Quantum Mechanics/Molecular Mechanics Simulations Distinguish Insulin-Regulated Aminopeptidase Substrate (Oxytocin) and Inhibitor (Angiotensin IV) and Reveal Determinants of Activity and Inhibition Article Swipe
 YOU?
·
· 2025
· Open Access
·
· DOI: https://doi.org/10.1021/acs.jcim.5c00869
YOU?
·
· 2025
· Open Access
·
· DOI: https://doi.org/10.1021/acs.jcim.5c00869
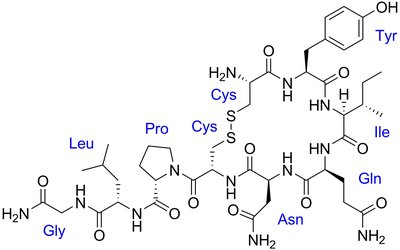
Insulin-regulated aminopeptidase (IRAP) is a zinc-dependent metalloenzyme identified as a novel target for combating diabetes-induced diseases due to its crucial role in glucose metabolism and insulin sensitivity regulation. IRAP's catalytic domain catalyzes the N-terminal peptide bond hydrolysis of natural substrate oxytocin, a neuroactive peptide linked to improved cognition and other elemental brain functions. Angiotensin IV and similar peptides are recognized as cognitive enhancers due to their ability to competitively inhibit IRAP's proteolytic activity, thereby mitigating natural neuropeptide degradation. Despite a very similar binding complex between the substrate and the inhibitor with IRAP, particularly around the scissile bond, it is unclear why the enzyme metabolizes oxytocin but does not efficiently degrade angiotensin IV. We employed enhanced sampling quantum mechanics/molecular mechanics (QM/MM) molecular dynamics simulations and higher-level QM/MM calculations to explore the reaction of these two peptides in IRAP. The calculated energy barrier for oxytocin cleavage was in very good agreement with the experimental data. A significantly higher energy barrier for the formation of the oxyanion tetrahedral intermediate (TI) and a higher overall barrier for the peptide cleavage were observed for the reaction with angiotensin IV. Comprehensive electronic structure analysis utilizing NBO and NCI methods unveiled the molecular basis for different reactivity, a stabilizing interaction between the sigma hole of the N-terminus disulfide bond and the hybridizing lone pair of the scissile peptide nitrogen in oxytocin. The interplay between a weak noncovalent spodium bond and strong bidentate coordination of the catalytic Zn2+ by angiotensin IV caused a larger deviation of the valine C-Cα-Cβ angle from ideal tetrahedral geometry, consequently destabilizing the TI. These results underscore the critical importance of analyzing the dynamics, interactions, and electronic properties of reaction intermediates and transition states in enzymatic processes. Our findings have significant implications for the rational design and development of IRAP inhibitors as potential therapeutic agents for memory disorders, neurodegenerative diseases, and diabetes.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.1021/acs.jcim.5c00869
- OA Status
- hybrid
- Cited By
- 1
- References
- 91
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4411218095
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4411218095Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1021/acs.jcim.5c00869Digital Object Identifier
- Title
-
Quantum Mechanics/Molecular Mechanics Simulations Distinguish Insulin-Regulated Aminopeptidase Substrate (Oxytocin) and Inhibitor (Angiotensin IV) and Reveal Determinants of Activity and InhibitionWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2025Year of publication
- Publication date
-
2025-06-11Full publication date if available
- Authors
-
Marko Hanževački, Rebecca M. Twidale, Eric J. M. Lang, W. Gerrard, David W. Wright, Vid Stojevic, Adrian J. MulhollandList of authors in order
- Landing page
-
https://doi.org/10.1021/acs.jcim.5c00869Publisher landing page
- Open access
-
YesWhether a free full text is available
- OA status
-
hybridOpen access status per OpenAlex
- OA URL
-
https://doi.org/10.1021/acs.jcim.5c00869Direct OA link when available
- Concepts
-
Molecular mechanics, Oxytocin, Aminopeptidase, Insulin, Substrate (aquarium), Molecular dynamics, Internal medicine, Chemistry, Physics, Biochemistry, Medicine, Biology, Computational chemistry, Amino acid, Ecology, LeucineTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
1Total citation count in OpenAlex
- Citations by year (recent)
-
2025: 1Per-year citation counts (last 5 years)
- References (count)
-
91Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4411218095 |
|---|---|
| doi | https://doi.org/10.1021/acs.jcim.5c00869 |
| ids.doi | https://doi.org/10.1021/acs.jcim.5c00869 |
| ids.pmid | https://pubmed.ncbi.nlm.nih.gov/40495656 |
| ids.openalex | https://openalex.org/W4411218095 |
| fwci | 7.13387101 |
| mesh[0].qualifier_ui | |
| mesh[0].descriptor_ui | D056004 |
| mesh[0].is_major_topic | True |
| mesh[0].qualifier_name | |
| mesh[0].descriptor_name | Molecular Dynamics Simulation |
| mesh[1].qualifier_ui | Q000037 |
| mesh[1].descriptor_ui | D010122 |
| mesh[1].is_major_topic | True |
| mesh[1].qualifier_name | antagonists & inhibitors |
| mesh[1].descriptor_name | Cystinyl Aminopeptidase |
| mesh[2].qualifier_ui | Q000378 |
| mesh[2].descriptor_ui | D010122 |
| mesh[2].is_major_topic | True |
| mesh[2].qualifier_name | metabolism |
| mesh[2].descriptor_name | Cystinyl Aminopeptidase |
| mesh[3].qualifier_ui | Q000737 |
| mesh[3].descriptor_ui | D010122 |
| mesh[3].is_major_topic | True |
| mesh[3].qualifier_name | chemistry |
| mesh[3].descriptor_name | Cystinyl Aminopeptidase |
| mesh[4].qualifier_ui | Q000378 |
| mesh[4].descriptor_ui | D010121 |
| mesh[4].is_major_topic | True |
| mesh[4].qualifier_name | metabolism |
| mesh[4].descriptor_name | Oxytocin |
| mesh[5].qualifier_ui | Q000737 |
| mesh[5].descriptor_ui | D010121 |
| mesh[5].is_major_topic | True |
| mesh[5].qualifier_name | chemistry |
| mesh[5].descriptor_name | Oxytocin |
| mesh[6].qualifier_ui | |
| mesh[6].descriptor_ui | D011789 |
| mesh[6].is_major_topic | True |
| mesh[6].qualifier_name | |
| mesh[6].descriptor_name | Quantum Theory |
| mesh[7].qualifier_ui | |
| mesh[7].descriptor_ui | D013379 |
| mesh[7].is_major_topic | False |
| mesh[7].qualifier_name | |
| mesh[7].descriptor_name | Substrate Specificity |
| mesh[8].qualifier_ui | Q000031 |
| mesh[8].descriptor_ui | D000804 |
| mesh[8].is_major_topic | True |
| mesh[8].qualifier_name | analogs & derivatives |
| mesh[8].descriptor_name | Angiotensin II |
| mesh[9].qualifier_ui | Q000737 |
| mesh[9].descriptor_ui | D000804 |
| mesh[9].is_major_topic | True |
| mesh[9].qualifier_name | chemistry |
| mesh[9].descriptor_name | Angiotensin II |
| mesh[10].qualifier_ui | Q000378 |
| mesh[10].descriptor_ui | D000804 |
| mesh[10].is_major_topic | True |
| mesh[10].qualifier_name | metabolism |
| mesh[10].descriptor_name | Angiotensin II |
| mesh[11].qualifier_ui | Q000494 |
| mesh[11].descriptor_ui | D000804 |
| mesh[11].is_major_topic | True |
| mesh[11].qualifier_name | pharmacology |
| mesh[11].descriptor_name | Angiotensin II |
| mesh[12].qualifier_ui | Q000494 |
| mesh[12].descriptor_ui | D004791 |
| mesh[12].is_major_topic | True |
| mesh[12].qualifier_name | pharmacology |
| mesh[12].descriptor_name | Enzyme Inhibitors |
| mesh[13].qualifier_ui | Q000737 |
| mesh[13].descriptor_ui | D004791 |
| mesh[13].is_major_topic | True |
| mesh[13].qualifier_name | chemistry |
| mesh[13].descriptor_name | Enzyme Inhibitors |
| mesh[14].qualifier_ui | Q000494 |
| mesh[14].descriptor_ui | D011480 |
| mesh[14].is_major_topic | True |
| mesh[14].qualifier_name | pharmacology |
| mesh[14].descriptor_name | Protease Inhibitors |
| mesh[15].qualifier_ui | Q000737 |
| mesh[15].descriptor_ui | D011480 |
| mesh[15].is_major_topic | True |
| mesh[15].qualifier_name | chemistry |
| mesh[15].descriptor_name | Protease Inhibitors |
| mesh[16].qualifier_ui | |
| mesh[16].descriptor_ui | D000098222 |
| mesh[16].is_major_topic | False |
| mesh[16].qualifier_name | |
| mesh[16].descriptor_name | Quantum Mechanics |
| mesh[17].qualifier_ui | |
| mesh[17].descriptor_ui | D056004 |
| mesh[17].is_major_topic | True |
| mesh[17].qualifier_name | |
| mesh[17].descriptor_name | Molecular Dynamics Simulation |
| mesh[18].qualifier_ui | Q000037 |
| mesh[18].descriptor_ui | D010122 |
| mesh[18].is_major_topic | True |
| mesh[18].qualifier_name | antagonists & inhibitors |
| mesh[18].descriptor_name | Cystinyl Aminopeptidase |
| mesh[19].qualifier_ui | Q000378 |
| mesh[19].descriptor_ui | D010122 |
| mesh[19].is_major_topic | True |
| mesh[19].qualifier_name | metabolism |
| mesh[19].descriptor_name | Cystinyl Aminopeptidase |
| mesh[20].qualifier_ui | Q000737 |
| mesh[20].descriptor_ui | D010122 |
| mesh[20].is_major_topic | True |
| mesh[20].qualifier_name | chemistry |
| mesh[20].descriptor_name | Cystinyl Aminopeptidase |
| mesh[21].qualifier_ui | Q000378 |
| mesh[21].descriptor_ui | D010121 |
| mesh[21].is_major_topic | True |
| mesh[21].qualifier_name | metabolism |
| mesh[21].descriptor_name | Oxytocin |
| mesh[22].qualifier_ui | Q000737 |
| mesh[22].descriptor_ui | D010121 |
| mesh[22].is_major_topic | True |
| mesh[22].qualifier_name | chemistry |
| mesh[22].descriptor_name | Oxytocin |
| mesh[23].qualifier_ui | |
| mesh[23].descriptor_ui | D011789 |
| mesh[23].is_major_topic | True |
| mesh[23].qualifier_name | |
| mesh[23].descriptor_name | Quantum Theory |
| mesh[24].qualifier_ui | |
| mesh[24].descriptor_ui | D013379 |
| mesh[24].is_major_topic | False |
| mesh[24].qualifier_name | |
| mesh[24].descriptor_name | Substrate Specificity |
| mesh[25].qualifier_ui | Q000031 |
| mesh[25].descriptor_ui | D000804 |
| mesh[25].is_major_topic | True |
| mesh[25].qualifier_name | analogs & derivatives |
| mesh[25].descriptor_name | Angiotensin II |
| mesh[26].qualifier_ui | Q000737 |
| mesh[26].descriptor_ui | D000804 |
| mesh[26].is_major_topic | True |
| mesh[26].qualifier_name | chemistry |
| mesh[26].descriptor_name | Angiotensin II |
| mesh[27].qualifier_ui | Q000378 |
| mesh[27].descriptor_ui | D000804 |
| mesh[27].is_major_topic | True |
| mesh[27].qualifier_name | metabolism |
| mesh[27].descriptor_name | Angiotensin II |
| mesh[28].qualifier_ui | Q000494 |
| mesh[28].descriptor_ui | D000804 |
| mesh[28].is_major_topic | True |
| mesh[28].qualifier_name | pharmacology |
| mesh[28].descriptor_name | Angiotensin II |
| mesh[29].qualifier_ui | Q000494 |
| mesh[29].descriptor_ui | D004791 |
| mesh[29].is_major_topic | True |
| mesh[29].qualifier_name | pharmacology |
| mesh[29].descriptor_name | Enzyme Inhibitors |
| mesh[30].qualifier_ui | Q000737 |
| mesh[30].descriptor_ui | D004791 |
| mesh[30].is_major_topic | True |
| mesh[30].qualifier_name | chemistry |
| mesh[30].descriptor_name | Enzyme Inhibitors |
| mesh[31].qualifier_ui | Q000494 |
| mesh[31].descriptor_ui | D011480 |
| mesh[31].is_major_topic | True |
| mesh[31].qualifier_name | pharmacology |
| mesh[31].descriptor_name | Protease Inhibitors |
| mesh[32].qualifier_ui | Q000737 |
| mesh[32].descriptor_ui | D011480 |
| mesh[32].is_major_topic | True |
| mesh[32].qualifier_name | chemistry |
| mesh[32].descriptor_name | Protease Inhibitors |
| mesh[33].qualifier_ui | |
| mesh[33].descriptor_ui | D000098222 |
| mesh[33].is_major_topic | False |
| mesh[33].qualifier_name | |
| mesh[33].descriptor_name | Quantum Mechanics |
| mesh[34].qualifier_ui | |
| mesh[34].descriptor_ui | D056004 |
| mesh[34].is_major_topic | True |
| mesh[34].qualifier_name | |
| mesh[34].descriptor_name | Molecular Dynamics Simulation |
| mesh[35].qualifier_ui | Q000037 |
| mesh[35].descriptor_ui | D010122 |
| mesh[35].is_major_topic | True |
| mesh[35].qualifier_name | antagonists & inhibitors |
| mesh[35].descriptor_name | Cystinyl Aminopeptidase |
| mesh[36].qualifier_ui | Q000378 |
| mesh[36].descriptor_ui | D010122 |
| mesh[36].is_major_topic | True |
| mesh[36].qualifier_name | metabolism |
| mesh[36].descriptor_name | Cystinyl Aminopeptidase |
| mesh[37].qualifier_ui | Q000737 |
| mesh[37].descriptor_ui | D010122 |
| mesh[37].is_major_topic | True |
| mesh[37].qualifier_name | chemistry |
| mesh[37].descriptor_name | Cystinyl Aminopeptidase |
| mesh[38].qualifier_ui | Q000378 |
| mesh[38].descriptor_ui | D010121 |
| mesh[38].is_major_topic | True |
| mesh[38].qualifier_name | metabolism |
| mesh[38].descriptor_name | Oxytocin |
| mesh[39].qualifier_ui | Q000737 |
| mesh[39].descriptor_ui | D010121 |
| mesh[39].is_major_topic | True |
| mesh[39].qualifier_name | chemistry |
| mesh[39].descriptor_name | Oxytocin |
| mesh[40].qualifier_ui | |
| mesh[40].descriptor_ui | D011789 |
| mesh[40].is_major_topic | True |
| mesh[40].qualifier_name | |
| mesh[40].descriptor_name | Quantum Theory |
| mesh[41].qualifier_ui | |
| mesh[41].descriptor_ui | D013379 |
| mesh[41].is_major_topic | False |
| mesh[41].qualifier_name | |
| mesh[41].descriptor_name | Substrate Specificity |
| mesh[42].qualifier_ui | Q000031 |
| mesh[42].descriptor_ui | D000804 |
| mesh[42].is_major_topic | True |
| mesh[42].qualifier_name | analogs & derivatives |
| mesh[42].descriptor_name | Angiotensin II |
| mesh[43].qualifier_ui | Q000737 |
| mesh[43].descriptor_ui | D000804 |
| mesh[43].is_major_topic | True |
| mesh[43].qualifier_name | chemistry |
| mesh[43].descriptor_name | Angiotensin II |
| mesh[44].qualifier_ui | Q000378 |
| mesh[44].descriptor_ui | D000804 |
| mesh[44].is_major_topic | True |
| mesh[44].qualifier_name | metabolism |
| mesh[44].descriptor_name | Angiotensin II |
| mesh[45].qualifier_ui | Q000494 |
| mesh[45].descriptor_ui | D000804 |
| mesh[45].is_major_topic | True |
| mesh[45].qualifier_name | pharmacology |
| mesh[45].descriptor_name | Angiotensin II |
| mesh[46].qualifier_ui | Q000494 |
| mesh[46].descriptor_ui | D004791 |
| mesh[46].is_major_topic | True |
| mesh[46].qualifier_name | pharmacology |
| mesh[46].descriptor_name | Enzyme Inhibitors |
| mesh[47].qualifier_ui | Q000737 |
| mesh[47].descriptor_ui | D004791 |
| mesh[47].is_major_topic | True |
| mesh[47].qualifier_name | chemistry |
| mesh[47].descriptor_name | Enzyme Inhibitors |
| mesh[48].qualifier_ui | Q000494 |
| mesh[48].descriptor_ui | D011480 |
| mesh[48].is_major_topic | True |
| mesh[48].qualifier_name | pharmacology |
| mesh[48].descriptor_name | Protease Inhibitors |
| mesh[49].qualifier_ui | Q000737 |
| mesh[49].descriptor_ui | D011480 |
| mesh[49].is_major_topic | True |
| mesh[49].qualifier_name | chemistry |
| mesh[49].descriptor_name | Protease Inhibitors |
| type | article |
| title | Quantum Mechanics/Molecular Mechanics Simulations Distinguish Insulin-Regulated Aminopeptidase Substrate (Oxytocin) and Inhibitor (Angiotensin IV) and Reveal Determinants of Activity and Inhibition |
| awards[0].id | https://openalex.org/G1198808071 |
| awards[0].funder_id | https://openalex.org/F4320334627 |
| awards[0].display_name | |
| awards[0].funder_award_id | EP/L015722/1 |
| awards[0].funder_display_name | Engineering and Physical Sciences Research Council |
| awards[1].id | https://openalex.org/G2564953149 |
| awards[1].funder_id | https://openalex.org/F4320334627 |
| awards[1].display_name | |
| awards[1].funder_award_id | EP/W013738/1 |
| awards[1].funder_display_name | Engineering and Physical Sciences Research Council |
| awards[2].id | https://openalex.org/G4817720962 |
| awards[2].funder_id | https://openalex.org/F4320338335 |
| awards[2].display_name | Predictive computational models for Enzyme Dynamics, Antimicrobial resistance, Catalysis and Thermoadaptation for Evolution and Design |
| awards[2].funder_award_id | 101021207 |
| awards[2].funder_display_name | H2020 European Research Council |
| awards[3].id | https://openalex.org/G1103163915 |
| awards[3].funder_id | https://openalex.org/F4320334629 |
| awards[3].display_name | |
| awards[3].funder_award_id | BB/L01386X1 |
| awards[3].funder_display_name | Biotechnology and Biological Sciences Research Council |
| biblio.issue | 12 |
| biblio.volume | 65 |
| biblio.last_page | 6272 |
| biblio.first_page | 6261 |
| topics[0].id | https://openalex.org/T11541 |
| topics[0].field.id | https://openalex.org/fields/32 |
| topics[0].field.display_name | Psychology |
| topics[0].score | 0.9987999796867371 |
| topics[0].domain.id | https://openalex.org/domains/2 |
| topics[0].domain.display_name | Social Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/3207 |
| topics[0].subfield.display_name | Social Psychology |
| topics[0].display_name | Neuroendocrine regulation and behavior |
| topics[1].id | https://openalex.org/T10814 |
| topics[1].field.id | https://openalex.org/fields/28 |
| topics[1].field.display_name | Neuroscience |
| topics[1].score | 0.9936000108718872 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/2804 |
| topics[1].subfield.display_name | Cellular and Molecular Neuroscience |
| topics[1].display_name | Neuropeptides and Animal Physiology |
| topics[2].id | https://openalex.org/T11178 |
| topics[2].field.id | https://openalex.org/fields/13 |
| topics[2].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[2].score | 0.9868999719619751 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1312 |
| topics[2].subfield.display_name | Molecular Biology |
| topics[2].display_name | Receptor Mechanisms and Signaling |
| funders[0].id | https://openalex.org/F4320334627 |
| funders[0].ror | https://ror.org/0439y7842 |
| funders[0].display_name | Engineering and Physical Sciences Research Council |
| funders[1].id | https://openalex.org/F4320334629 |
| funders[1].ror | https://ror.org/00cwqg982 |
| funders[1].display_name | Biotechnology and Biological Sciences Research Council |
| funders[2].id | https://openalex.org/F4320338335 |
| funders[2].ror | https://ror.org/0472cxd90 |
| funders[2].display_name | H2020 European Research Council |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C25057246 |
| concepts[0].level | 3 |
| concepts[0].score | 0.9217821359634399 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q1344441 |
| concepts[0].display_name | Molecular mechanics |
| concepts[1].id | https://openalex.org/C2776176026 |
| concepts[1].level | 2 |
| concepts[1].score | 0.6361129879951477 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q169960 |
| concepts[1].display_name | Oxytocin |
| concepts[2].id | https://openalex.org/C2780985610 |
| concepts[2].level | 4 |
| concepts[2].score | 0.5564090013504028 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q419527 |
| concepts[2].display_name | Aminopeptidase |
| concepts[3].id | https://openalex.org/C2779306644 |
| concepts[3].level | 2 |
| concepts[3].score | 0.4725610613822937 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q2002370 |
| concepts[3].display_name | Insulin |
| concepts[4].id | https://openalex.org/C2777289219 |
| concepts[4].level | 2 |
| concepts[4].score | 0.4420173466205597 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q7632154 |
| concepts[4].display_name | Substrate (aquarium) |
| concepts[5].id | https://openalex.org/C59593255 |
| concepts[5].level | 2 |
| concepts[5].score | 0.351955771446228 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q901663 |
| concepts[5].display_name | Molecular dynamics |
| concepts[6].id | https://openalex.org/C126322002 |
| concepts[6].level | 1 |
| concepts[6].score | 0.34475600719451904 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q11180 |
| concepts[6].display_name | Internal medicine |
| concepts[7].id | https://openalex.org/C185592680 |
| concepts[7].level | 0 |
| concepts[7].score | 0.34338194131851196 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q2329 |
| concepts[7].display_name | Chemistry |
| concepts[8].id | https://openalex.org/C121332964 |
| concepts[8].level | 0 |
| concepts[8].score | 0.3306429982185364 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q413 |
| concepts[8].display_name | Physics |
| concepts[9].id | https://openalex.org/C55493867 |
| concepts[9].level | 1 |
| concepts[9].score | 0.2826350927352905 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q7094 |
| concepts[9].display_name | Biochemistry |
| concepts[10].id | https://openalex.org/C71924100 |
| concepts[10].level | 0 |
| concepts[10].score | 0.2766672968864441 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q11190 |
| concepts[10].display_name | Medicine |
| concepts[11].id | https://openalex.org/C86803240 |
| concepts[11].level | 0 |
| concepts[11].score | 0.2450850009918213 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[11].display_name | Biology |
| concepts[12].id | https://openalex.org/C147597530 |
| concepts[12].level | 1 |
| concepts[12].score | 0.20014700293540955 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q369472 |
| concepts[12].display_name | Computational chemistry |
| concepts[13].id | https://openalex.org/C515207424 |
| concepts[13].level | 2 |
| concepts[13].score | 0.11498028039932251 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q8066 |
| concepts[13].display_name | Amino acid |
| concepts[14].id | https://openalex.org/C18903297 |
| concepts[14].level | 1 |
| concepts[14].score | 0.09380388259887695 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q7150 |
| concepts[14].display_name | Ecology |
| concepts[15].id | https://openalex.org/C2776580952 |
| concepts[15].level | 3 |
| concepts[15].score | 0.0 |
| concepts[15].wikidata | https://www.wikidata.org/wiki/Q483745 |
| concepts[15].display_name | Leucine |
| keywords[0].id | https://openalex.org/keywords/molecular-mechanics |
| keywords[0].score | 0.9217821359634399 |
| keywords[0].display_name | Molecular mechanics |
| keywords[1].id | https://openalex.org/keywords/oxytocin |
| keywords[1].score | 0.6361129879951477 |
| keywords[1].display_name | Oxytocin |
| keywords[2].id | https://openalex.org/keywords/aminopeptidase |
| keywords[2].score | 0.5564090013504028 |
| keywords[2].display_name | Aminopeptidase |
| keywords[3].id | https://openalex.org/keywords/insulin |
| keywords[3].score | 0.4725610613822937 |
| keywords[3].display_name | Insulin |
| keywords[4].id | https://openalex.org/keywords/substrate |
| keywords[4].score | 0.4420173466205597 |
| keywords[4].display_name | Substrate (aquarium) |
| keywords[5].id | https://openalex.org/keywords/molecular-dynamics |
| keywords[5].score | 0.351955771446228 |
| keywords[5].display_name | Molecular dynamics |
| keywords[6].id | https://openalex.org/keywords/internal-medicine |
| keywords[6].score | 0.34475600719451904 |
| keywords[6].display_name | Internal medicine |
| keywords[7].id | https://openalex.org/keywords/chemistry |
| keywords[7].score | 0.34338194131851196 |
| keywords[7].display_name | Chemistry |
| keywords[8].id | https://openalex.org/keywords/physics |
| keywords[8].score | 0.3306429982185364 |
| keywords[8].display_name | Physics |
| keywords[9].id | https://openalex.org/keywords/biochemistry |
| keywords[9].score | 0.2826350927352905 |
| keywords[9].display_name | Biochemistry |
| keywords[10].id | https://openalex.org/keywords/medicine |
| keywords[10].score | 0.2766672968864441 |
| keywords[10].display_name | Medicine |
| keywords[11].id | https://openalex.org/keywords/biology |
| keywords[11].score | 0.2450850009918213 |
| keywords[11].display_name | Biology |
| keywords[12].id | https://openalex.org/keywords/computational-chemistry |
| keywords[12].score | 0.20014700293540955 |
| keywords[12].display_name | Computational chemistry |
| keywords[13].id | https://openalex.org/keywords/amino-acid |
| keywords[13].score | 0.11498028039932251 |
| keywords[13].display_name | Amino acid |
| keywords[14].id | https://openalex.org/keywords/ecology |
| keywords[14].score | 0.09380388259887695 |
| keywords[14].display_name | Ecology |
| language | en |
| locations[0].id | doi:10.1021/acs.jcim.5c00869 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S167262187 |
| locations[0].source.issn | 1549-9596, 1549-960X |
| locations[0].source.type | journal |
| locations[0].source.is_oa | False |
| locations[0].source.issn_l | 1549-9596 |
| locations[0].source.is_core | True |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | Journal of Chemical Information and Modeling |
| locations[0].source.host_organization | https://openalex.org/P4310320006 |
| locations[0].source.host_organization_name | American Chemical Society |
| locations[0].source.host_organization_lineage | https://openalex.org/P4310320006 |
| locations[0].source.host_organization_lineage_names | American Chemical Society |
| locations[0].license | cc-by |
| locations[0].pdf_url | |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | Journal of Chemical Information and Modeling |
| locations[0].landing_page_url | https://doi.org/10.1021/acs.jcim.5c00869 |
| locations[1].id | pmid:40495656 |
| locations[1].is_oa | False |
| locations[1].source.id | https://openalex.org/S4306525036 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | PubMed |
| locations[1].source.host_organization | https://openalex.org/I1299303238 |
| locations[1].source.host_organization_name | National Institutes of Health |
| locations[1].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[1].license | |
| locations[1].pdf_url | |
| locations[1].version | publishedVersion |
| locations[1].raw_type | |
| locations[1].license_id | |
| locations[1].is_accepted | True |
| locations[1].is_published | True |
| locations[1].raw_source_name | Journal of chemical information and modeling |
| locations[1].landing_page_url | https://pubmed.ncbi.nlm.nih.gov/40495656 |
| locations[2].id | pmh:oai:pubmedcentral.nih.gov:12199304 |
| locations[2].is_oa | True |
| locations[2].source.id | https://openalex.org/S2764455111 |
| locations[2].source.issn | |
| locations[2].source.type | repository |
| locations[2].source.is_oa | False |
| locations[2].source.issn_l | |
| locations[2].source.is_core | False |
| locations[2].source.is_in_doaj | False |
| locations[2].source.display_name | PubMed Central |
| locations[2].source.host_organization | https://openalex.org/I1299303238 |
| locations[2].source.host_organization_name | National Institutes of Health |
| locations[2].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[2].license | cc-by |
| locations[2].pdf_url | |
| locations[2].version | submittedVersion |
| locations[2].raw_type | Text |
| locations[2].license_id | https://openalex.org/licenses/cc-by |
| locations[2].is_accepted | False |
| locations[2].is_published | False |
| locations[2].raw_source_name | J Chem Inf Model |
| locations[2].landing_page_url | https://www.ncbi.nlm.nih.gov/pmc/articles/12199304 |
| indexed_in | crossref, pubmed |
| authorships[0].author.id | https://openalex.org/A5085897064 |
| authorships[0].author.orcid | https://orcid.org/0000-0002-0834-871X |
| authorships[0].author.display_name | Marko Hanževački |
| authorships[0].countries | GB |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I36234482 |
| authorships[0].affiliations[0].raw_affiliation_string | University of Bristol |
| authorships[0].affiliations[1].raw_affiliation_string | Centre for Computational Chemistry, School of Chemistry |
| authorships[0].institutions[0].id | https://openalex.org/I36234482 |
| authorships[0].institutions[0].ror | https://ror.org/0524sp257 |
| authorships[0].institutions[0].type | education |
| authorships[0].institutions[0].lineage | https://openalex.org/I36234482 |
| authorships[0].institutions[0].country_code | GB |
| authorships[0].institutions[0].display_name | University of Bristol |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Marko Hanževački |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Centre for Computational Chemistry, School of Chemistry, University of Bristol |
| authorships[1].author.id | https://openalex.org/A5027864754 |
| authorships[1].author.orcid | https://orcid.org/0000-0002-4730-2220 |
| authorships[1].author.display_name | Rebecca M. Twidale |
| authorships[1].countries | GB |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I36234482 |
| authorships[1].affiliations[0].raw_affiliation_string | University of Bristol |
| authorships[1].affiliations[1].raw_affiliation_string | Centre for Computational Chemistry, School of Chemistry |
| authorships[1].institutions[0].id | https://openalex.org/I36234482 |
| authorships[1].institutions[0].ror | https://ror.org/0524sp257 |
| authorships[1].institutions[0].type | education |
| authorships[1].institutions[0].lineage | https://openalex.org/I36234482 |
| authorships[1].institutions[0].country_code | GB |
| authorships[1].institutions[0].display_name | University of Bristol |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Rebecca M. Twidale |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | Centre for Computational Chemistry, School of Chemistry, University of Bristol |
| authorships[2].author.id | https://openalex.org/A5033135680 |
| authorships[2].author.orcid | https://orcid.org/0000-0002-3808-054X |
| authorships[2].author.display_name | Eric J. M. Lang |
| authorships[2].countries | GB |
| authorships[2].affiliations[0].raw_affiliation_string | Centre for Computational Chemistry, School of Chemistry |
| authorships[2].affiliations[1].institution_ids | https://openalex.org/I36234482 |
| authorships[2].affiliations[1].raw_affiliation_string | University of Bristol |
| authorships[2].institutions[0].id | https://openalex.org/I36234482 |
| authorships[2].institutions[0].ror | https://ror.org/0524sp257 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I36234482 |
| authorships[2].institutions[0].country_code | GB |
| authorships[2].institutions[0].display_name | University of Bristol |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Eric J.M. Lang |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | Centre for Computational Chemistry, School of Chemistry, University of Bristol |
| authorships[3].author.id | https://openalex.org/A5028874930 |
| authorships[3].author.orcid | https://orcid.org/0000-0002-8499-8597 |
| authorships[3].author.display_name | W. Gerrard |
| authorships[3].affiliations[0].raw_affiliation_string | Kuano, Hauxton House, Mill Scitech Park, Mill Lane, Cambridge CB22 5HX, United Kingdom |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Will Gerrard |
| authorships[3].is_corresponding | False |
| authorships[3].raw_affiliation_strings | Kuano, Hauxton House, Mill Scitech Park, Mill Lane, Cambridge CB22 5HX, United Kingdom |
| authorships[4].author.id | https://openalex.org/A5048093507 |
| authorships[4].author.orcid | https://orcid.org/0000-0002-7145-9105 |
| authorships[4].author.display_name | David W. Wright |
| authorships[4].affiliations[0].raw_affiliation_string | Kuano, Hauxton House, Mill Scitech Park, Mill Lane, Cambridge CB22 5HX, United Kingdom |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | David W. Wright |
| authorships[4].is_corresponding | False |
| authorships[4].raw_affiliation_strings | Kuano, Hauxton House, Mill Scitech Park, Mill Lane, Cambridge CB22 5HX, United Kingdom |
| authorships[5].author.id | https://openalex.org/A5106576559 |
| authorships[5].author.orcid | |
| authorships[5].author.display_name | Vid Stojevic |
| authorships[5].affiliations[0].raw_affiliation_string | Kuano, Hauxton House, Mill Scitech Park, Mill Lane, Cambridge CB22 5HX, United Kingdom |
| authorships[5].author_position | middle |
| authorships[5].raw_author_name | Vid Stojevic |
| authorships[5].is_corresponding | False |
| authorships[5].raw_affiliation_strings | Kuano, Hauxton House, Mill Scitech Park, Mill Lane, Cambridge CB22 5HX, United Kingdom |
| authorships[6].author.id | https://openalex.org/A5044048108 |
| authorships[6].author.orcid | https://orcid.org/0000-0003-1015-4567 |
| authorships[6].author.display_name | Adrian J. Mulholland |
| authorships[6].countries | GB |
| authorships[6].affiliations[0].institution_ids | https://openalex.org/I36234482 |
| authorships[6].affiliations[0].raw_affiliation_string | University of Bristol |
| authorships[6].affiliations[1].raw_affiliation_string | Centre for Computational Chemistry, School of Chemistry |
| authorships[6].institutions[0].id | https://openalex.org/I36234482 |
| authorships[6].institutions[0].ror | https://ror.org/0524sp257 |
| authorships[6].institutions[0].type | education |
| authorships[6].institutions[0].lineage | https://openalex.org/I36234482 |
| authorships[6].institutions[0].country_code | GB |
| authorships[6].institutions[0].display_name | University of Bristol |
| authorships[6].author_position | last |
| authorships[6].raw_author_name | Adrian J. Mulholland |
| authorships[6].is_corresponding | True |
| authorships[6].raw_affiliation_strings | Centre for Computational Chemistry, School of Chemistry, University of Bristol |
| has_content.pdf | False |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://doi.org/10.1021/acs.jcim.5c00869 |
| open_access.oa_status | hybrid |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Quantum Mechanics/Molecular Mechanics Simulations Distinguish Insulin-Regulated Aminopeptidase Substrate (Oxytocin) and Inhibitor (Angiotensin IV) and Reveal Determinants of Activity and Inhibition |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T11541 |
| primary_topic.field.id | https://openalex.org/fields/32 |
| primary_topic.field.display_name | Psychology |
| primary_topic.score | 0.9987999796867371 |
| primary_topic.domain.id | https://openalex.org/domains/2 |
| primary_topic.domain.display_name | Social Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/3207 |
| primary_topic.subfield.display_name | Social Psychology |
| primary_topic.display_name | Neuroendocrine regulation and behavior |
| related_works | https://openalex.org/W2013503606, https://openalex.org/W4234389666, https://openalex.org/W3156080391, https://openalex.org/W2805000670, https://openalex.org/W4315498887, https://openalex.org/W2034563028, https://openalex.org/W2000096903, https://openalex.org/W1990241706, https://openalex.org/W1980551862, https://openalex.org/W4380998201 |
| cited_by_count | 1 |
| counts_by_year[0].year | 2025 |
| counts_by_year[0].cited_by_count | 1 |
| locations_count | 3 |
| best_oa_location.id | doi:10.1021/acs.jcim.5c00869 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S167262187 |
| best_oa_location.source.issn | 1549-9596, 1549-960X |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | False |
| best_oa_location.source.issn_l | 1549-9596 |
| best_oa_location.source.is_core | True |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | Journal of Chemical Information and Modeling |
| best_oa_location.source.host_organization | https://openalex.org/P4310320006 |
| best_oa_location.source.host_organization_name | American Chemical Society |
| best_oa_location.source.host_organization_lineage | https://openalex.org/P4310320006 |
| best_oa_location.source.host_organization_lineage_names | American Chemical Society |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | journal-article |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | Journal of Chemical Information and Modeling |
| best_oa_location.landing_page_url | https://doi.org/10.1021/acs.jcim.5c00869 |
| primary_location.id | doi:10.1021/acs.jcim.5c00869 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S167262187 |
| primary_location.source.issn | 1549-9596, 1549-960X |
| primary_location.source.type | journal |
| primary_location.source.is_oa | False |
| primary_location.source.issn_l | 1549-9596 |
| primary_location.source.is_core | True |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | Journal of Chemical Information and Modeling |
| primary_location.source.host_organization | https://openalex.org/P4310320006 |
| primary_location.source.host_organization_name | American Chemical Society |
| primary_location.source.host_organization_lineage | https://openalex.org/P4310320006 |
| primary_location.source.host_organization_lineage_names | American Chemical Society |
| primary_location.license | cc-by |
| primary_location.pdf_url | |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | Journal of Chemical Information and Modeling |
| primary_location.landing_page_url | https://doi.org/10.1021/acs.jcim.5c00869 |
| publication_date | 2025-06-11 |
| publication_year | 2025 |
| referenced_works | https://openalex.org/W1940439926, https://openalex.org/W2120717049, https://openalex.org/W2128092437, https://openalex.org/W2100723009, https://openalex.org/W3090783903, https://openalex.org/W3093956419, https://openalex.org/W4377143233, https://openalex.org/W2005900492, https://openalex.org/W2607678445, https://openalex.org/W2156997485, https://openalex.org/W2912540153, https://openalex.org/W3007155626, https://openalex.org/W2590947056, https://openalex.org/W2577382579, https://openalex.org/W2049609626, https://openalex.org/W2140202716, https://openalex.org/W2068670295, https://openalex.org/W2021036812, https://openalex.org/W4388638569, https://openalex.org/W2947284788, https://openalex.org/W2166273490, https://openalex.org/W2168161193, https://openalex.org/W3208413900, https://openalex.org/W2142459107, https://openalex.org/W1987255508, https://openalex.org/W2013646394, https://openalex.org/W1547520663, https://openalex.org/W2031017779, https://openalex.org/W3088140707, https://openalex.org/W2170711881, https://openalex.org/W3138524854, https://openalex.org/W1832541910, https://openalex.org/W1964156630, https://openalex.org/W2011605939, https://openalex.org/W1988724422, https://openalex.org/W1989281340, https://openalex.org/W2319465148, https://openalex.org/W1988517119, https://openalex.org/W1995022799, https://openalex.org/W2020217296, https://openalex.org/W4320800236, https://openalex.org/W1997834593, https://openalex.org/W2048521920, https://openalex.org/W1515510017, https://openalex.org/W3088065968, https://openalex.org/W2053015924, https://openalex.org/W2055982079, https://openalex.org/W2022114028, https://openalex.org/W2016775336, https://openalex.org/W2975999623, https://openalex.org/W3092766219, https://openalex.org/W4394567953, https://openalex.org/W3013754213, https://openalex.org/W2799421578, https://openalex.org/W3033781256, https://openalex.org/W3022815335, https://openalex.org/W4390971924, https://openalex.org/W2599860559, https://openalex.org/W3177828909, https://openalex.org/W3211795435, https://openalex.org/W2117458899, https://openalex.org/W2799876761, https://openalex.org/W2050456292, https://openalex.org/W2404280981, https://openalex.org/W2074386125, https://openalex.org/W1976499671, https://openalex.org/W1968166922, https://openalex.org/W2333538638, https://openalex.org/W2772641670, https://openalex.org/W4361017438, https://openalex.org/W4395030775, https://openalex.org/W2132525235, https://openalex.org/W1587563234, https://openalex.org/W3034846184, https://openalex.org/W3190366419, https://openalex.org/W4405060807, https://openalex.org/W3158379422, https://openalex.org/W2254827003, https://openalex.org/W2039051836, https://openalex.org/W2057201665, https://openalex.org/W1982521448, https://openalex.org/W2010830164, https://openalex.org/W2030286780, https://openalex.org/W2511416709, https://openalex.org/W2925800094, https://openalex.org/W1990317939, https://openalex.org/W4366548649, https://openalex.org/W3046110940, https://openalex.org/W2122976061, https://openalex.org/W2049876452, https://openalex.org/W2140794227 |
| referenced_works_count | 91 |
| abstract_inverted_index.A | 153 |
| abstract_inverted_index.a | 4, 9, 41, 79, 168, 200, 227, 244 |
| abstract_inverted_index.IV | 54, 242 |
| abstract_inverted_index.We | 112 |
| abstract_inverted_index.as | 8, 60, 297 |
| abstract_inverted_index.by | 240 |
| abstract_inverted_index.in | 21, 135, 145, 222, 280 |
| abstract_inverted_index.is | 3, 98 |
| abstract_inverted_index.it | 97 |
| abstract_inverted_index.of | 37, 131, 161, 207, 217, 236, 247, 266, 274, 294 |
| abstract_inverted_index.to | 17, 45, 64, 67, 127 |
| abstract_inverted_index.IV. | 111, 183 |
| abstract_inverted_index.NBO | 189 |
| abstract_inverted_index.NCI | 191 |
| abstract_inverted_index.Our | 283 |
| abstract_inverted_index.TI. | 259 |
| abstract_inverted_index.The | 137, 224 |
| abstract_inverted_index.and | 24, 48, 55, 87, 123, 167, 190, 212, 232, 271, 277, 292, 306 |
| abstract_inverted_index.are | 58 |
| abstract_inverted_index.but | 105 |
| abstract_inverted_index.due | 16, 63 |
| abstract_inverted_index.for | 12, 141, 158, 172, 178, 197, 288, 301 |
| abstract_inverted_index.its | 18 |
| abstract_inverted_index.not | 107 |
| abstract_inverted_index.the | 32, 85, 88, 94, 101, 129, 150, 159, 162, 173, 179, 194, 204, 208, 213, 218, 237, 248, 258, 263, 268, 289 |
| abstract_inverted_index.two | 133 |
| abstract_inverted_index.was | 144 |
| abstract_inverted_index.why | 100 |
| abstract_inverted_index.(TI) | 166 |
| abstract_inverted_index.IRAP | 295 |
| abstract_inverted_index.bond | 35, 211, 231 |
| abstract_inverted_index.does | 106 |
| abstract_inverted_index.from | 252 |
| abstract_inverted_index.good | 147 |
| abstract_inverted_index.have | 285 |
| abstract_inverted_index.hole | 206 |
| abstract_inverted_index.lone | 215 |
| abstract_inverted_index.pair | 216 |
| abstract_inverted_index.role | 20 |
| abstract_inverted_index.very | 80, 146 |
| abstract_inverted_index.weak | 228 |
| abstract_inverted_index.were | 176 |
| abstract_inverted_index.with | 90, 149, 181 |
| abstract_inverted_index.IRAP, | 91 |
| abstract_inverted_index.IRAP. | 136 |
| abstract_inverted_index.QM/MM | 125 |
| abstract_inverted_index.These | 260 |
| abstract_inverted_index.angle | 251 |
| abstract_inverted_index.basis | 196 |
| abstract_inverted_index.bond, | 96 |
| abstract_inverted_index.brain | 51 |
| abstract_inverted_index.data. | 152 |
| abstract_inverted_index.ideal | 253 |
| abstract_inverted_index.novel | 10 |
| abstract_inverted_index.other | 49 |
| abstract_inverted_index.sigma | 205 |
| abstract_inverted_index.their | 65 |
| abstract_inverted_index.these | 132 |
| abstract_inverted_index.(IRAP) | 2 |
| abstract_inverted_index.IRAP's | 28, 70 |
| abstract_inverted_index.agents | 300 |
| abstract_inverted_index.around | 93 |
| abstract_inverted_index.caused | 243 |
| abstract_inverted_index.design | 291 |
| abstract_inverted_index.domain | 30 |
| abstract_inverted_index.energy | 139, 156 |
| abstract_inverted_index.enzyme | 102 |
| abstract_inverted_index.higher | 155, 169 |
| abstract_inverted_index.larger | 245 |
| abstract_inverted_index.linked | 44 |
| abstract_inverted_index.memory | 302 |
| abstract_inverted_index.states | 279 |
| abstract_inverted_index.strong | 233 |
| abstract_inverted_index.target | 11 |
| abstract_inverted_index.valine | 249 |
| abstract_inverted_index.(QM/MM) | 119 |
| abstract_inverted_index.Despite | 78 |
| abstract_inverted_index.ability | 66 |
| abstract_inverted_index.barrier | 140, 157, 171 |
| abstract_inverted_index.between | 84, 203, 226 |
| abstract_inverted_index.binding | 82 |
| abstract_inverted_index.complex | 83 |
| abstract_inverted_index.crucial | 19 |
| abstract_inverted_index.degrade | 109 |
| abstract_inverted_index.explore | 128 |
| abstract_inverted_index.glucose | 22 |
| abstract_inverted_index.inhibit | 69 |
| abstract_inverted_index.insulin | 25 |
| abstract_inverted_index.methods | 192 |
| abstract_inverted_index.natural | 38, 75 |
| abstract_inverted_index.overall | 170 |
| abstract_inverted_index.peptide | 34, 43, 174, 220 |
| abstract_inverted_index.quantum | 116 |
| abstract_inverted_index.results | 261 |
| abstract_inverted_index.similar | 56, 81 |
| abstract_inverted_index.spodium | 230 |
| abstract_inverted_index.thereby | 73 |
| abstract_inverted_index.unclear | 99 |
| abstract_inverted_index.analysis | 187 |
| abstract_inverted_index.cleavage | 143, 175 |
| abstract_inverted_index.critical | 264 |
| abstract_inverted_index.diseases | 15 |
| abstract_inverted_index.dynamics | 121 |
| abstract_inverted_index.employed | 113 |
| abstract_inverted_index.enhanced | 114 |
| abstract_inverted_index.findings | 284 |
| abstract_inverted_index.improved | 46 |
| abstract_inverted_index.nitrogen | 221 |
| abstract_inverted_index.observed | 177 |
| abstract_inverted_index.oxyanion | 163 |
| abstract_inverted_index.oxytocin | 104, 142 |
| abstract_inverted_index.peptides | 57, 134 |
| abstract_inverted_index.rational | 290 |
| abstract_inverted_index.reaction | 130, 180, 275 |
| abstract_inverted_index.sampling | 115 |
| abstract_inverted_index.scissile | 95, 219 |
| abstract_inverted_index.unveiled | 193 |
| abstract_inverted_index.C-Cα-Cβ | 250 |
| abstract_inverted_index.activity, | 72 |
| abstract_inverted_index.agreement | 148 |
| abstract_inverted_index.analyzing | 267 |
| abstract_inverted_index.bidentate | 234 |
| abstract_inverted_index.catalytic | 29, 238 |
| abstract_inverted_index.catalyzes | 31 |
| abstract_inverted_index.cognition | 47 |
| abstract_inverted_index.cognitive | 61 |
| abstract_inverted_index.combating | 13 |
| abstract_inverted_index.deviation | 246 |
| abstract_inverted_index.diabetes. | 307 |
| abstract_inverted_index.different | 198 |
| abstract_inverted_index.diseases, | 305 |
| abstract_inverted_index.disulfide | 210 |
| abstract_inverted_index.dynamics, | 269 |
| abstract_inverted_index.elemental | 50 |
| abstract_inverted_index.enhancers | 62 |
| abstract_inverted_index.enzymatic | 281 |
| abstract_inverted_index.formation | 160 |
| abstract_inverted_index.geometry, | 255 |
| abstract_inverted_index.inhibitor | 89 |
| abstract_inverted_index.interplay | 225 |
| abstract_inverted_index.mechanics | 118 |
| abstract_inverted_index.molecular | 120, 195 |
| abstract_inverted_index.oxytocin, | 40 |
| abstract_inverted_index.oxytocin. | 223 |
| abstract_inverted_index.potential | 298 |
| abstract_inverted_index.structure | 186 |
| abstract_inverted_index.substrate | 39, 86 |
| abstract_inverted_index.utilizing | 188 |
| abstract_inverted_index.N-terminal | 33 |
| abstract_inverted_index.N-terminus | 209 |
| abstract_inverted_index.calculated | 138 |
| abstract_inverted_index.disorders, | 303 |
| abstract_inverted_index.electronic | 185, 272 |
| abstract_inverted_index.functions. | 52 |
| abstract_inverted_index.hydrolysis | 36 |
| abstract_inverted_index.identified | 7 |
| abstract_inverted_index.importance | 265 |
| abstract_inverted_index.inhibitors | 296 |
| abstract_inverted_index.metabolism | 23 |
| abstract_inverted_index.mitigating | 74 |
| abstract_inverted_index.processes. | 282 |
| abstract_inverted_index.properties | 273 |
| abstract_inverted_index.recognized | 59 |
| abstract_inverted_index.transition | 278 |
| abstract_inverted_index.underscore | 262 |
| abstract_inverted_index.Angiotensin | 53 |
| abstract_inverted_index.angiotensin | 110, 182, 241 |
| abstract_inverted_index.development | 293 |
| abstract_inverted_index.efficiently | 108 |
| abstract_inverted_index.hybridizing | 214 |
| abstract_inverted_index.interaction | 202 |
| abstract_inverted_index.metabolizes | 103 |
| abstract_inverted_index.neuroactive | 42 |
| abstract_inverted_index.noncovalent | 229 |
| abstract_inverted_index.proteolytic | 71 |
| abstract_inverted_index.reactivity, | 199 |
| abstract_inverted_index.regulation. | 27 |
| abstract_inverted_index.sensitivity | 26 |
| abstract_inverted_index.significant | 286 |
| abstract_inverted_index.simulations | 122 |
| abstract_inverted_index.stabilizing | 201 |
| abstract_inverted_index.tetrahedral | 164, 254 |
| abstract_inverted_index.therapeutic | 299 |
| abstract_inverted_index.calculations | 126 |
| abstract_inverted_index.consequently | 256 |
| abstract_inverted_index.coordination | 235 |
| abstract_inverted_index.degradation. | 77 |
| abstract_inverted_index.experimental | 151 |
| abstract_inverted_index.higher-level | 124 |
| abstract_inverted_index.implications | 287 |
| abstract_inverted_index.intermediate | 165 |
| abstract_inverted_index.neuropeptide | 76 |
| abstract_inverted_index.particularly | 92 |
| abstract_inverted_index.Comprehensive | 184 |
| abstract_inverted_index.competitively | 68 |
| abstract_inverted_index.destabilizing | 257 |
| abstract_inverted_index.interactions, | 270 |
| abstract_inverted_index.intermediates | 276 |
| abstract_inverted_index.metalloenzyme | 6 |
| abstract_inverted_index.significantly | 154 |
| abstract_inverted_index.aminopeptidase | 1 |
| abstract_inverted_index.zinc-dependent | 5 |
| abstract_inverted_index.Zn<sup>2+</sup> | 239 |
| abstract_inverted_index.diabetes-induced | 14 |
| abstract_inverted_index.Insulin-regulated | 0 |
| abstract_inverted_index.neurodegenerative | 304 |
| abstract_inverted_index.mechanics/molecular | 117 |
| cited_by_percentile_year.max | 95 |
| cited_by_percentile_year.min | 91 |
| corresponding_author_ids | https://openalex.org/A5044048108 |
| countries_distinct_count | 1 |
| institutions_distinct_count | 7 |
| corresponding_institution_ids | https://openalex.org/I36234482 |
| citation_normalized_percentile.value | 0.91962838 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | True |