Quantitative Fragmentation Model for Bottom-Up Shotgun Lipidomics Article Swipe
 YOU?
·
· 2019
· Open Access
·
· DOI: https://doi.org/10.1021/acs.analchem.9b03270
YOU?
·
· 2019
· Open Access
·
· DOI: https://doi.org/10.1021/acs.analchem.9b03270
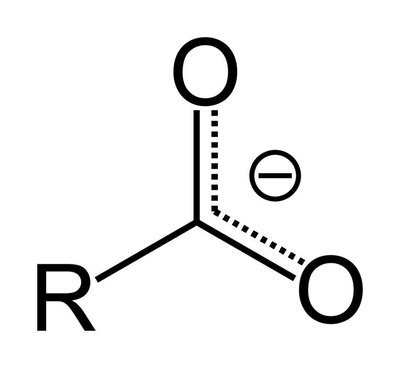
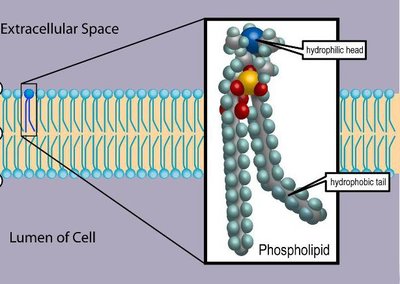
Quantitative bottom-up shotgun lipidomics relies on molecular species-specific "signature" fragments consistently detectable in tandem mass spectra of analytes and standards. Molecular species of glycerophospholipids are typically quantified using carboxylate fragments of their fatty acid moieties produced by higher-energy collisional dissociation of their molecular anions. However, employing standards whose fatty acids moieties are similar, yet not identical, to the target lipids could severely compromise their quantification. We developed a generic and portable fragmentation model implemented in the open-source LipidXte software that harmonizes the abundances of carboxylate anion fragments originating from fatty acid moieties having different sn-1/2 positions at the glycerol backbone, length of the hydrocarbon chain, and number and location of double bonds. The postacquisition adjustment enables unbiased absolute (molar) quantification of glycerophospholipid species independent of instrument settings, collision energy, and employed internal standards.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.1021/acs.analchem.9b03270
- OA Status
- hybrid
- Cited By
- 25
- References
- 32
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W2969231064
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W2969231064Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1021/acs.analchem.9b03270Digital Object Identifier
- Title
-
Quantitative Fragmentation Model for Bottom-Up Shotgun LipidomicsWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2019Year of publication
- Publication date
-
2019-08-23Full publication date if available
- Authors
-
Kai Schuhmann, HongKee Moon, Henrik Thomas, Jacobo Miranda Ackerman, Michael Groessl, Nicolai Wagner, Markus Kellmann, Ian Henry, André Nadler, Andrej ShevchenkoList of authors in order
- Landing page
-
https://doi.org/10.1021/acs.analchem.9b03270Publisher landing page
- Open access
-
YesWhether a free full text is available
- OA status
-
hybridOpen access status per OpenAlex
- OA URL
-
https://doi.org/10.1021/acs.analchem.9b03270Direct OA link when available
- Concepts
-
Chemistry, Glycerophospholipids, Glycerophospholipid, Lipidomics, Fragmentation (computing), Tandem mass spectrometry, Carboxylate, Collision-induced dissociation, Mass spectrometry, Chromatography, Analytical Chemistry (journal), Stereochemistry, Phospholipid, Biochemistry, Membrane, Operating system, Computer scienceTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
25Total citation count in OpenAlex
- Citations by year (recent)
-
2025: 3, 2024: 1, 2023: 4, 2022: 10, 2021: 3Per-year citation counts (last 5 years)
- References (count)
-
32Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W2969231064 |
|---|---|
| doi | https://doi.org/10.1021/acs.analchem.9b03270 |
| ids.doi | https://doi.org/10.1021/acs.analchem.9b03270 |
| ids.mag | 2969231064 |
| ids.pmid | https://pubmed.ncbi.nlm.nih.gov/31441640 |
| ids.openalex | https://openalex.org/W2969231064 |
| fwci | 1.75186713 |
| mesh[0].qualifier_ui | Q000032 |
| mesh[0].descriptor_ui | D020404 |
| mesh[0].is_major_topic | False |
| mesh[0].qualifier_name | analysis |
| mesh[0].descriptor_name | Glycerophospholipids |
| mesh[1].qualifier_ui | |
| mesh[1].descriptor_ui | D000081362 |
| mesh[1].is_major_topic | True |
| mesh[1].qualifier_name | |
| mesh[1].descriptor_name | Lipidomics |
| mesh[2].qualifier_ui | |
| mesh[2].descriptor_ui | D008958 |
| mesh[2].is_major_topic | False |
| mesh[2].qualifier_name | |
| mesh[2].descriptor_name | Models, Molecular |
| mesh[3].qualifier_ui | |
| mesh[3].descriptor_ui | D012984 |
| mesh[3].is_major_topic | False |
| mesh[3].qualifier_name | |
| mesh[3].descriptor_name | Software |
| mesh[4].qualifier_ui | |
| mesh[4].descriptor_ui | D053719 |
| mesh[4].is_major_topic | False |
| mesh[4].qualifier_name | |
| mesh[4].descriptor_name | Tandem Mass Spectrometry |
| mesh[5].qualifier_ui | Q000032 |
| mesh[5].descriptor_ui | D020404 |
| mesh[5].is_major_topic | False |
| mesh[5].qualifier_name | analysis |
| mesh[5].descriptor_name | Glycerophospholipids |
| mesh[6].qualifier_ui | |
| mesh[6].descriptor_ui | D000081362 |
| mesh[6].is_major_topic | True |
| mesh[6].qualifier_name | |
| mesh[6].descriptor_name | Lipidomics |
| mesh[7].qualifier_ui | |
| mesh[7].descriptor_ui | D008958 |
| mesh[7].is_major_topic | False |
| mesh[7].qualifier_name | |
| mesh[7].descriptor_name | Models, Molecular |
| mesh[8].qualifier_ui | |
| mesh[8].descriptor_ui | D012984 |
| mesh[8].is_major_topic | False |
| mesh[8].qualifier_name | |
| mesh[8].descriptor_name | Software |
| mesh[9].qualifier_ui | |
| mesh[9].descriptor_ui | D053719 |
| mesh[9].is_major_topic | False |
| mesh[9].qualifier_name | |
| mesh[9].descriptor_name | Tandem Mass Spectrometry |
| type | article |
| title | Quantitative Fragmentation Model for Bottom-Up Shotgun Lipidomics |
| awards[0].id | https://openalex.org/G3632341369 |
| awards[0].funder_id | https://openalex.org/F4320338335 |
| awards[0].funder_award_id | GA758334 ASYMMEM |
| awards[0].funder_display_name | H2020 European Research Council |
| awards[1].id | https://openalex.org/G5542608249 |
| awards[1].funder_id | https://openalex.org/F4320320879 |
| awards[1].funder_award_id | TRR83 |
| awards[1].funder_display_name | Deutsche Forschungsgemeinschaft |
| biblio.issue | 18 |
| biblio.volume | 91 |
| biblio.last_page | 12093 |
| biblio.first_page | 12085 |
| grants[0].funder | https://openalex.org/F4320320879 |
| grants[0].award_id | TRR83 |
| grants[0].funder_display_name | Deutsche Forschungsgemeinschaft |
| grants[1].funder | https://openalex.org/F4320322434 |
| grants[1].award_id | |
| grants[1].funder_display_name | Max-Planck-Gesellschaft |
| grants[2].funder | https://openalex.org/F4320338335 |
| grants[2].award_id | GA758334 ASYMMEM |
| grants[2].funder_display_name | H2020 European Research Council |
| topics[0].id | https://openalex.org/T10836 |
| topics[0].field.id | https://openalex.org/fields/13 |
| topics[0].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[0].score | 1.0 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1312 |
| topics[0].subfield.display_name | Molecular Biology |
| topics[0].display_name | Metabolomics and Mass Spectrometry Studies |
| topics[1].id | https://openalex.org/T10683 |
| topics[1].field.id | https://openalex.org/fields/16 |
| topics[1].field.display_name | Chemistry |
| topics[1].score | 0.9994999766349792 |
| topics[1].domain.id | https://openalex.org/domains/3 |
| topics[1].domain.display_name | Physical Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1607 |
| topics[1].subfield.display_name | Spectroscopy |
| topics[1].display_name | Mass Spectrometry Techniques and Applications |
| topics[2].id | https://openalex.org/T10519 |
| topics[2].field.id | https://openalex.org/fields/16 |
| topics[2].field.display_name | Chemistry |
| topics[2].score | 0.9958999752998352 |
| topics[2].domain.id | https://openalex.org/domains/3 |
| topics[2].domain.display_name | Physical Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1607 |
| topics[2].subfield.display_name | Spectroscopy |
| topics[2].display_name | Advanced Proteomics Techniques and Applications |
| funders[0].id | https://openalex.org/F4320320879 |
| funders[0].ror | https://ror.org/018mejw64 |
| funders[0].display_name | Deutsche Forschungsgemeinschaft |
| funders[1].id | https://openalex.org/F4320322434 |
| funders[1].ror | https://ror.org/01hhn8329 |
| funders[1].display_name | Max-Planck-Gesellschaft |
| funders[2].id | https://openalex.org/F4320338335 |
| funders[2].ror | https://ror.org/0472cxd90 |
| funders[2].display_name | H2020 European Research Council |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C185592680 |
| concepts[0].level | 0 |
| concepts[0].score | 0.9405179619789124 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q2329 |
| concepts[0].display_name | Chemistry |
| concepts[1].id | https://openalex.org/C2909522894 |
| concepts[1].level | 4 |
| concepts[1].score | 0.83428955078125 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q420004 |
| concepts[1].display_name | Glycerophospholipids |
| concepts[2].id | https://openalex.org/C2776952329 |
| concepts[2].level | 4 |
| concepts[2].score | 0.8052918910980225 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q420004 |
| concepts[2].display_name | Glycerophospholipid |
| concepts[3].id | https://openalex.org/C58017582 |
| concepts[3].level | 2 |
| concepts[3].score | 0.7643468379974365 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q6556376 |
| concepts[3].display_name | Lipidomics |
| concepts[4].id | https://openalex.org/C191015642 |
| concepts[4].level | 2 |
| concepts[4].score | 0.7042391896247864 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q1132459 |
| concepts[4].display_name | Fragmentation (computing) |
| concepts[5].id | https://openalex.org/C31827203 |
| concepts[5].level | 3 |
| concepts[5].score | 0.5801004767417908 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q874296 |
| concepts[5].display_name | Tandem mass spectrometry |
| concepts[6].id | https://openalex.org/C2778074193 |
| concepts[6].level | 2 |
| concepts[6].score | 0.4900566339492798 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q3498942 |
| concepts[6].display_name | Carboxylate |
| concepts[7].id | https://openalex.org/C176107965 |
| concepts[7].level | 4 |
| concepts[7].score | 0.42172950506210327 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q3710336 |
| concepts[7].display_name | Collision-induced dissociation |
| concepts[8].id | https://openalex.org/C162356407 |
| concepts[8].level | 2 |
| concepts[8].score | 0.4061531126499176 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q180809 |
| concepts[8].display_name | Mass spectrometry |
| concepts[9].id | https://openalex.org/C43617362 |
| concepts[9].level | 1 |
| concepts[9].score | 0.39681071043014526 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q170050 |
| concepts[9].display_name | Chromatography |
| concepts[10].id | https://openalex.org/C113196181 |
| concepts[10].level | 2 |
| concepts[10].score | 0.3227241635322571 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q485223 |
| concepts[10].display_name | Analytical Chemistry (journal) |
| concepts[11].id | https://openalex.org/C71240020 |
| concepts[11].level | 1 |
| concepts[11].score | 0.28539782762527466 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q186011 |
| concepts[11].display_name | Stereochemistry |
| concepts[12].id | https://openalex.org/C2778918659 |
| concepts[12].level | 3 |
| concepts[12].score | 0.2279486358165741 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q186915 |
| concepts[12].display_name | Phospholipid |
| concepts[13].id | https://openalex.org/C55493867 |
| concepts[13].level | 1 |
| concepts[13].score | 0.19425731897354126 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q7094 |
| concepts[13].display_name | Biochemistry |
| concepts[14].id | https://openalex.org/C41625074 |
| concepts[14].level | 2 |
| concepts[14].score | 0.08692878484725952 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q176088 |
| concepts[14].display_name | Membrane |
| concepts[15].id | https://openalex.org/C111919701 |
| concepts[15].level | 1 |
| concepts[15].score | 0.0 |
| concepts[15].wikidata | https://www.wikidata.org/wiki/Q9135 |
| concepts[15].display_name | Operating system |
| concepts[16].id | https://openalex.org/C41008148 |
| concepts[16].level | 0 |
| concepts[16].score | 0.0 |
| concepts[16].wikidata | https://www.wikidata.org/wiki/Q21198 |
| concepts[16].display_name | Computer science |
| keywords[0].id | https://openalex.org/keywords/chemistry |
| keywords[0].score | 0.9405179619789124 |
| keywords[0].display_name | Chemistry |
| keywords[1].id | https://openalex.org/keywords/glycerophospholipids |
| keywords[1].score | 0.83428955078125 |
| keywords[1].display_name | Glycerophospholipids |
| keywords[2].id | https://openalex.org/keywords/glycerophospholipid |
| keywords[2].score | 0.8052918910980225 |
| keywords[2].display_name | Glycerophospholipid |
| keywords[3].id | https://openalex.org/keywords/lipidomics |
| keywords[3].score | 0.7643468379974365 |
| keywords[3].display_name | Lipidomics |
| keywords[4].id | https://openalex.org/keywords/fragmentation |
| keywords[4].score | 0.7042391896247864 |
| keywords[4].display_name | Fragmentation (computing) |
| keywords[5].id | https://openalex.org/keywords/tandem-mass-spectrometry |
| keywords[5].score | 0.5801004767417908 |
| keywords[5].display_name | Tandem mass spectrometry |
| keywords[6].id | https://openalex.org/keywords/carboxylate |
| keywords[6].score | 0.4900566339492798 |
| keywords[6].display_name | Carboxylate |
| keywords[7].id | https://openalex.org/keywords/collision-induced-dissociation |
| keywords[7].score | 0.42172950506210327 |
| keywords[7].display_name | Collision-induced dissociation |
| keywords[8].id | https://openalex.org/keywords/mass-spectrometry |
| keywords[8].score | 0.4061531126499176 |
| keywords[8].display_name | Mass spectrometry |
| keywords[9].id | https://openalex.org/keywords/chromatography |
| keywords[9].score | 0.39681071043014526 |
| keywords[9].display_name | Chromatography |
| keywords[10].id | https://openalex.org/keywords/analytical-chemistry |
| keywords[10].score | 0.3227241635322571 |
| keywords[10].display_name | Analytical Chemistry (journal) |
| keywords[11].id | https://openalex.org/keywords/stereochemistry |
| keywords[11].score | 0.28539782762527466 |
| keywords[11].display_name | Stereochemistry |
| keywords[12].id | https://openalex.org/keywords/phospholipid |
| keywords[12].score | 0.2279486358165741 |
| keywords[12].display_name | Phospholipid |
| keywords[13].id | https://openalex.org/keywords/biochemistry |
| keywords[13].score | 0.19425731897354126 |
| keywords[13].display_name | Biochemistry |
| keywords[14].id | https://openalex.org/keywords/membrane |
| keywords[14].score | 0.08692878484725952 |
| keywords[14].display_name | Membrane |
| language | en |
| locations[0].id | doi:10.1021/acs.analchem.9b03270 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S191963794 |
| locations[0].source.issn | 0003-2700, 1520-6882 |
| locations[0].source.type | journal |
| locations[0].source.is_oa | False |
| locations[0].source.issn_l | 0003-2700 |
| locations[0].source.is_core | True |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | Analytical Chemistry |
| locations[0].source.host_organization | https://openalex.org/P4310320006 |
| locations[0].source.host_organization_name | American Chemical Society |
| locations[0].source.host_organization_lineage | https://openalex.org/P4310320006 |
| locations[0].license | cc-by |
| locations[0].pdf_url | |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | Analytical Chemistry |
| locations[0].landing_page_url | https://doi.org/10.1021/acs.analchem.9b03270 |
| locations[1].id | pmid:31441640 |
| locations[1].is_oa | False |
| locations[1].source.id | https://openalex.org/S4306525036 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | PubMed |
| locations[1].source.host_organization | https://openalex.org/I1299303238 |
| locations[1].source.host_organization_name | National Institutes of Health |
| locations[1].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[1].license | |
| locations[1].pdf_url | |
| locations[1].version | publishedVersion |
| locations[1].raw_type | |
| locations[1].license_id | |
| locations[1].is_accepted | True |
| locations[1].is_published | True |
| locations[1].raw_source_name | Analytical chemistry |
| locations[1].landing_page_url | https://pubmed.ncbi.nlm.nih.gov/31441640 |
| locations[2].id | pmh:oai:pubmedcentral.nih.gov:6751524 |
| locations[2].is_oa | True |
| locations[2].source.id | https://openalex.org/S2764455111 |
| locations[2].source.issn | |
| locations[2].source.type | repository |
| locations[2].source.is_oa | False |
| locations[2].source.issn_l | |
| locations[2].source.is_core | False |
| locations[2].source.is_in_doaj | False |
| locations[2].source.display_name | PubMed Central |
| locations[2].source.host_organization | https://openalex.org/I1299303238 |
| locations[2].source.host_organization_name | National Institutes of Health |
| locations[2].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[2].license | |
| locations[2].pdf_url | |
| locations[2].version | submittedVersion |
| locations[2].raw_type | Text |
| locations[2].license_id | |
| locations[2].is_accepted | False |
| locations[2].is_published | False |
| locations[2].raw_source_name | Anal Chem |
| locations[2].landing_page_url | https://www.ncbi.nlm.nih.gov/pmc/articles/6751524 |
| indexed_in | crossref, pubmed |
| authorships[0].author.id | https://openalex.org/A5073546882 |
| authorships[0].author.orcid | https://orcid.org/0000-0001-9776-7141 |
| authorships[0].author.display_name | Kai Schuhmann |
| authorships[0].countries | DE |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I4210159854 |
| authorships[0].affiliations[0].raw_affiliation_string | Max Planck Institute of Molecular Cell Biology and Genetics, Pfotenhauerstr. 108, 01307 Dresden, Germany |
| authorships[0].institutions[0].id | https://openalex.org/I4210159854 |
| authorships[0].institutions[0].ror | https://ror.org/05b8d3w18 |
| authorships[0].institutions[0].type | facility |
| authorships[0].institutions[0].lineage | https://openalex.org/I149899117, https://openalex.org/I4210159854 |
| authorships[0].institutions[0].country_code | DE |
| authorships[0].institutions[0].display_name | Max Planck Institute of Molecular Cell Biology and Genetics |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Kai Schuhmann |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Max Planck Institute of Molecular Cell Biology and Genetics, Pfotenhauerstr. 108, 01307 Dresden, Germany |
| authorships[1].author.id | https://openalex.org/A5015600934 |
| authorships[1].author.orcid | https://orcid.org/0000-0001-7159-924X |
| authorships[1].author.display_name | HongKee Moon |
| authorships[1].countries | DE |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I4210159854 |
| authorships[1].affiliations[0].raw_affiliation_string | Max Planck Institute of Molecular Cell Biology and Genetics, Pfotenhauerstr. 108, 01307 Dresden, Germany |
| authorships[1].institutions[0].id | https://openalex.org/I4210159854 |
| authorships[1].institutions[0].ror | https://ror.org/05b8d3w18 |
| authorships[1].institutions[0].type | facility |
| authorships[1].institutions[0].lineage | https://openalex.org/I149899117, https://openalex.org/I4210159854 |
| authorships[1].institutions[0].country_code | DE |
| authorships[1].institutions[0].display_name | Max Planck Institute of Molecular Cell Biology and Genetics |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | HongKee Moon |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | Max Planck Institute of Molecular Cell Biology and Genetics, Pfotenhauerstr. 108, 01307 Dresden, Germany |
| authorships[2].author.id | https://openalex.org/A5102092363 |
| authorships[2].author.orcid | |
| authorships[2].author.display_name | Henrik Thomas |
| authorships[2].countries | DE |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I4210159854 |
| authorships[2].affiliations[0].raw_affiliation_string | Max Planck Institute of Molecular Cell Biology and Genetics, Pfotenhauerstr. 108, 01307 Dresden, Germany |
| authorships[2].institutions[0].id | https://openalex.org/I4210159854 |
| authorships[2].institutions[0].ror | https://ror.org/05b8d3w18 |
| authorships[2].institutions[0].type | facility |
| authorships[2].institutions[0].lineage | https://openalex.org/I149899117, https://openalex.org/I4210159854 |
| authorships[2].institutions[0].country_code | DE |
| authorships[2].institutions[0].display_name | Max Planck Institute of Molecular Cell Biology and Genetics |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Henrik Thomas |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | Max Planck Institute of Molecular Cell Biology and Genetics, Pfotenhauerstr. 108, 01307 Dresden, Germany |
| authorships[3].author.id | https://openalex.org/A5108667414 |
| authorships[3].author.orcid | |
| authorships[3].author.display_name | Jacobo Miranda Ackerman |
| authorships[3].countries | DE |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I4210159854 |
| authorships[3].affiliations[0].raw_affiliation_string | Max Planck Institute of Molecular Cell Biology and Genetics, Pfotenhauerstr. 108, 01307 Dresden, Germany |
| authorships[3].institutions[0].id | https://openalex.org/I4210159854 |
| authorships[3].institutions[0].ror | https://ror.org/05b8d3w18 |
| authorships[3].institutions[0].type | facility |
| authorships[3].institutions[0].lineage | https://openalex.org/I149899117, https://openalex.org/I4210159854 |
| authorships[3].institutions[0].country_code | DE |
| authorships[3].institutions[0].display_name | Max Planck Institute of Molecular Cell Biology and Genetics |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Jacobo Miranda Ackerman |
| authorships[3].is_corresponding | False |
| authorships[3].raw_affiliation_strings | Max Planck Institute of Molecular Cell Biology and Genetics, Pfotenhauerstr. 108, 01307 Dresden, Germany |
| authorships[4].author.id | https://openalex.org/A5070110597 |
| authorships[4].author.orcid | https://orcid.org/0000-0002-6740-5980 |
| authorships[4].author.display_name | Michael Groessl |
| authorships[4].countries | CH |
| authorships[4].affiliations[0].institution_ids | https://openalex.org/I2801112126 |
| authorships[4].affiliations[0].raw_affiliation_string | Department of Nephrology and Hypertension, Inselspital, Bern University Hospital, Freiburgstr. 15, 3010 Bern, Switzerland |
| authorships[4].affiliations[1].institution_ids | https://openalex.org/I118564535 |
| authorships[4].affiliations[1].raw_affiliation_string | Department for BioMedical Research, University of Bern, Murtenstr. 35, 3010 Bern, Switzerland |
| authorships[4].institutions[0].id | https://openalex.org/I2801112126 |
| authorships[4].institutions[0].ror | https://ror.org/01q9sj412 |
| authorships[4].institutions[0].type | healthcare |
| authorships[4].institutions[0].lineage | https://openalex.org/I2801112126 |
| authorships[4].institutions[0].country_code | CH |
| authorships[4].institutions[0].display_name | University Hospital of Bern |
| authorships[4].institutions[1].id | https://openalex.org/I118564535 |
| authorships[4].institutions[1].ror | https://ror.org/02k7v4d05 |
| authorships[4].institutions[1].type | education |
| authorships[4].institutions[1].lineage | https://openalex.org/I118564535 |
| authorships[4].institutions[1].country_code | CH |
| authorships[4].institutions[1].display_name | University of Bern |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Michael Groessl |
| authorships[4].is_corresponding | False |
| authorships[4].raw_affiliation_strings | Department for BioMedical Research, University of Bern, Murtenstr. 35, 3010 Bern, Switzerland, Department of Nephrology and Hypertension, Inselspital, Bern University Hospital, Freiburgstr. 15, 3010 Bern, Switzerland |
| authorships[5].author.id | https://openalex.org/A5088421650 |
| authorships[5].author.orcid | |
| authorships[5].author.display_name | Nicolai Wagner |
| authorships[5].countries | DE |
| authorships[5].affiliations[0].institution_ids | https://openalex.org/I4210159854 |
| authorships[5].affiliations[0].raw_affiliation_string | Max Planck Institute of Molecular Cell Biology and Genetics, Pfotenhauerstr. 108, 01307 Dresden, Germany |
| authorships[5].institutions[0].id | https://openalex.org/I4210159854 |
| authorships[5].institutions[0].ror | https://ror.org/05b8d3w18 |
| authorships[5].institutions[0].type | facility |
| authorships[5].institutions[0].lineage | https://openalex.org/I149899117, https://openalex.org/I4210159854 |
| authorships[5].institutions[0].country_code | DE |
| authorships[5].institutions[0].display_name | Max Planck Institute of Molecular Cell Biology and Genetics |
| authorships[5].author_position | middle |
| authorships[5].raw_author_name | Nicolai Wagner |
| authorships[5].is_corresponding | False |
| authorships[5].raw_affiliation_strings | Max Planck Institute of Molecular Cell Biology and Genetics, Pfotenhauerstr. 108, 01307 Dresden, Germany |
| authorships[6].author.id | https://openalex.org/A5103429933 |
| authorships[6].author.orcid | |
| authorships[6].author.display_name | Markus Kellmann |
| authorships[6].countries | DE |
| authorships[6].affiliations[0].institution_ids | https://openalex.org/I4210153897 |
| authorships[6].affiliations[0].raw_affiliation_string | Thermo Fisher Scientific, Hanna-Kunath-Str. 11, 28199 Bremen, Germany |
| authorships[6].institutions[0].id | https://openalex.org/I4210153897 |
| authorships[6].institutions[0].ror | https://ror.org/04428zn91 |
| authorships[6].institutions[0].type | company |
| authorships[6].institutions[0].lineage | https://openalex.org/I4210142480, https://openalex.org/I4210153897 |
| authorships[6].institutions[0].country_code | DE |
| authorships[6].institutions[0].display_name | Thermo Fisher Scientific (Germany) |
| authorships[6].author_position | middle |
| authorships[6].raw_author_name | Markus Kellmann |
| authorships[6].is_corresponding | False |
| authorships[6].raw_affiliation_strings | Thermo Fisher Scientific, Hanna-Kunath-Str. 11, 28199 Bremen, Germany |
| authorships[7].author.id | https://openalex.org/A5015089975 |
| authorships[7].author.orcid | https://orcid.org/0000-0002-7134-3110 |
| authorships[7].author.display_name | Ian Henry |
| authorships[7].countries | DE |
| authorships[7].affiliations[0].institution_ids | https://openalex.org/I4210159854 |
| authorships[7].affiliations[0].raw_affiliation_string | Max Planck Institute of Molecular Cell Biology and Genetics, Pfotenhauerstr. 108, 01307 Dresden, Germany |
| authorships[7].institutions[0].id | https://openalex.org/I4210159854 |
| authorships[7].institutions[0].ror | https://ror.org/05b8d3w18 |
| authorships[7].institutions[0].type | facility |
| authorships[7].institutions[0].lineage | https://openalex.org/I149899117, https://openalex.org/I4210159854 |
| authorships[7].institutions[0].country_code | DE |
| authorships[7].institutions[0].display_name | Max Planck Institute of Molecular Cell Biology and Genetics |
| authorships[7].author_position | middle |
| authorships[7].raw_author_name | Ian Henry |
| authorships[7].is_corresponding | False |
| authorships[7].raw_affiliation_strings | Max Planck Institute of Molecular Cell Biology and Genetics, Pfotenhauerstr. 108, 01307 Dresden, Germany |
| authorships[8].author.id | https://openalex.org/A5091292168 |
| authorships[8].author.orcid | https://orcid.org/0000-0001-6329-3910 |
| authorships[8].author.display_name | André Nadler |
| authorships[8].countries | DE |
| authorships[8].affiliations[0].institution_ids | https://openalex.org/I4210159854 |
| authorships[8].affiliations[0].raw_affiliation_string | Max Planck Institute of Molecular Cell Biology and Genetics, Pfotenhauerstr. 108, 01307 Dresden, Germany |
| authorships[8].institutions[0].id | https://openalex.org/I4210159854 |
| authorships[8].institutions[0].ror | https://ror.org/05b8d3w18 |
| authorships[8].institutions[0].type | facility |
| authorships[8].institutions[0].lineage | https://openalex.org/I149899117, https://openalex.org/I4210159854 |
| authorships[8].institutions[0].country_code | DE |
| authorships[8].institutions[0].display_name | Max Planck Institute of Molecular Cell Biology and Genetics |
| authorships[8].author_position | middle |
| authorships[8].raw_author_name | André Nadler |
| authorships[8].is_corresponding | False |
| authorships[8].raw_affiliation_strings | Max Planck Institute of Molecular Cell Biology and Genetics, Pfotenhauerstr. 108, 01307 Dresden, Germany |
| authorships[9].author.id | https://openalex.org/A5000266633 |
| authorships[9].author.orcid | https://orcid.org/0000-0002-5079-1109 |
| authorships[9].author.display_name | Andrej Shevchenko |
| authorships[9].countries | DE |
| authorships[9].affiliations[0].institution_ids | https://openalex.org/I4210159854 |
| authorships[9].affiliations[0].raw_affiliation_string | Max Planck Institute of Molecular Cell Biology and Genetics, Pfotenhauerstr. 108, 01307 Dresden, Germany |
| authorships[9].institutions[0].id | https://openalex.org/I4210159854 |
| authorships[9].institutions[0].ror | https://ror.org/05b8d3w18 |
| authorships[9].institutions[0].type | facility |
| authorships[9].institutions[0].lineage | https://openalex.org/I149899117, https://openalex.org/I4210159854 |
| authorships[9].institutions[0].country_code | DE |
| authorships[9].institutions[0].display_name | Max Planck Institute of Molecular Cell Biology and Genetics |
| authorships[9].author_position | last |
| authorships[9].raw_author_name | Andrej Shevchenko |
| authorships[9].is_corresponding | True |
| authorships[9].raw_affiliation_strings | Max Planck Institute of Molecular Cell Biology and Genetics, Pfotenhauerstr. 108, 01307 Dresden, Germany |
| has_content.pdf | False |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://doi.org/10.1021/acs.analchem.9b03270 |
| open_access.oa_status | hybrid |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Quantitative Fragmentation Model for Bottom-Up Shotgun Lipidomics |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T10836 |
| primary_topic.field.id | https://openalex.org/fields/13 |
| primary_topic.field.display_name | Biochemistry, Genetics and Molecular Biology |
| primary_topic.score | 1.0 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1312 |
| primary_topic.subfield.display_name | Molecular Biology |
| primary_topic.display_name | Metabolomics and Mass Spectrometry Studies |
| related_works | https://openalex.org/W2613145611, https://openalex.org/W1722882657, https://openalex.org/W1973617509, https://openalex.org/W1554365828, https://openalex.org/W2107948216, https://openalex.org/W2122184160, https://openalex.org/W2414929044, https://openalex.org/W2099724648, https://openalex.org/W4296545149, https://openalex.org/W2041235251 |
| cited_by_count | 25 |
| counts_by_year[0].year | 2025 |
| counts_by_year[0].cited_by_count | 3 |
| counts_by_year[1].year | 2024 |
| counts_by_year[1].cited_by_count | 1 |
| counts_by_year[2].year | 2023 |
| counts_by_year[2].cited_by_count | 4 |
| counts_by_year[3].year | 2022 |
| counts_by_year[3].cited_by_count | 10 |
| counts_by_year[4].year | 2021 |
| counts_by_year[4].cited_by_count | 3 |
| counts_by_year[5].year | 2020 |
| counts_by_year[5].cited_by_count | 4 |
| locations_count | 3 |
| best_oa_location.id | doi:10.1021/acs.analchem.9b03270 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S191963794 |
| best_oa_location.source.issn | 0003-2700, 1520-6882 |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | False |
| best_oa_location.source.issn_l | 0003-2700 |
| best_oa_location.source.is_core | True |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | Analytical Chemistry |
| best_oa_location.source.host_organization | https://openalex.org/P4310320006 |
| best_oa_location.source.host_organization_name | American Chemical Society |
| best_oa_location.source.host_organization_lineage | https://openalex.org/P4310320006 |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | journal-article |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | Analytical Chemistry |
| best_oa_location.landing_page_url | https://doi.org/10.1021/acs.analchem.9b03270 |
| primary_location.id | doi:10.1021/acs.analchem.9b03270 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S191963794 |
| primary_location.source.issn | 0003-2700, 1520-6882 |
| primary_location.source.type | journal |
| primary_location.source.is_oa | False |
| primary_location.source.issn_l | 0003-2700 |
| primary_location.source.is_core | True |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | Analytical Chemistry |
| primary_location.source.host_organization | https://openalex.org/P4310320006 |
| primary_location.source.host_organization_name | American Chemical Society |
| primary_location.source.host_organization_lineage | https://openalex.org/P4310320006 |
| primary_location.license | cc-by |
| primary_location.pdf_url | |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | Analytical Chemistry |
| primary_location.landing_page_url | https://doi.org/10.1021/acs.analchem.9b03270 |
| publication_date | 2019-08-23 |
| publication_year | 2019 |
| referenced_works | https://openalex.org/W2896503758, https://openalex.org/W2237957677, https://openalex.org/W2522015218, https://openalex.org/W2912375479, https://openalex.org/W2220707375, https://openalex.org/W2769815973, https://openalex.org/W2420961370, https://openalex.org/W2145029774, https://openalex.org/W1984954439, https://openalex.org/W1569454591, https://openalex.org/W2043775257, https://openalex.org/W2335725835, https://openalex.org/W2087431894, https://openalex.org/W2147621341, https://openalex.org/W2017109633, https://openalex.org/W1980303483, https://openalex.org/W2081380820, https://openalex.org/W2041235251, https://openalex.org/W2764166773, https://openalex.org/W2886084326, https://openalex.org/W2045950965, https://openalex.org/W1840351579, https://openalex.org/W2620867558, https://openalex.org/W1908250124, https://openalex.org/W2054391202, https://openalex.org/W2158138282, https://openalex.org/W1514397955, https://openalex.org/W2783500810, https://openalex.org/W2908586978, https://openalex.org/W2570411824, https://openalex.org/W2761019098, https://openalex.org/W2935456378 |
| referenced_works_count | 32 |
| abstract_inverted_index.a | 67 |
| abstract_inverted_index.We | 65 |
| abstract_inverted_index.at | 96 |
| abstract_inverted_index.by | 36 |
| abstract_inverted_index.in | 12, 74 |
| abstract_inverted_index.of | 16, 22, 30, 40, 83, 101, 109, 120, 124 |
| abstract_inverted_index.on | 5 |
| abstract_inverted_index.to | 56 |
| abstract_inverted_index.The | 112 |
| abstract_inverted_index.and | 18, 69, 105, 107, 129 |
| abstract_inverted_index.are | 24, 51 |
| abstract_inverted_index.not | 54 |
| abstract_inverted_index.the | 57, 75, 81, 97, 102 |
| abstract_inverted_index.yet | 53 |
| abstract_inverted_index.acid | 33, 90 |
| abstract_inverted_index.from | 88 |
| abstract_inverted_index.mass | 14 |
| abstract_inverted_index.that | 79 |
| abstract_inverted_index.acids | 49 |
| abstract_inverted_index.anion | 85 |
| abstract_inverted_index.could | 60 |
| abstract_inverted_index.fatty | 32, 48, 89 |
| abstract_inverted_index.model | 72 |
| abstract_inverted_index.their | 31, 41, 63 |
| abstract_inverted_index.using | 27 |
| abstract_inverted_index.whose | 47 |
| abstract_inverted_index.bonds. | 111 |
| abstract_inverted_index.chain, | 104 |
| abstract_inverted_index.double | 110 |
| abstract_inverted_index.having | 92 |
| abstract_inverted_index.length | 100 |
| abstract_inverted_index.lipids | 59 |
| abstract_inverted_index.number | 106 |
| abstract_inverted_index.relies | 4 |
| abstract_inverted_index.sn-1/2 | 94 |
| abstract_inverted_index.tandem | 13 |
| abstract_inverted_index.target | 58 |
| abstract_inverted_index.(molar) | 118 |
| abstract_inverted_index.anions. | 43 |
| abstract_inverted_index.enables | 115 |
| abstract_inverted_index.energy, | 128 |
| abstract_inverted_index.generic | 68 |
| abstract_inverted_index.shotgun | 2 |
| abstract_inverted_index.species | 21, 122 |
| abstract_inverted_index.spectra | 15 |
| abstract_inverted_index.However, | 44 |
| abstract_inverted_index.LipidXte | 77 |
| abstract_inverted_index.absolute | 117 |
| abstract_inverted_index.analytes | 17 |
| abstract_inverted_index.employed | 130 |
| abstract_inverted_index.glycerol | 98 |
| abstract_inverted_index.internal | 131 |
| abstract_inverted_index.location | 108 |
| abstract_inverted_index.moieties | 34, 50, 91 |
| abstract_inverted_index.portable | 70 |
| abstract_inverted_index.produced | 35 |
| abstract_inverted_index.severely | 61 |
| abstract_inverted_index.similar, | 52 |
| abstract_inverted_index.software | 78 |
| abstract_inverted_index.unbiased | 116 |
| abstract_inverted_index.Molecular | 20 |
| abstract_inverted_index.backbone, | 99 |
| abstract_inverted_index.bottom-up | 1 |
| abstract_inverted_index.collision | 127 |
| abstract_inverted_index.developed | 66 |
| abstract_inverted_index.different | 93 |
| abstract_inverted_index.employing | 45 |
| abstract_inverted_index.fragments | 9, 29, 86 |
| abstract_inverted_index.molecular | 6, 42 |
| abstract_inverted_index.positions | 95 |
| abstract_inverted_index.settings, | 126 |
| abstract_inverted_index.standards | 46 |
| abstract_inverted_index.typically | 25 |
| abstract_inverted_index.abundances | 82 |
| abstract_inverted_index.adjustment | 114 |
| abstract_inverted_index.compromise | 62 |
| abstract_inverted_index.detectable | 11 |
| abstract_inverted_index.harmonizes | 80 |
| abstract_inverted_index.identical, | 55 |
| abstract_inverted_index.instrument | 125 |
| abstract_inverted_index.lipidomics | 3 |
| abstract_inverted_index.quantified | 26 |
| abstract_inverted_index.standards. | 19, 132 |
| abstract_inverted_index."signature" | 8 |
| abstract_inverted_index.carboxylate | 28, 84 |
| abstract_inverted_index.collisional | 38 |
| abstract_inverted_index.hydrocarbon | 103 |
| abstract_inverted_index.implemented | 73 |
| abstract_inverted_index.independent | 123 |
| abstract_inverted_index.open-source | 76 |
| abstract_inverted_index.originating | 87 |
| abstract_inverted_index.Quantitative | 0 |
| abstract_inverted_index.consistently | 10 |
| abstract_inverted_index.dissociation | 39 |
| abstract_inverted_index.fragmentation | 71 |
| abstract_inverted_index.higher-energy | 37 |
| abstract_inverted_index.quantification | 119 |
| abstract_inverted_index.postacquisition | 113 |
| abstract_inverted_index.quantification. | 64 |
| abstract_inverted_index.species-specific | 7 |
| abstract_inverted_index.glycerophospholipid | 121 |
| abstract_inverted_index.glycerophospholipids | 23 |
| cited_by_percentile_year.max | 99 |
| cited_by_percentile_year.min | 90 |
| corresponding_author_ids | https://openalex.org/A5000266633 |
| countries_distinct_count | 2 |
| institutions_distinct_count | 10 |
| corresponding_institution_ids | https://openalex.org/I4210159854 |
| citation_normalized_percentile.value | 0.82780157 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |