authorships[1].institutions[0].type | education |
| authorships[1].institutions[0].lineage | https://openalex.org/I113940042 |
| authorships[1].institutions[0].country_code | CN |
| authorships[1].institutions[0].display_name | Shanghai University |
| authorships[1].author_position | last |
| authorships[1].raw_author_name | Zhenhai Li |
| authorships[1].is_corresponding | True |
| authorships[1].raw_affiliation_strings | Shanghai Key Laboratory of Mechanics in Energy Engineering, Shanghai Institute of Applied Mathematics and Mechanics, School of Mechanics and Engineering Science, Shanghai University, Shanghai 200072, China |
| has_content.pdf | True |
| has_content.grobid_xml | True |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://www.biorxiv.org/content/biorxiv/early/2023/05/30/2023.05.27.542552.full.pdf |
| open_access.oa_status | green |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Protein-protein interaction prediction via structure-based deep learning |
| has_fulltext | True |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T10211 |
| primary_topic.field.id | https://openalex.org/fields/17 |
| primary_topic.field.display_name | Computer Science |
| primary_topic.score | 0.9990000128746033 |
| primary_topic.domain.id | https://openalex.org/domains/3 |
| primary_topic.domain.display_name | Physical Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1703 |
| primary_topic.subfield.display_name | Computational Theory and Mathematics |
| primary_topic.display_name | Computational Drug Discovery Methods |
| related_works | https://openalex.org/W2022849497, https://openalex.org/W2407190427, https://openalex.org/W3081299480, https://openalex.org/W2919210741, https://openalex.org/W2907584218, https://openalex.org/W3002446410, https://openalex.org/W4390224712, https://openalex.org/W4322096758, https://openalex.org/W4387310732, https://openalex.org/W2034313881 |
| cited_by_count | 3 |
| counts_by_year[0].year | 2025 |
| counts_by_year[0].cited_by_count | 1 |
| counts_by_year[1].year | 2024 |
| counts_by_year[1].cited_by_count | 2 |
| locations_count | 1 |
| best_oa_location.id | doi:10.1101/2023.05.27.542552 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4306402567 |
| best_oa_location.source.issn | |
| best_oa_location.source.type | repository |
| best_oa_location.source.is_oa | False |
| best_oa_location.source.issn_l | |
| best_oa_location.source.is_core | False |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| best_oa_location.source.host_organization | https://openalex.org/I2750212522 |
| best_oa_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| best_oa_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| best_oa_location.license | cc-by-nc-nd |
| best_oa_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2023/05/30/2023.05.27.542552.full.pdf |
| best_oa_location.version | acceptedVersion |
| best_oa_location.raw_type | posted-content |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by-nc-nd |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | False |
| best_oa_location.raw_source_name | |
| best_oa_location.landing_page_url | https://doi.org/10.1101/2023.05.27.542552 |
| primary_location.id | doi:10.1101/2023.05.27.542552 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4306402567 |
| primary_location.source.issn | |
| primary_location.source.type | repository |
| primary_location.source.is_oa | False |
| primary_location.source.issn_l | |
| primary_location.source.is_core | False |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| primary_location.source.host_organization | https://openalex.org/I2750212522 |
| primary_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| primary_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| primary_location.license | cc-by-nc-nd |
| primary_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2023/05/30/2023.05.27.542552.full.pdf |
| primary_location.version | acceptedVersion |
| primary_location.raw_type | posted-content |
| primary_location.license_id | https://openalex.org/licenses/cc-by-nc-nd |
| primary_location.is_accepted | True |
| primary_location.is_published | False |
| primary_location.raw_source_name | |
| primary_location.landing_page_url | https://doi.org/10.1101/2023.05.27.542552 |
| publication_date | 2023-05-30 |
| publication_year | 2023 |
| referenced_works | https://openalex.org/W2079595896, https://openalex.org/W2616561032, https://openalex.org/W2167869241, https://openalex.org/W2099982459, https://openalex.org/W2067752117, https://openalex.org/W1977310857, https://openalex.org/W2085989567, https://openalex.org/W2029759111, https://openalex.org/W3112376646, https://openalex.org/W76924599, https://openalex.org/W1977391294, https://openalex.org/W2152705149, https://openalex.org/W2890911678, https://openalex.org/W3127830150, https://openalex.org/W3034703136, https://openalex.org/W2042301222, https://openalex.org/W2786213090, https://openalex.org/W1918391659, https://openalex.org/W2134197776, https://openalex.org/W2167851075, https://openalex.org/W2106612397, https://openalex.org/W2096166549, https://openalex.org/W2194775991, https://openalex.org/W2109255472, https://openalex.org/W2097356092, https://openalex.org/W2114850508, https://openalex.org/W4317703013, https://openalex.org/W2991877021, https://openalex.org/W2502312327, https://openalex.org/W2336829316, https://openalex.org/W2119583098, https://openalex.org/W2155006702, https://openalex.org/W2948220209, https://openalex.org/W3099395032 |
| referenced_works_count | 34 |
| abstract_inverted_index.a | 49, 112, 148 |
| abstract_inverted_index.3D | 97 |
| abstract_inverted_index.AI | 155 |
| abstract_inverted_index.In | 44, 80 |
| abstract_inverted_index.an | 5, 154 |
| abstract_inverted_index.in | 8, 140 |
| abstract_inverted_index.on | 16, 70, 134 |
| abstract_inverted_index.or | 137 |
| abstract_inverted_index.to | 23, 66, 87, 107, 111, 152 |
| abstract_inverted_index.we | 47 |
| abstract_inverted_index.PPI | 156 |
| abstract_inverted_index.SPP | 103 |
| abstract_inverted_index.The | 120, 144 |
| abstract_inverted_index.and | 36, 41, 61, 76, 91, 99, 115, 127 |
| abstract_inverted_index.low | 39 |
| abstract_inverted_index.the | 71, 81, 89, 95, 102, 117 |
| abstract_inverted_index.was | 85, 105 |
| abstract_inverted_index.Many | 11 |
| abstract_inverted_index.PPIs | 68 |
| abstract_inverted_index.been | 21 |
| abstract_inverted_index.deep | 52 |
| abstract_inverted_index.from | 38, 94 |
| abstract_inverted_index.gene | 138 |
| abstract_inverted_index.have | 20, 29 |
| abstract_inverted_index.life | 9 |
| abstract_inverted_index.maps | 110 |
| abstract_inverted_index.most | 141 |
| abstract_inverted_index.play | 4 |
| abstract_inverted_index.role | 7 |
| abstract_inverted_index.this | 45 |
| abstract_inverted_index.used | 106 |
| abstract_inverted_index.with | 32 |
| abstract_inverted_index.PPIs. | 25 |
| abstract_inverted_index.RSPPI | 82, 121, 145 |
| abstract_inverted_index.avoid | 116 |
| abstract_inverted_index.based | 15, 69, 132 |
| abstract_inverted_index.layer | 104 |
| abstract_inverted_index.model | 122, 146 |
| abstract_inverted_index.novel | 50, 149 |
| abstract_inverted_index.these | 27 |
| abstract_inverted_index.(PPIs) | 3 |
| abstract_inverted_index.(SPP), | 65 |
| abstract_inverted_index.Neural | 58 |
| abstract_inverted_index.RSPPI, | 55 |
| abstract_inverted_index.ResNet | 84 |
| abstract_inverted_index.either | 133 |
| abstract_inverted_index.model, | 83 |
| abstract_inverted_index.models | 28 |
| abstract_inverted_index.single | 113 |
| abstract_inverted_index.study, | 46 |
| abstract_inverted_index.suffer | 37 |
| abstract_inverted_index.vector | 114 |
| abstract_inverted_index.Network | 59 |
| abstract_inverted_index.Pooling | 64 |
| abstract_inverted_index.Pyramid | 63 |
| abstract_inverted_index.Spatial | 62 |
| abstract_inverted_index.dealing | 31 |
| abstract_inverted_index.develop | 153 |
| abstract_inverted_index.extract | 88 |
| abstract_inverted_index.feature | 109 |
| abstract_inverted_index.lengths | 35 |
| abstract_inverted_index.machine | 12 |
| abstract_inverted_index.methods | 131 |
| abstract_inverted_index.predict | 24, 67 |
| abstract_inverted_index.primary | 100 |
| abstract_inverted_index.protein | 17, 72, 96, 135 |
| abstract_inverted_index.several | 129 |
| abstract_inverted_index.spatial | 77 |
| abstract_inverted_index.various | 33 |
| abstract_inverted_index.(ResNet) | 60 |
| abstract_inverted_index.Abstract | 0 |
| abstract_inverted_index.However, | 26 |
| abstract_inverted_index.Residual | 57 |
| abstract_inverted_index.employed | 86 |
| abstract_inverted_index.learning | 13, 53 |
| abstract_inverted_index.metrics. | 143 |
| abstract_inverted_index.ontology | 139 |
| abstract_inverted_index.proposed | 48 |
| abstract_inverted_index.provides | 147 |
| abstract_inverted_index.sequence | 18, 34, 73, 136 |
| abstract_inverted_index.accuracy. | 43 |
| abstract_inverted_index.combining | 56 |
| abstract_inverted_index.developed | 22 |
| abstract_inverted_index.direction | 151 |
| abstract_inverted_index.essential | 6 |
| abstract_inverted_index.excellent | 124 |
| abstract_inverted_index.possessed | 123 |
| abstract_inverted_index.sequence; | 101 |
| abstract_inverted_index.strategic | 150 |
| abstract_inverted_index.structure | 98 |
| abstract_inverted_index.transform | 108 |
| abstract_inverted_index.algorithm. | 158 |
| abstract_inverted_index.algorithms | 14 |
| abstract_inverted_index.difficulty | 30 |
| abstract_inverted_index.end-to-end | 51 |
| abstract_inverted_index.evaluation | 142 |
| abstract_inverted_index.framework, | 54 |
| abstract_inverted_index.prediction | 42, 157 |
| abstract_inverted_index.properties | 75 |
| abstract_inverted_index.structural | 78, 90 |
| abstract_inverted_index.activities. | 10 |
| abstract_inverted_index.information | 19, 93 |
| abstract_inverted_index.performance | 126 |
| abstract_inverted_index.fixed-length | 118 |
| abstract_inverted_index.information. | 79 |
| abstract_inverted_index.interactions | 2 |
| abstract_inverted_index.outperformed | 128 |
| abstract_inverted_index.requirement. | 119 |
| abstract_inverted_index.cross-species | 125 |
| abstract_inverted_index.generalization | 40 |
| abstract_inverted_index.Protein-protein | 1 |
| abstract_inverted_index.physicochemical | 92 |
| abstract_inverted_index.physicochemistry | 74 |
| abstract_inverted_index.state-of-the-art | 130 |
| cited_by_percentile_year.max | 96 |
| cited_by_percentile_year.min | 91 |
| corresponding_author_ids | https://openalex.org/A5102814477 |
| countries_distinct_count | 1 |
| institutions_distinct_count | 2 |
| corresponding_institution_ids | https://openalex.org/I113940042 |
| sustainable_development_goals[0].id | https://metadata.un.org/sdg/12 |
| sustainable_development_goals[0].score | 0.5600000023841858 |
| sustainable_development_goals[0].display_name | Responsible consumption and production |
| citation_normalized_percentile.value | 0.73592611 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |