Rapamycin-inspired macrocycles with new target specificity Article Swipe
 YOU?
·
· 2023
· Open Access
·
· DOI: https://doi.org/10.32920/21979724.v1
YOU?
·
· 2023
· Open Access
·
· DOI: https://doi.org/10.32920/21979724.v1
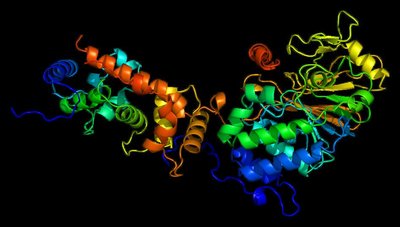
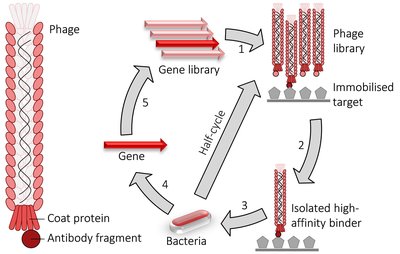
Rapamycin and FK506 are macrocyclic natural products with an extraordinary mode of action, in which they form binary complexes with FK506-binding protein (FKBP) through a shared FKBP-binding domain before forming ternary complexes with their respective targets, mechanistic target of rapamycin (mTOR) and calcineurin, respectively. Inspired by this, we sought to build a rapamycin-like macromolecule library to target new cellular proteins by replacing the effector domain of rapamycin with a combinatorial library of oligopeptides. We developed a robust macrocyclization method using ring-closing metathesis and synthesized a 45,000-compound library of hybrid macrocycles (named rapafucins) using optimized FKBP-binding domains. Screening of the rapafucin library in human cells led to the discovery of rapadocin, an inhibitor of nucleoside uptake. Rapadocin is a potent, isoform-specific and FKBP-dependent inhibitor of the equilibrative nucleoside transporter 1 and is efficacious in an animal model of kidney ischaemia reperfusion injury. Together, these results demonstrate that rapafucins are a new class of chemical probes and drug leads that can expand the repertoire of protein targets well beyond mTOR and calcineurin.
Related Topics
- Type
- preprint
- Language
- en
- Landing Page
- https://doi.org/10.32920/21979724.v1
- OA Status
- green
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4318574254
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4318574254Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.32920/21979724.v1Digital Object Identifier
- Title
-
Rapamycin-inspired macrocycles with new target specificityWork title
- Type
-
preprintOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2023Year of publication
- Publication date
-
2023-01-30Full publication date if available
- Authors
-
Zufeng Guo, Sam Y. Hong, Jingxin Wang, Shahid Rehan, Wukun Liu, Hanjing Peng, Manisha Das, Wei Li, Shridhar Bhat, Brandon Peiffer, Brett R. Ullman, Chung‐Ming Tse, Zlatina Tarmakova, Cordelia Schiene‐Fischer, Gunter Fischer, Imogen R. Coe, Ville O. Paavilainen, Zhaoli Sun, Jun O. LiuList of authors in order
- Landing page
-
https://doi.org/10.32920/21979724.v1Publisher landing page
- Open access
-
YesWhether a free full text is available
- OA status
-
greenOpen access status per OpenAlex
- OA URL
-
https://figshare.com/articles/journal_contribution/Rapamycin-inspired_macrocycles_with_new_target_specificity/21979724Direct OA link when available
- Concepts
-
FKBP, Effector, Calcineurin, Chemistry, Phage display, Drug discovery, Chemical biology, HEK 293 cells, Computational biology, Cell biology, Biochemistry, Biology, Receptor, Peptide, Medicine, Surgery, TransplantationTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
0Total citation count in OpenAlex
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4318574254 |
|---|---|
| doi | https://doi.org/10.32920/21979724.v1 |
| ids.doi | https://doi.org/10.32920/21979724.v1 |
| ids.openalex | https://openalex.org/W4318574254 |
| fwci | 0.0 |
| type | preprint |
| title | Rapamycin-inspired macrocycles with new target specificity |
| biblio.issue | |
| biblio.volume | |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T12827 |
| topics[0].field.id | https://openalex.org/fields/13 |
| topics[0].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[0].score | 0.9610000252723694 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1312 |
| topics[0].subfield.display_name | Molecular Biology |
| topics[0].display_name | Biochemical and Molecular Research |
| topics[1].id | https://openalex.org/T10409 |
| topics[1].field.id | https://openalex.org/fields/22 |
| topics[1].field.display_name | Engineering |
| topics[1].score | 0.9592999815940857 |
| topics[1].domain.id | https://openalex.org/domains/3 |
| topics[1].domain.display_name | Physical Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/2208 |
| topics[1].subfield.display_name | Electrical and Electronic Engineering |
| topics[1].display_name | Fuel Cells and Related Materials |
| topics[2].id | https://openalex.org/T11193 |
| topics[2].field.id | https://openalex.org/fields/13 |
| topics[2].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[2].score | 0.9251000285148621 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1314 |
| topics[2].subfield.display_name | Physiology |
| topics[2].display_name | Adenosine and Purinergic Signaling |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C133956753 |
| concepts[0].level | 2 |
| concepts[0].score | 0.9570474624633789 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q583864 |
| concepts[0].display_name | FKBP |
| concepts[1].id | https://openalex.org/C51785407 |
| concepts[1].level | 2 |
| concepts[1].score | 0.6048033237457275 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q2627568 |
| concepts[1].display_name | Effector |
| concepts[2].id | https://openalex.org/C128057223 |
| concepts[2].level | 3 |
| concepts[2].score | 0.5609540939331055 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q420566 |
| concepts[2].display_name | Calcineurin |
| concepts[3].id | https://openalex.org/C185592680 |
| concepts[3].level | 0 |
| concepts[3].score | 0.5499581098556519 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q2329 |
| concepts[3].display_name | Chemistry |
| concepts[4].id | https://openalex.org/C186268636 |
| concepts[4].level | 3 |
| concepts[4].score | 0.4732390344142914 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q896217 |
| concepts[4].display_name | Phage display |
| concepts[5].id | https://openalex.org/C74187038 |
| concepts[5].level | 2 |
| concepts[5].score | 0.46968916058540344 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q1418791 |
| concepts[5].display_name | Drug discovery |
| concepts[6].id | https://openalex.org/C171852809 |
| concepts[6].level | 2 |
| concepts[6].score | 0.4642399549484253 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q731154 |
| concepts[6].display_name | Chemical biology |
| concepts[7].id | https://openalex.org/C174510640 |
| concepts[7].level | 3 |
| concepts[7].score | 0.45878273248672485 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q489618 |
| concepts[7].display_name | HEK 293 cells |
| concepts[8].id | https://openalex.org/C70721500 |
| concepts[8].level | 1 |
| concepts[8].score | 0.3706929087638855 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q177005 |
| concepts[8].display_name | Computational biology |
| concepts[9].id | https://openalex.org/C95444343 |
| concepts[9].level | 1 |
| concepts[9].score | 0.3670744001865387 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q7141 |
| concepts[9].display_name | Cell biology |
| concepts[10].id | https://openalex.org/C55493867 |
| concepts[10].level | 1 |
| concepts[10].score | 0.3570631742477417 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q7094 |
| concepts[10].display_name | Biochemistry |
| concepts[11].id | https://openalex.org/C86803240 |
| concepts[11].level | 0 |
| concepts[11].score | 0.32958322763442993 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[11].display_name | Biology |
| concepts[12].id | https://openalex.org/C170493617 |
| concepts[12].level | 2 |
| concepts[12].score | 0.2807224988937378 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q208467 |
| concepts[12].display_name | Receptor |
| concepts[13].id | https://openalex.org/C2779281246 |
| concepts[13].level | 2 |
| concepts[13].score | 0.12469631433486938 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q172847 |
| concepts[13].display_name | Peptide |
| concepts[14].id | https://openalex.org/C71924100 |
| concepts[14].level | 0 |
| concepts[14].score | 0.09983479976654053 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q11190 |
| concepts[14].display_name | Medicine |
| concepts[15].id | https://openalex.org/C141071460 |
| concepts[15].level | 1 |
| concepts[15].score | 0.0 |
| concepts[15].wikidata | https://www.wikidata.org/wiki/Q40821 |
| concepts[15].display_name | Surgery |
| concepts[16].id | https://openalex.org/C2911091166 |
| concepts[16].level | 2 |
| concepts[16].score | 0.0 |
| concepts[16].wikidata | https://www.wikidata.org/wiki/Q106419912 |
| concepts[16].display_name | Transplantation |
| keywords[0].id | https://openalex.org/keywords/fkbp |
| keywords[0].score | 0.9570474624633789 |
| keywords[0].display_name | FKBP |
| keywords[1].id | https://openalex.org/keywords/effector |
| keywords[1].score | 0.6048033237457275 |
| keywords[1].display_name | Effector |
| keywords[2].id | https://openalex.org/keywords/calcineurin |
| keywords[2].score | 0.5609540939331055 |
| keywords[2].display_name | Calcineurin |
| keywords[3].id | https://openalex.org/keywords/chemistry |
| keywords[3].score | 0.5499581098556519 |
| keywords[3].display_name | Chemistry |
| keywords[4].id | https://openalex.org/keywords/phage-display |
| keywords[4].score | 0.4732390344142914 |
| keywords[4].display_name | Phage display |
| keywords[5].id | https://openalex.org/keywords/drug-discovery |
| keywords[5].score | 0.46968916058540344 |
| keywords[5].display_name | Drug discovery |
| keywords[6].id | https://openalex.org/keywords/chemical-biology |
| keywords[6].score | 0.4642399549484253 |
| keywords[6].display_name | Chemical biology |
| keywords[7].id | https://openalex.org/keywords/hek-293-cells |
| keywords[7].score | 0.45878273248672485 |
| keywords[7].display_name | HEK 293 cells |
| keywords[8].id | https://openalex.org/keywords/computational-biology |
| keywords[8].score | 0.3706929087638855 |
| keywords[8].display_name | Computational biology |
| keywords[9].id | https://openalex.org/keywords/cell-biology |
| keywords[9].score | 0.3670744001865387 |
| keywords[9].display_name | Cell biology |
| keywords[10].id | https://openalex.org/keywords/biochemistry |
| keywords[10].score | 0.3570631742477417 |
| keywords[10].display_name | Biochemistry |
| keywords[11].id | https://openalex.org/keywords/biology |
| keywords[11].score | 0.32958322763442993 |
| keywords[11].display_name | Biology |
| keywords[12].id | https://openalex.org/keywords/receptor |
| keywords[12].score | 0.2807224988937378 |
| keywords[12].display_name | Receptor |
| keywords[13].id | https://openalex.org/keywords/peptide |
| keywords[13].score | 0.12469631433486938 |
| keywords[13].display_name | Peptide |
| keywords[14].id | https://openalex.org/keywords/medicine |
| keywords[14].score | 0.09983479976654053 |
| keywords[14].display_name | Medicine |
| language | en |
| locations[0].id | doi:10.32920/21979724.v1 |
| locations[0].is_oa | False |
| locations[0].source | |
| locations[0].license | |
| locations[0].pdf_url | |
| locations[0].version | acceptedVersion |
| locations[0].raw_type | posted-content |
| locations[0].license_id | |
| locations[0].is_accepted | True |
| locations[0].is_published | False |
| locations[0].raw_source_name | |
| locations[0].landing_page_url | https://doi.org/10.32920/21979724.v1 |
| locations[1].id | pmh:oai:figshare.com:article/21979724 |
| locations[1].is_oa | True |
| locations[1].source.id | https://openalex.org/S4306400572 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | OPAL (Open@LaTrobe) (La Trobe University) |
| locations[1].source.host_organization | https://openalex.org/I196829312 |
| locations[1].source.host_organization_name | La Trobe University |
| locations[1].source.host_organization_lineage | https://openalex.org/I196829312 |
| locations[1].license | cc-by |
| locations[1].pdf_url | |
| locations[1].version | submittedVersion |
| locations[1].raw_type | Text |
| locations[1].license_id | https://openalex.org/licenses/cc-by |
| locations[1].is_accepted | False |
| locations[1].is_published | False |
| locations[1].raw_source_name | |
| locations[1].landing_page_url | https://figshare.com/articles/journal_contribution/Rapamycin-inspired_macrocycles_with_new_target_specificity/21979724 |
| indexed_in | crossref |
| authorships[0].author.id | https://openalex.org/A5101984952 |
| authorships[0].author.orcid | https://orcid.org/0000-0002-4370-9445 |
| authorships[0].author.display_name | Zufeng Guo |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Zufeng Guo |
| authorships[0].is_corresponding | False |
| authorships[1].author.id | https://openalex.org/A5059085968 |
| authorships[1].author.orcid | https://orcid.org/0000-0001-5968-9795 |
| authorships[1].author.display_name | Sam Y. Hong |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Sam Y. Hong |
| authorships[1].is_corresponding | False |
| authorships[2].author.id | https://openalex.org/A5100686065 |
| authorships[2].author.orcid | https://orcid.org/0000-0002-9414-4093 |
| authorships[2].author.display_name | Jingxin Wang |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Jingxin Wang |
| authorships[2].is_corresponding | False |
| authorships[3].author.id | https://openalex.org/A5084834526 |
| authorships[3].author.orcid | https://orcid.org/0000-0003-1329-8056 |
| authorships[3].author.display_name | Shahid Rehan |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Shahid Rehan |
| authorships[3].is_corresponding | False |
| authorships[4].author.id | https://openalex.org/A5077280501 |
| authorships[4].author.orcid | https://orcid.org/0000-0002-0469-907X |
| authorships[4].author.display_name | Wukun Liu |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Wukun Liu |
| authorships[4].is_corresponding | False |
| authorships[5].author.id | https://openalex.org/A5032737990 |
| authorships[5].author.orcid | |
| authorships[5].author.display_name | Hanjing Peng |
| authorships[5].author_position | middle |
| authorships[5].raw_author_name | Hanjing Peng |
| authorships[5].is_corresponding | False |
| authorships[6].author.id | https://openalex.org/A5102819351 |
| authorships[6].author.orcid | https://orcid.org/0000-0002-0067-9186 |
| authorships[6].author.display_name | Manisha Das |
| authorships[6].author_position | middle |
| authorships[6].raw_author_name | Manisha Das |
| authorships[6].is_corresponding | False |
| authorships[7].author.id | https://openalex.org/A5100318346 |
| authorships[7].author.orcid | https://orcid.org/0000-0003-0388-1467 |
| authorships[7].author.display_name | Wei Li |
| authorships[7].author_position | middle |
| authorships[7].raw_author_name | Wei Li |
| authorships[7].is_corresponding | False |
| authorships[8].author.id | https://openalex.org/A5111163749 |
| authorships[8].author.orcid | |
| authorships[8].author.display_name | Shridhar Bhat |
| authorships[8].author_position | middle |
| authorships[8].raw_author_name | Shridhar Bhat |
| authorships[8].is_corresponding | False |
| authorships[9].author.id | https://openalex.org/A5043521467 |
| authorships[9].author.orcid | https://orcid.org/0000-0002-6783-2923 |
| authorships[9].author.display_name | Brandon Peiffer |
| authorships[9].author_position | middle |
| authorships[9].raw_author_name | Brandon J. Peiffer |
| authorships[9].is_corresponding | False |
| authorships[10].author.id | https://openalex.org/A5007385115 |
| authorships[10].author.orcid | |
| authorships[10].author.display_name | Brett R. Ullman |
| authorships[10].author_position | middle |
| authorships[10].raw_author_name | Brett R. Ullman |
| authorships[10].is_corresponding | False |
| authorships[11].author.id | https://openalex.org/A5021443559 |
| authorships[11].author.orcid | https://orcid.org/0000-0002-1873-2029 |
| authorships[11].author.display_name | Chung‐Ming Tse |
| authorships[11].author_position | middle |
| authorships[11].raw_author_name | Chung Ming Tse |
| authorships[11].is_corresponding | False |
| authorships[12].author.id | https://openalex.org/A5054004770 |
| authorships[12].author.orcid | |
| authorships[12].author.display_name | Zlatina Tarmakova |
| authorships[12].author_position | middle |
| authorships[12].raw_author_name | Zlatina Tarmakova |
| authorships[12].is_corresponding | False |
| authorships[13].author.id | https://openalex.org/A5022178172 |
| authorships[13].author.orcid | |
| authorships[13].author.display_name | Cordelia Schiene‐Fischer |
| authorships[13].author_position | middle |
| authorships[13].raw_author_name | Cordelia Schiene-Fischer |
| authorships[13].is_corresponding | False |
| authorships[14].author.id | https://openalex.org/A5058816586 |
| authorships[14].author.orcid | |
| authorships[14].author.display_name | Gunter Fischer |
| authorships[14].author_position | middle |
| authorships[14].raw_author_name | Gunter Fischer |
| authorships[14].is_corresponding | False |
| authorships[15].author.id | https://openalex.org/A5083019990 |
| authorships[15].author.orcid | https://orcid.org/0000-0002-9054-1817 |
| authorships[15].author.display_name | Imogen R. Coe |
| authorships[15].author_position | middle |
| authorships[15].raw_author_name | Imogen Coe |
| authorships[15].is_corresponding | False |
| authorships[16].author.id | https://openalex.org/A5022915896 |
| authorships[16].author.orcid | https://orcid.org/0000-0002-3160-7767 |
| authorships[16].author.display_name | Ville O. Paavilainen |
| authorships[16].author_position | middle |
| authorships[16].raw_author_name | Ville O. Paavilainen |
| authorships[16].is_corresponding | False |
| authorships[17].author.id | https://openalex.org/A5101270036 |
| authorships[17].author.orcid | |
| authorships[17].author.display_name | Zhaoli Sun |
| authorships[17].author_position | middle |
| authorships[17].raw_author_name | Zhaoli Sun |
| authorships[17].is_corresponding | False |
| authorships[18].author.id | https://openalex.org/A5015479608 |
| authorships[18].author.orcid | https://orcid.org/0000-0003-3842-9841 |
| authorships[18].author.display_name | Jun O. Liu |
| authorships[18].author_position | last |
| authorships[18].raw_author_name | Jun O. Liu |
| authorships[18].is_corresponding | False |
| has_content.pdf | False |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://figshare.com/articles/journal_contribution/Rapamycin-inspired_macrocycles_with_new_target_specificity/21979724 |
| open_access.oa_status | green |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Rapamycin-inspired macrocycles with new target specificity |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T12827 |
| primary_topic.field.id | https://openalex.org/fields/13 |
| primary_topic.field.display_name | Biochemistry, Genetics and Molecular Biology |
| primary_topic.score | 0.9610000252723694 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1312 |
| primary_topic.subfield.display_name | Molecular Biology |
| primary_topic.display_name | Biochemical and Molecular Research |
| related_works | https://openalex.org/W2089579242, https://openalex.org/W1984754431, https://openalex.org/W2023166792, https://openalex.org/W2007540790, https://openalex.org/W1967603109, https://openalex.org/W1756176550, https://openalex.org/W2047106546, https://openalex.org/W1843531984, https://openalex.org/W2593713688, https://openalex.org/W1982869248 |
| cited_by_count | 0 |
| locations_count | 2 |
| best_oa_location.id | pmh:oai:figshare.com:article/21979724 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4306400572 |
| best_oa_location.source.issn | |
| best_oa_location.source.type | repository |
| best_oa_location.source.is_oa | False |
| best_oa_location.source.issn_l | |
| best_oa_location.source.is_core | False |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | OPAL (Open@LaTrobe) (La Trobe University) |
| best_oa_location.source.host_organization | https://openalex.org/I196829312 |
| best_oa_location.source.host_organization_name | La Trobe University |
| best_oa_location.source.host_organization_lineage | https://openalex.org/I196829312 |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | |
| best_oa_location.version | submittedVersion |
| best_oa_location.raw_type | Text |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | False |
| best_oa_location.is_published | False |
| best_oa_location.raw_source_name | |
| best_oa_location.landing_page_url | https://figshare.com/articles/journal_contribution/Rapamycin-inspired_macrocycles_with_new_target_specificity/21979724 |
| primary_location.id | doi:10.32920/21979724.v1 |
| primary_location.is_oa | False |
| primary_location.source | |
| primary_location.license | |
| primary_location.pdf_url | |
| primary_location.version | acceptedVersion |
| primary_location.raw_type | posted-content |
| primary_location.license_id | |
| primary_location.is_accepted | True |
| primary_location.is_published | False |
| primary_location.raw_source_name | |
| primary_location.landing_page_url | https://doi.org/10.32920/21979724.v1 |
| publication_date | 2023-01-30 |
| publication_year | 2023 |
| referenced_works_count | 0 |
| abstract_inverted_index.1 | 128 |
| abstract_inverted_index.a | 24, 51, 68, 75, 84, 117, 148 |
| abstract_inverted_index.We | 73 |
| abstract_inverted_index.an | 8, 110, 133 |
| abstract_inverted_index.by | 45, 60 |
| abstract_inverted_index.in | 13, 101, 132 |
| abstract_inverted_index.is | 116, 130 |
| abstract_inverted_index.of | 11, 38, 65, 71, 87, 97, 108, 112, 123, 136, 151, 162 |
| abstract_inverted_index.to | 49, 55, 105 |
| abstract_inverted_index.we | 47 |
| abstract_inverted_index.and | 1, 41, 82, 120, 129, 154, 168 |
| abstract_inverted_index.are | 3, 147 |
| abstract_inverted_index.can | 158 |
| abstract_inverted_index.led | 104 |
| abstract_inverted_index.new | 57, 149 |
| abstract_inverted_index.the | 62, 98, 106, 124, 160 |
| abstract_inverted_index.drug | 155 |
| abstract_inverted_index.form | 16 |
| abstract_inverted_index.mTOR | 167 |
| abstract_inverted_index.mode | 10 |
| abstract_inverted_index.that | 145, 157 |
| abstract_inverted_index.they | 15 |
| abstract_inverted_index.well | 165 |
| abstract_inverted_index.with | 7, 19, 32, 67 |
| abstract_inverted_index.FK506 | 2 |
| abstract_inverted_index.build | 50 |
| abstract_inverted_index.cells | 103 |
| abstract_inverted_index.class | 150 |
| abstract_inverted_index.human | 102 |
| abstract_inverted_index.leads | 156 |
| abstract_inverted_index.model | 135 |
| abstract_inverted_index.their | 33 |
| abstract_inverted_index.these | 142 |
| abstract_inverted_index.this, | 46 |
| abstract_inverted_index.using | 79, 92 |
| abstract_inverted_index.which | 14 |
| abstract_inverted_index.(FKBP) | 22 |
| abstract_inverted_index.(mTOR) | 40 |
| abstract_inverted_index.(named | 90 |
| abstract_inverted_index.animal | 134 |
| abstract_inverted_index.before | 28 |
| abstract_inverted_index.beyond | 166 |
| abstract_inverted_index.binary | 17 |
| abstract_inverted_index.domain | 27, 64 |
| abstract_inverted_index.expand | 159 |
| abstract_inverted_index.hybrid | 88 |
| abstract_inverted_index.kidney | 137 |
| abstract_inverted_index.method | 78 |
| abstract_inverted_index.probes | 153 |
| abstract_inverted_index.robust | 76 |
| abstract_inverted_index.shared | 25 |
| abstract_inverted_index.sought | 48 |
| abstract_inverted_index.target | 37, 56 |
| abstract_inverted_index.action, | 12 |
| abstract_inverted_index.forming | 29 |
| abstract_inverted_index.injury. | 140 |
| abstract_inverted_index.library | 54, 70, 86, 100 |
| abstract_inverted_index.natural | 5 |
| abstract_inverted_index.potent, | 118 |
| abstract_inverted_index.protein | 21, 163 |
| abstract_inverted_index.results | 143 |
| abstract_inverted_index.targets | 164 |
| abstract_inverted_index.ternary | 30 |
| abstract_inverted_index.through | 23 |
| abstract_inverted_index.uptake. | 114 |
| abstract_inverted_index.Inspired | 44 |
| abstract_inverted_index.cellular | 58 |
| abstract_inverted_index.chemical | 152 |
| abstract_inverted_index.domains. | 95 |
| abstract_inverted_index.effector | 63 |
| abstract_inverted_index.products | 6 |
| abstract_inverted_index.proteins | 59 |
| abstract_inverted_index.targets, | 35 |
| abstract_inverted_index.Rapadocin | 115 |
| abstract_inverted_index.Screening | 96 |
| abstract_inverted_index.Together, | 141 |
| abstract_inverted_index.complexes | 18, 31 |
| abstract_inverted_index.developed | 74 |
| abstract_inverted_index.discovery | 107 |
| abstract_inverted_index.inhibitor | 111, 122 |
| abstract_inverted_index.ischaemia | 138 |
| abstract_inverted_index.optimized | 93 |
| abstract_inverted_index.rapafucin | 99 |
| abstract_inverted_index.rapamycin | 39, 66 |
| abstract_inverted_index.replacing | 61 |
| abstract_inverted_index.metathesis | 81 |
| abstract_inverted_index.nucleoside | 113, 126 |
| abstract_inverted_index.rapadocin, | 109 |
| abstract_inverted_index.rapafucins | 146 |
| abstract_inverted_index.repertoire | 161 |
| abstract_inverted_index.respective | 34 |
| abstract_inverted_index.demonstrate | 144 |
| abstract_inverted_index.efficacious | 131 |
| abstract_inverted_index.macrocycles | 89 |
| abstract_inverted_index.macrocyclic | 4 |
| abstract_inverted_index.mechanistic | 36 |
| abstract_inverted_index.rapafucins) | 91 |
| abstract_inverted_index.reperfusion | 139 |
| abstract_inverted_index.synthesized | 83 |
| abstract_inverted_index.transporter | 127 |
| abstract_inverted_index.FKBP-binding | 26, 94 |
| abstract_inverted_index.calcineurin, | 42 |
| abstract_inverted_index.ring-closing | 80 |
| abstract_inverted_index.FK506-binding | 20 |
| abstract_inverted_index.combinatorial | 69 |
| abstract_inverted_index.equilibrative | 125 |
| abstract_inverted_index.extraordinary | 9 |
| abstract_inverted_index.macromolecule | 53 |
| abstract_inverted_index.respectively. | 43 |
| abstract_inverted_index.FKBP-dependent | 121 |
| abstract_inverted_index.oligopeptides. | 72 |
| abstract_inverted_index.rapamycin-like | 52 |
| abstract_inverted_index.45,000-compound | 85 |
| abstract_inverted_index.isoform-specific | 119 |
| abstract_inverted_index.macrocyclization | 77 |
| abstract_inverted_index.<p>Rapamycin | 0 |
| abstract_inverted_index.calcineurin.</p> | 169 |
| cited_by_percentile_year | |
| countries_distinct_count | 0 |
| institutions_distinct_count | 19 |
| citation_normalized_percentile.value | 0.00725284 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |