Robust detection of SARS-CoV-2 exposure in population using T-cell repertoire profiling Article Swipe
 YOU?
·
· 2025
· Open Access
·
· DOI: https://doi.org/10.5281/zenodo.17780804
YOU?
·
· 2025
· Open Access
·
· DOI: https://doi.org/10.5281/zenodo.17780804
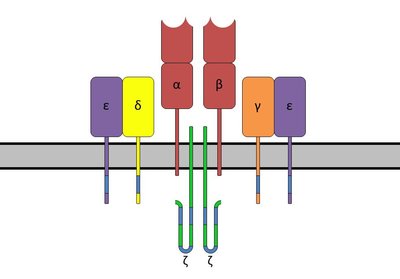
The dataset contains processed T-cell receptor repertoire sequencing data from >1200 individuals of different sex and age. Note that only samples with good sequencing coverage are published (>10^5 reads per file). The main aim of our study is to find TCR sequence biomarkers and develop a bioinformatic pipeline that allows building an accurate and robust classifier that distinguishes COVID-19-convalescent donors from unexposed individuals. We performed immunosequencing of the rearranged TCR α and β regions for PBMCs. For the cohort described in this study (Cohort-I) we sequence both chains of the TCR heterodimer as both of these chains are required to properly predict antigen recognition26. We ran conventional T-cell repertoire data analysis and pre-processed data to remove low-coverage samples. Of samples in Cohort-I which passed read count threshold, 383/377 TCR α/β samples were from healthy donors (SARS-CoV-2 PCR test negative or obtained prior to pandemic) and 890/848 were from COVID-19-positive patients. The majority of samples were accompanied by information on HLA class I and II alleles. Samples were prepared and sequenced in nine batches. The metadata for both TCR alpha and beta repertoires contains the following information: batch_name - one of the 9 unique batch identifiers sample_id, patient_id - information on sample identifier and donor identifier COVID_status, COVID_IgG, COVID_IgM - information on COVID-19 status HLA-A.1, HLA-A.2, HLA-B.1, HLA-B.2, HLA-C.1, HLA-C.2 - MHC class I alleles HLA-DPB1.1, HLA-DPB1.2, HLA-DQB1.1, HLA-DQB1.2, HLA-DRB1.1, HLA-DRB1.2 - MHC class II alleles Each file in TRA.zip/TRB.zip archive stores the information on either TCR alpha or beta repertoire. Each line in a file corresponds to the unique clonotype and each clonotype is accompanied with the following information: count - number of reads where the clonotype was detected freq - count of reads with the clonotype divided by thw whole number of reads in a sample cdr3nt, cdr3aa - nucleotide and amino acid sequences of TCR's CDR3 sequence v, d, j - the V/D/J segment name which was used for the clonotype's rearrangement VEnd, DStart, DEnd, JStart - information on VDJ junction positions distance_from_biomarker - information of the Levenstein distance to the closest COVID-19 biomarker, a cutoff of distance <= 1 for CDR3aa was used We proceed with selecting a set of CDR3 sequences that can serve as biomarkers and form a feature list for COVID-19 status classifier. We also validate the resulting set of clonotypes in several ways. Co-occurence of specific TCR α and β clonotypes can serve as an independent validation for biomarkers and their co-association with some specific pathogen. Additional information on donor HLAs is provided to filter the set of biomarkers based on HLA restriction: association with donor HLA serves as an additional evidence for TCR specificity to a specific set of antigens presented in a given donor and allows detecting the fingerprint of past and present infection. Furthermore, clonotypes with similar sequences can be aggregated into 'metaclonotype' biomarkers based on clonotype graph analysis. Finally, we train various COVID-19 status classifiers on selected batches from Cohort-I data using different algorithms and incorporating different feature sets. Verification of the robustness of our results was performed using independent batches of the Cohort-I and data from Cohort-II published previously.
Related Topics
- Type
- dataset
- Landing Page
- https://doi.org/10.5281/zenodo.17780804
- OA Status
- green
- OpenAlex ID
- https://openalex.org/W7108215880
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W7108215880Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.5281/zenodo.17780804Digital Object Identifier
- Title
-
Robust detection of SARS-CoV-2 exposure in population using T-cell repertoire profilingWork title
- Type
-
datasetOpenAlex work type
- Publication year
-
2025Year of publication
- Publication date
-
2025-12-01Full publication date if available
- Authors
-
Genomics Of Adaptive Immunity LaboratoryList of authors in order
- Landing page
-
https://doi.org/10.5281/zenodo.17780804Publisher landing page
- Open access
-
YesWhether a free full text is available
- OA status
-
greenOpen access status per OpenAlex
- OA URL
-
https://doi.org/10.5281/zenodo.17780804Direct OA link when available
- Concepts
-
T-cell receptor, Repertoire, Biology, Computational biology, Population, Major histocompatibility complex, Identifier, Genetics, Human leukocyte antigen, Classifier (UML), Computer science, Allele, Population stratification, Sequence analysis, Annotation, Phylogenetic tree, False positive paradox, Metadata, MHC class I, Polymerase chain reaction, DNA sequencingTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
0Total citation count in OpenAlex
Full payload
| id | https://openalex.org/W7108215880 |
|---|---|
| doi | https://doi.org/10.5281/zenodo.17780804 |
| ids.doi | https://doi.org/10.5281/zenodo.17780804 |
| ids.openalex | https://openalex.org/W7108215880 |
| fwci | |
| type | dataset |
| title | Robust detection of SARS-CoV-2 exposure in population using T-cell repertoire profiling |
| biblio.issue | |
| biblio.volume | |
| biblio.last_page | |
| biblio.first_page | |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C19317047 |
| concepts[0].level | 4 |
| concepts[0].score | 0.7478038668632507 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q412037 |
| concepts[0].display_name | T-cell receptor |
| concepts[1].id | https://openalex.org/C2778473898 |
| concepts[1].level | 2 |
| concepts[1].score | 0.71787428855896 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q2145110 |
| concepts[1].display_name | Repertoire |
| concepts[2].id | https://openalex.org/C86803240 |
| concepts[2].level | 0 |
| concepts[2].score | 0.5615885257720947 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[2].display_name | Biology |
| concepts[3].id | https://openalex.org/C70721500 |
| concepts[3].level | 1 |
| concepts[3].score | 0.5593005418777466 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q177005 |
| concepts[3].display_name | Computational biology |
| concepts[4].id | https://openalex.org/C2908647359 |
| concepts[4].level | 2 |
| concepts[4].score | 0.505902111530304 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q2625603 |
| concepts[4].display_name | Population |
| concepts[5].id | https://openalex.org/C207936829 |
| concepts[5].level | 3 |
| concepts[5].score | 0.48665663599967957 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q423163 |
| concepts[5].display_name | Major histocompatibility complex |
| concepts[6].id | https://openalex.org/C154504017 |
| concepts[6].level | 2 |
| concepts[6].score | 0.4629485607147217 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q853614 |
| concepts[6].display_name | Identifier |
| concepts[7].id | https://openalex.org/C54355233 |
| concepts[7].level | 1 |
| concepts[7].score | 0.45738494396209717 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[7].display_name | Genetics |
| concepts[8].id | https://openalex.org/C188280979 |
| concepts[8].level | 3 |
| concepts[8].score | 0.44162410497665405 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q911125 |
| concepts[8].display_name | Human leukocyte antigen |
| concepts[9].id | https://openalex.org/C95623464 |
| concepts[9].level | 2 |
| concepts[9].score | 0.37191760540008545 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q1096149 |
| concepts[9].display_name | Classifier (UML) |
| concepts[10].id | https://openalex.org/C41008148 |
| concepts[10].level | 0 |
| concepts[10].score | 0.350879043340683 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q21198 |
| concepts[10].display_name | Computer science |
| concepts[11].id | https://openalex.org/C180754005 |
| concepts[11].level | 3 |
| concepts[11].score | 0.3497229814529419 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q80726 |
| concepts[11].display_name | Allele |
| concepts[12].id | https://openalex.org/C166976648 |
| concepts[12].level | 5 |
| concepts[12].score | 0.33223021030426025 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q7229824 |
| concepts[12].display_name | Population stratification |
| concepts[13].id | https://openalex.org/C61053724 |
| concepts[13].level | 3 |
| concepts[13].score | 0.32761862874031067 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q1154615 |
| concepts[13].display_name | Sequence analysis |
| concepts[14].id | https://openalex.org/C2776321320 |
| concepts[14].level | 2 |
| concepts[14].score | 0.29113098978996277 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q857525 |
| concepts[14].display_name | Annotation |
| concepts[15].id | https://openalex.org/C193252679 |
| concepts[15].level | 3 |
| concepts[15].score | 0.28803595900535583 |
| concepts[15].wikidata | https://www.wikidata.org/wiki/Q242125 |
| concepts[15].display_name | Phylogenetic tree |
| concepts[16].id | https://openalex.org/C64869954 |
| concepts[16].level | 2 |
| concepts[16].score | 0.28523585200309753 |
| concepts[16].wikidata | https://www.wikidata.org/wiki/Q1859747 |
| concepts[16].display_name | False positive paradox |
| concepts[17].id | https://openalex.org/C93518851 |
| concepts[17].level | 2 |
| concepts[17].score | 0.27350759506225586 |
| concepts[17].wikidata | https://www.wikidata.org/wiki/Q180160 |
| concepts[17].display_name | Metadata |
| concepts[18].id | https://openalex.org/C170627219 |
| concepts[18].level | 4 |
| concepts[18].score | 0.26560306549072266 |
| concepts[18].wikidata | https://www.wikidata.org/wiki/Q13218497 |
| concepts[18].display_name | MHC class I |
| concepts[19].id | https://openalex.org/C49105822 |
| concepts[19].level | 3 |
| concepts[19].score | 0.26485979557037354 |
| concepts[19].wikidata | https://www.wikidata.org/wiki/Q176996 |
| concepts[19].display_name | Polymerase chain reaction |
| concepts[20].id | https://openalex.org/C51679486 |
| concepts[20].level | 3 |
| concepts[20].score | 0.25053006410598755 |
| concepts[20].wikidata | https://www.wikidata.org/wiki/Q380546 |
| concepts[20].display_name | DNA sequencing |
| keywords[0].id | https://openalex.org/keywords/t-cell-receptor |
| keywords[0].score | 0.7478038668632507 |
| keywords[0].display_name | T-cell receptor |
| keywords[1].id | https://openalex.org/keywords/repertoire |
| keywords[1].score | 0.71787428855896 |
| keywords[1].display_name | Repertoire |
| keywords[2].id | https://openalex.org/keywords/population |
| keywords[2].score | 0.505902111530304 |
| keywords[2].display_name | Population |
| keywords[3].id | https://openalex.org/keywords/major-histocompatibility-complex |
| keywords[3].score | 0.48665663599967957 |
| keywords[3].display_name | Major histocompatibility complex |
| keywords[4].id | https://openalex.org/keywords/identifier |
| keywords[4].score | 0.4629485607147217 |
| keywords[4].display_name | Identifier |
| keywords[5].id | https://openalex.org/keywords/human-leukocyte-antigen |
| keywords[5].score | 0.44162410497665405 |
| keywords[5].display_name | Human leukocyte antigen |
| keywords[6].id | https://openalex.org/keywords/classifier |
| keywords[6].score | 0.37191760540008545 |
| keywords[6].display_name | Classifier (UML) |
| keywords[7].id | https://openalex.org/keywords/allele |
| keywords[7].score | 0.3497229814529419 |
| keywords[7].display_name | Allele |
| language | |
| locations[0].id | doi:10.5281/zenodo.17780804 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4306400562 |
| locations[0].source.issn | |
| locations[0].source.type | repository |
| locations[0].source.is_oa | True |
| locations[0].source.issn_l | |
| locations[0].source.is_core | False |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | Zenodo (CERN European Organization for Nuclear Research) |
| locations[0].source.host_organization | https://openalex.org/I67311998 |
| locations[0].source.host_organization_name | European Organization for Nuclear Research |
| locations[0].source.host_organization_lineage | https://openalex.org/I67311998 |
| locations[0].license | |
| locations[0].pdf_url | |
| locations[0].version | |
| locations[0].raw_type | dataset |
| locations[0].license_id | |
| locations[0].is_accepted | False |
| locations[0].is_published | |
| locations[0].raw_source_name | |
| locations[0].landing_page_url | https://doi.org/10.5281/zenodo.17780804 |
| indexed_in | datacite |
| authorships[0].author.id | https://openalex.org/A5094320479 |
| authorships[0].author.orcid | |
| authorships[0].author.display_name | Genomics Of Adaptive Immunity Laboratory |
| authorships[0].affiliations[0].raw_affiliation_string | Institute of Bioorganic Chemistry |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Genomics of Adaptive Immunity Laboratory |
| authorships[0].is_corresponding | True |
| authorships[0].raw_affiliation_strings | Institute of Bioorganic Chemistry |
| has_content.pdf | False |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://doi.org/10.5281/zenodo.17780804 |
| open_access.oa_status | green |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-12-03T00:00:00 |
| display_name | Robust detection of SARS-CoV-2 exposure in population using T-cell repertoire profiling |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-12-03T00:07:38.036990 |
| primary_topic | |
| cited_by_count | 0 |
| locations_count | 1 |
| best_oa_location.id | doi:10.5281/zenodo.17780804 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4306400562 |
| best_oa_location.source.issn | |
| best_oa_location.source.type | repository |
| best_oa_location.source.is_oa | True |
| best_oa_location.source.issn_l | |
| best_oa_location.source.is_core | False |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | Zenodo (CERN European Organization for Nuclear Research) |
| best_oa_location.source.host_organization | https://openalex.org/I67311998 |
| best_oa_location.source.host_organization_name | European Organization for Nuclear Research |
| best_oa_location.source.host_organization_lineage | https://openalex.org/I67311998 |
| best_oa_location.license | |
| best_oa_location.pdf_url | |
| best_oa_location.version | |
| best_oa_location.raw_type | dataset |
| best_oa_location.license_id | |
| best_oa_location.is_accepted | False |
| best_oa_location.is_published | False |
| best_oa_location.raw_source_name | |
| best_oa_location.landing_page_url | https://doi.org/10.5281/zenodo.17780804 |
| primary_location.id | doi:10.5281/zenodo.17780804 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4306400562 |
| primary_location.source.issn | |
| primary_location.source.type | repository |
| primary_location.source.is_oa | True |
| primary_location.source.issn_l | |
| primary_location.source.is_core | False |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | Zenodo (CERN European Organization for Nuclear Research) |
| primary_location.source.host_organization | https://openalex.org/I67311998 |
| primary_location.source.host_organization_name | European Organization for Nuclear Research |
| primary_location.source.host_organization_lineage | https://openalex.org/I67311998 |
| primary_location.license | |
| primary_location.pdf_url | |
| primary_location.version | |
| primary_location.raw_type | dataset |
| primary_location.license_id | |
| primary_location.is_accepted | False |
| primary_location.is_published | False |
| primary_location.raw_source_name | |
| primary_location.landing_page_url | https://doi.org/10.5281/zenodo.17780804 |
| publication_date | 2025-12-01 |
| publication_year | 2025 |
| referenced_works_count | 0 |
| abstract_inverted_index.- | 187, 197, 208, 219, 230, 270, 280, 299, 312, 328, 335 |
| abstract_inverted_index.1 | 351 |
| abstract_inverted_index.9 | 191 |
| abstract_inverted_index.I | 161, 222 |
| abstract_inverted_index.a | 45, 253, 295, 346, 360, 372, 443, 450 |
| abstract_inverted_index.j | 311 |
| abstract_inverted_index.<= | 350 |
| abstract_inverted_index.II | 163, 233 |
| abstract_inverted_index.Of | 118 |
| abstract_inverted_index.We | 63, 104, 356, 379 |
| abstract_inverted_index.an | 51, 401, 436 |
| abstract_inverted_index.as | 92, 368, 400, 435 |
| abstract_inverted_index.be | 469 |
| abstract_inverted_index.by | 156, 288 |
| abstract_inverted_index.d, | 310 |
| abstract_inverted_index.in | 80, 120, 170, 237, 252, 294, 387, 449 |
| abstract_inverted_index.is | 37, 263, 418 |
| abstract_inverted_index.of | 12, 34, 66, 88, 94, 152, 189, 272, 282, 292, 305, 337, 348, 362, 385, 391, 424, 446, 458, 501, 504, 512 |
| abstract_inverted_index.on | 158, 199, 210, 243, 330, 415, 427, 475, 486 |
| abstract_inverted_index.or | 139, 247 |
| abstract_inverted_index.to | 38, 99, 114, 142, 256, 341, 420, 442 |
| abstract_inverted_index.v, | 309 |
| abstract_inverted_index.we | 84, 480 |
| abstract_inverted_index.α | 70, 394 |
| abstract_inverted_index.β | 72, 396 |
| abstract_inverted_index.For | 76 |
| abstract_inverted_index.HLA | 159, 428, 433 |
| abstract_inverted_index.MHC | 220, 231 |
| abstract_inverted_index.PCR | 136 |
| abstract_inverted_index.TCR | 40, 69, 90, 128, 177, 245, 393, 440 |
| abstract_inverted_index.The | 0, 31, 150, 173 |
| abstract_inverted_index.VDJ | 331 |
| abstract_inverted_index.aim | 33 |
| abstract_inverted_index.and | 15, 43, 53, 71, 111, 144, 162, 168, 179, 202, 260, 301, 370, 395, 406, 453, 460, 495, 515 |
| abstract_inverted_index.are | 25, 97 |
| abstract_inverted_index.can | 366, 398, 468 |
| abstract_inverted_index.for | 74, 175, 320, 352, 375, 404, 439 |
| abstract_inverted_index.one | 188 |
| abstract_inverted_index.our | 35, 505 |
| abstract_inverted_index.per | 29 |
| abstract_inverted_index.ran | 105 |
| abstract_inverted_index.set | 361, 384, 423, 445 |
| abstract_inverted_index.sex | 14 |
| abstract_inverted_index.the | 67, 77, 89, 183, 190, 241, 257, 266, 275, 285, 313, 321, 338, 342, 382, 422, 456, 502, 513 |
| abstract_inverted_index.thw | 289 |
| abstract_inverted_index.was | 277, 318, 354, 507 |
| abstract_inverted_index.CDR3 | 307, 363 |
| abstract_inverted_index.Each | 235, 250 |
| abstract_inverted_index.HLAs | 417 |
| abstract_inverted_index.Note | 17 |
| abstract_inverted_index.acid | 303 |
| abstract_inverted_index.age. | 16 |
| abstract_inverted_index.also | 380 |
| abstract_inverted_index.beta | 180, 248 |
| abstract_inverted_index.both | 86, 93, 176 |
| abstract_inverted_index.data | 8, 109, 113, 491, 516 |
| abstract_inverted_index.each | 261 |
| abstract_inverted_index.file | 236, 254 |
| abstract_inverted_index.find | 39 |
| abstract_inverted_index.form | 371 |
| abstract_inverted_index.freq | 279 |
| abstract_inverted_index.from | 9, 60, 132, 147, 489, 517 |
| abstract_inverted_index.good | 22 |
| abstract_inverted_index.into | 471 |
| abstract_inverted_index.line | 251 |
| abstract_inverted_index.list | 374 |
| abstract_inverted_index.main | 32 |
| abstract_inverted_index.name | 316 |
| abstract_inverted_index.nine | 171 |
| abstract_inverted_index.only | 19 |
| abstract_inverted_index.past | 459 |
| abstract_inverted_index.read | 124 |
| abstract_inverted_index.some | 410 |
| abstract_inverted_index.test | 137 |
| abstract_inverted_index.that | 18, 48, 56, 365 |
| abstract_inverted_index.this | 81 |
| abstract_inverted_index.used | 319, 355 |
| abstract_inverted_index.were | 131, 146, 154, 166 |
| abstract_inverted_index.with | 21, 265, 284, 358, 409, 431, 465 |
| abstract_inverted_index.>1200 | 10 |
| abstract_inverted_index.DEnd, | 326 |
| abstract_inverted_index.TCR's | 306 |
| abstract_inverted_index.V/D/J | 314 |
| abstract_inverted_index.VEnd, | 324 |
| abstract_inverted_index.alpha | 178, 246 |
| abstract_inverted_index.amino | 302 |
| abstract_inverted_index.based | 426, 474 |
| abstract_inverted_index.batch | 193 |
| abstract_inverted_index.class | 160, 221, 232 |
| abstract_inverted_index.count | 125, 269, 281 |
| abstract_inverted_index.donor | 203, 416, 432, 452 |
| abstract_inverted_index.given | 451 |
| abstract_inverted_index.graph | 477 |
| abstract_inverted_index.prior | 141 |
| abstract_inverted_index.reads | 28, 273, 283, 293 |
| abstract_inverted_index.serve | 367, 399 |
| abstract_inverted_index.sets. | 499 |
| abstract_inverted_index.study | 36, 82 |
| abstract_inverted_index.their | 407 |
| abstract_inverted_index.these | 95 |
| abstract_inverted_index.train | 481 |
| abstract_inverted_index.using | 492, 509 |
| abstract_inverted_index.ways. | 389 |
| abstract_inverted_index.where | 274 |
| abstract_inverted_index.which | 122, 317 |
| abstract_inverted_index.whole | 290 |
| abstract_inverted_index.α/β | 129 |
| abstract_inverted_index.(>10^5 | 27 |
| abstract_inverted_index.CDR3aa | 353 |
| abstract_inverted_index.JStart | 327 |
| abstract_inverted_index.PBMCs. | 75 |
| abstract_inverted_index.T-cell | 4, 107 |
| abstract_inverted_index.allows | 49, 454 |
| abstract_inverted_index.cdr3aa | 298 |
| abstract_inverted_index.chains | 87, 96 |
| abstract_inverted_index.cohort | 78 |
| abstract_inverted_index.cutoff | 347 |
| abstract_inverted_index.donors | 59, 134 |
| abstract_inverted_index.either | 244 |
| abstract_inverted_index.file). | 30 |
| abstract_inverted_index.filter | 421 |
| abstract_inverted_index.number | 271, 291 |
| abstract_inverted_index.passed | 123 |
| abstract_inverted_index.remove | 115 |
| abstract_inverted_index.robust | 54 |
| abstract_inverted_index.sample | 200, 296 |
| abstract_inverted_index.serves | 434 |
| abstract_inverted_index.status | 212, 377, 484 |
| abstract_inverted_index.stores | 240 |
| abstract_inverted_index.unique | 192, 258 |
| abstract_inverted_index.383/377 | 127 |
| abstract_inverted_index.890/848 | 145 |
| abstract_inverted_index.DStart, | 325 |
| abstract_inverted_index.HLA-C.2 | 218 |
| abstract_inverted_index.Samples | 165 |
| abstract_inverted_index.alleles | 223, 234 |
| abstract_inverted_index.antigen | 102 |
| abstract_inverted_index.archive | 239 |
| abstract_inverted_index.batches | 488, 511 |
| abstract_inverted_index.cdr3nt, | 297 |
| abstract_inverted_index.closest | 343 |
| abstract_inverted_index.dataset | 1 |
| abstract_inverted_index.develop | 44 |
| abstract_inverted_index.divided | 287 |
| abstract_inverted_index.feature | 373, 498 |
| abstract_inverted_index.healthy | 133 |
| abstract_inverted_index.predict | 101 |
| abstract_inverted_index.present | 461 |
| abstract_inverted_index.proceed | 357 |
| abstract_inverted_index.regions | 73 |
| abstract_inverted_index.results | 506 |
| abstract_inverted_index.samples | 20, 119, 130, 153 |
| abstract_inverted_index.segment | 315 |
| abstract_inverted_index.several | 388 |
| abstract_inverted_index.similar | 466 |
| abstract_inverted_index.various | 482 |
| abstract_inverted_index.COVID-19 | 211, 344, 376, 483 |
| abstract_inverted_index.Cohort-I | 121, 490, 514 |
| abstract_inverted_index.Finally, | 479 |
| abstract_inverted_index.HLA-A.1, | 213 |
| abstract_inverted_index.HLA-A.2, | 214 |
| abstract_inverted_index.HLA-B.1, | 215 |
| abstract_inverted_index.HLA-B.2, | 216 |
| abstract_inverted_index.HLA-C.1, | 217 |
| abstract_inverted_index.accurate | 52 |
| abstract_inverted_index.alleles. | 164 |
| abstract_inverted_index.analysis | 110 |
| abstract_inverted_index.antigens | 447 |
| abstract_inverted_index.batches. | 172 |
| abstract_inverted_index.building | 50 |
| abstract_inverted_index.contains | 2, 182 |
| abstract_inverted_index.coverage | 24 |
| abstract_inverted_index.detected | 278 |
| abstract_inverted_index.distance | 340, 349 |
| abstract_inverted_index.evidence | 438 |
| abstract_inverted_index.junction | 332 |
| abstract_inverted_index.majority | 151 |
| abstract_inverted_index.metadata | 174 |
| abstract_inverted_index.negative | 138 |
| abstract_inverted_index.obtained | 140 |
| abstract_inverted_index.pipeline | 47 |
| abstract_inverted_index.prepared | 167 |
| abstract_inverted_index.properly | 100 |
| abstract_inverted_index.provided | 419 |
| abstract_inverted_index.receptor | 5 |
| abstract_inverted_index.required | 98 |
| abstract_inverted_index.samples. | 117 |
| abstract_inverted_index.selected | 487 |
| abstract_inverted_index.sequence | 41, 85, 308 |
| abstract_inverted_index.specific | 392, 411, 444 |
| abstract_inverted_index.validate | 381 |
| abstract_inverted_index.COVID_IgM | 207 |
| abstract_inverted_index.Cohort-II | 518 |
| abstract_inverted_index.analysis. | 478 |
| abstract_inverted_index.clonotype | 259, 262, 276, 286, 476 |
| abstract_inverted_index.described | 79 |
| abstract_inverted_index.detecting | 455 |
| abstract_inverted_index.different | 13, 493, 497 |
| abstract_inverted_index.following | 184, 267 |
| abstract_inverted_index.pandemic) | 143 |
| abstract_inverted_index.pathogen. | 412 |
| abstract_inverted_index.patients. | 149 |
| abstract_inverted_index.performed | 64, 508 |
| abstract_inverted_index.positions | 333 |
| abstract_inverted_index.presented | 448 |
| abstract_inverted_index.processed | 3 |
| abstract_inverted_index.published | 26, 519 |
| abstract_inverted_index.resulting | 383 |
| abstract_inverted_index.selecting | 359 |
| abstract_inverted_index.sequenced | 169 |
| abstract_inverted_index.sequences | 304, 364, 467 |
| abstract_inverted_index.unexposed | 61 |
| abstract_inverted_index.(Cohort-I) | 83 |
| abstract_inverted_index.Additional | 413 |
| abstract_inverted_index.COVID_IgG, | 206 |
| abstract_inverted_index.HLA-DRB1.2 | 229 |
| abstract_inverted_index.Levenstein | 339 |
| abstract_inverted_index.additional | 437 |
| abstract_inverted_index.aggregated | 470 |
| abstract_inverted_index.algorithms | 494 |
| abstract_inverted_index.batch_name | 186 |
| abstract_inverted_index.biomarker, | 345 |
| abstract_inverted_index.biomarkers | 42, 369, 405, 425, 473 |
| abstract_inverted_index.classifier | 55 |
| abstract_inverted_index.clonotypes | 386, 397, 464 |
| abstract_inverted_index.identifier | 201, 204 |
| abstract_inverted_index.infection. | 462 |
| abstract_inverted_index.nucleotide | 300 |
| abstract_inverted_index.patient_id | 196 |
| abstract_inverted_index.rearranged | 68 |
| abstract_inverted_index.repertoire | 6, 108 |
| abstract_inverted_index.robustness | 503 |
| abstract_inverted_index.sample_id, | 195 |
| abstract_inverted_index.sequencing | 7, 23 |
| abstract_inverted_index.threshold, | 126 |
| abstract_inverted_index.validation | 403 |
| abstract_inverted_index.(SARS-CoV-2 | 135 |
| abstract_inverted_index.HLA-DPB1.1, | 224 |
| abstract_inverted_index.HLA-DPB1.2, | 225 |
| abstract_inverted_index.HLA-DQB1.1, | 226 |
| abstract_inverted_index.HLA-DQB1.2, | 227 |
| abstract_inverted_index.HLA-DRB1.1, | 228 |
| abstract_inverted_index.accompanied | 155, 264 |
| abstract_inverted_index.association | 430 |
| abstract_inverted_index.classifier. | 378 |
| abstract_inverted_index.classifiers | 485 |
| abstract_inverted_index.clonotype's | 322 |
| abstract_inverted_index.corresponds | 255 |
| abstract_inverted_index.fingerprint | 457 |
| abstract_inverted_index.heterodimer | 91 |
| abstract_inverted_index.identifiers | 194 |
| abstract_inverted_index.independent | 402, 510 |
| abstract_inverted_index.individuals | 11 |
| abstract_inverted_index.information | 157, 198, 209, 242, 329, 336, 414 |
| abstract_inverted_index.previously. | 520 |
| abstract_inverted_index.repertoire. | 249 |
| abstract_inverted_index.repertoires | 181 |
| abstract_inverted_index.specificity | 441 |
| abstract_inverted_index.Co-occurence | 390 |
| abstract_inverted_index.Furthermore, | 463 |
| abstract_inverted_index.Verification | 500 |
| abstract_inverted_index.conventional | 106 |
| abstract_inverted_index.individuals. | 62 |
| abstract_inverted_index.information: | 185, 268 |
| abstract_inverted_index.low-coverage | 116 |
| abstract_inverted_index.restriction: | 429 |
| abstract_inverted_index.COVID_status, | 205 |
| abstract_inverted_index.bioinformatic | 46 |
| abstract_inverted_index.distinguishes | 57 |
| abstract_inverted_index.incorporating | 496 |
| abstract_inverted_index.pre-processed | 112 |
| abstract_inverted_index.rearrangement | 323 |
| abstract_inverted_index.co-association | 408 |
| abstract_inverted_index.recognition26. | 103 |
| abstract_inverted_index.'metaclonotype' | 472 |
| abstract_inverted_index.TRA.zip/TRB.zip | 238 |
| abstract_inverted_index.immunosequencing | 65 |
| abstract_inverted_index.COVID-19-positive | 148 |
| abstract_inverted_index.COVID-19-convalescent | 58 |
| abstract_inverted_index.distance_from_biomarker | 334 |
| cited_by_percentile_year | |
| countries_distinct_count | 0 |
| institutions_distinct_count | 1 |
| citation_normalized_percentile |