Scope of Liquid Biopsy in Urological Malignancies Article Swipe
 YOU?
·
· 2021
· Open Access
·
· DOI: https://doi.org/10.4103/jss.jss_23_21
YOU?
·
· 2021
· Open Access
·
· DOI: https://doi.org/10.4103/jss.jss_23_21
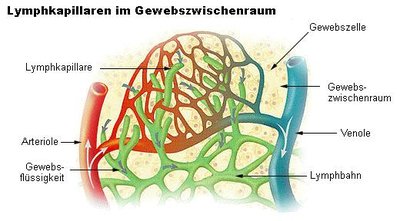
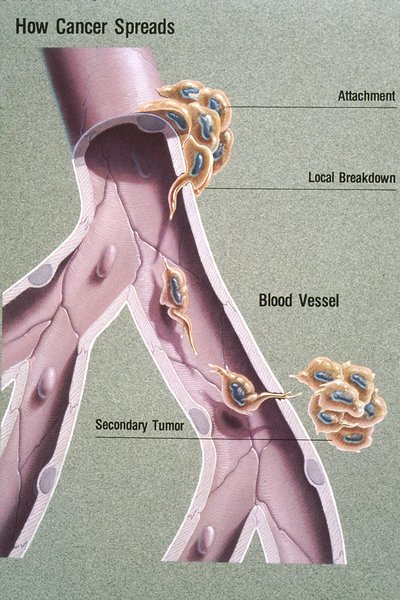
Due to the recent rise in the trend of urological malignancies, noninvasive tumor biomarkers are being researched and used for many clinical settings. Thus, the identification of specific and effective biomarkers in the form of liquid biopsy has become a point of major focus. The term liquid biopsy refers to the use of body fluids and/or blood as a surrogate of tissue samples for diagnostic purposes. The rationale for using liquid biopsies instead of actual tissue samples is to avoid redundant invasive procedures when providing the same diagnostic information.[1] With the advancement of personalized cancer therapy, samples obtained from the cancerous tissues are not only diagnostic but also serve as predictive biomarkers. Thus, there is increasing demand for the detection of the ever-expanding repertoire of biomarkers. Furthermore, when patients develop resistance to targeted therapy, obtaining additional diagnostic samples, preferably from the resistant clone, is critical to delineate the resistance mechanism in guiding appropriate treatment. Liquid biopsy involves cancer categorization by obtaining biomolecules such as cell-free DNA (cfDNA), circulating tumor cells (CTCs), RNAs (microRNAs, long noncoding RNAs [lncRNAs], and messenger RNAs), cell-free proteins, peptides, and exosomes in body fluids. The myriad of body fluids include peripheral blood, urine, cerebrospinal fluid, or effusion fluids. Tumor cells can reach the bloodstream from their original location through blood and/or lymphatic vessels located in the tumor stroma or by tumor vascular invasion. Via the lymphatics, tumor cells may reach the bloodstream, hence rendering CTCs and cfDNA detectable.[2] Tumor itself induces angiogenesis and also forms sinusoidal spaces where tumor cells protrude into the lumen. Cell components, including ctDNA, are shed by the tumor itself into the bloodstream. Tumor cells, their proteins and nucleic acids become detectable in urine when these are shed from tumors like urothelial neoplasm arising from renal pelvis, ureter, bladder and urethra. RCCs and prostate carcinomas too shed these.[3] The kidney barrier is permeable only to low-molecular-weight DNA fragments and therefore the mean size of the cfDNA fragments released in the urine is approximately 50–100 base pairs (70 KDa). This information is relevant to assess the source of urinary free DNA. The high-molecular-weight DNA found in urine speculates DNA derived from the whole cells released by the urothelium or renal tubules.[4] Since prostate gland drains into the urethra, the genetic material from prostate cancer that has spread within preexisting ducts and acini is easily detected in voided urine. Tumors release DNA fragments into circulation that contain tumor-specific alterations including point mutations, copy-number variation, and DNA methylation. Casadio et al.[5] showed elevated urine cfDNA integrity in patients with prostate adenocarcinoma as 79% sensitive and 84% specific. Classic methods for cfDNA assessment include polymerase chain reaction (PCR)-based approaches. More recently, digital PCR has emerged as a sensitive tool for the detection of point mutations in cfDNA. Targeted and whole genome sequencing technologies are increasingly being applied to cfDNA analysis. CTCs in peripheral blood may occur as one cell/100 million cells that are circulating in the blood originating from the primary tumor or metastatic foci. CTC counts predict disease burden, whereas changes in CTC counts during the course of systemic therapy indicate treatment response. Patients of renal cell carcinoma with advanced stage and aggressive phenotypes showed elevated blood CTC levels correlating with lymph node status and presence of metastasis.[6] The Food and Drug Administration has approved the CELLSEARCH® CTC Test to monitor patients of metastatic prostate cancer. This test counts CTCs of epithelial origin (CD45-, EPCAM+, and cytokeratins 8, 18+, and/or 19+) in whole blood. LncRNAs are >200 nucleotides long that regulate gene expression at transcriptional, posttranscriptional, and/or epigenetic levels. These get altered in cancers promoting tumor formation, progression, and metastasis. Urinary sediments with elevated UCA1 levels have been identified as a potential diagnostic marker for bladder tumors. Precise molecular biomarkers are, hence, urgently needed in the era of personalized medicine to support clinical decision-making. Excisional biopsies do not truly demonstrate tumor heterogeneity, both within a tumor and between primary tumor and metastases. Moreover, during monitoring/therapy, repeated biopsy is often not feasible, risky, and painful for the patient. In contrast, a liquid biopsy reflects the genetic profile of all tumor subclones present in a patient.[7] It also gives an opportunity to take serial samples to monitor tumoral changes. Meanwhile, it allows the clinician to administer a personalized therapy to the patients.[8] Liquid biopsy is capable of replacing or at least augmenting the use of invasive biopsy which has limited success and associated complications. It is clear that we are moving into an era of precision medicine, where treatment is tailored based on tumor behavior rather than the average response to therapy. Molecular profiling, the global analysis of genomic, transcriptomic, and/or proteomic profiles, represents a critical prerequisite for the successful development of individualized treatment strategies. Liquid biopsy is a promising noninvasive tool for molecular profiling, enabling assessment of cfDNA and other circulating molecules in various biological fluids for biomarker discovery. So far, the most exciting applications of liquid biopsies seem to be prognosis and early assessment of treatment failure.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.4103/jss.jss_23_21
- OA Status
- diamond
- References
- 9
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W3168165204
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W3168165204Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.4103/jss.jss_23_21Digital Object Identifier
- Title
-
Scope of Liquid Biopsy in Urological MalignanciesWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2021Year of publication
- Publication date
-
2021-01-01Full publication date if available
- Authors
-
RajendraB Nerli, Shreya ChandraList of authors in order
- Landing page
-
https://doi.org/10.4103/jss.jss_23_21Publisher landing page
- Open access
-
YesWhether a free full text is available
- OA status
-
diamondOpen access status per OpenAlex
- OA URL
-
https://doi.org/10.4103/jss.jss_23_21Direct OA link when available
- Concepts
-
Liquid biopsy, Medicine, Circulating tumor cell, Cell-free fetal DNA, Pathology, Biopsy, Lymphatic system, Exosome, Cancer, Microvesicles, microRNA, Biology, Internal medicine, Metastasis, Gene, Genetics, Prenatal diagnosis, Fetus, Pregnancy, BiochemistryTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
0Total citation count in OpenAlex
- References (count)
-
9Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W3168165204 |
|---|---|
| doi | https://doi.org/10.4103/jss.jss_23_21 |
| ids.doi | https://doi.org/10.4103/jss.jss_23_21 |
| ids.mag | 3168165204 |
| ids.openalex | https://openalex.org/W3168165204 |
| fwci | 0.0 |
| type | article |
| title | Scope of Liquid Biopsy in Urological Malignancies |
| biblio.issue | 1 |
| biblio.volume | 48 |
| biblio.last_page | 2 |
| biblio.first_page | 1 |
| topics[0].id | https://openalex.org/T11287 |
| topics[0].field.id | https://openalex.org/fields/13 |
| topics[0].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[0].score | 0.9993000030517578 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1306 |
| topics[0].subfield.display_name | Cancer Research |
| topics[0].display_name | Cancer Genomics and Diagnostics |
| topics[1].id | https://openalex.org/T10449 |
| topics[1].field.id | https://openalex.org/fields/27 |
| topics[1].field.display_name | Medicine |
| topics[1].score | 0.998199999332428 |
| topics[1].domain.id | https://openalex.org/domains/4 |
| topics[1].domain.display_name | Health Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/2740 |
| topics[1].subfield.display_name | Pulmonary and Respiratory Medicine |
| topics[1].display_name | Renal cell carcinoma treatment |
| topics[2].id | https://openalex.org/T10458 |
| topics[2].field.id | https://openalex.org/fields/27 |
| topics[2].field.display_name | Medicine |
| topics[2].score | 0.980400025844574 |
| topics[2].domain.id | https://openalex.org/domains/4 |
| topics[2].domain.display_name | Health Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/2746 |
| topics[2].subfield.display_name | Surgery |
| topics[2].display_name | Bladder and Urothelial Cancer Treatments |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C2779529041 |
| concepts[0].level | 3 |
| concepts[0].score | 0.857001781463623 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q22349364 |
| concepts[0].display_name | Liquid biopsy |
| concepts[1].id | https://openalex.org/C71924100 |
| concepts[1].level | 0 |
| concepts[1].score | 0.7554597854614258 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q11190 |
| concepts[1].display_name | Medicine |
| concepts[2].id | https://openalex.org/C61238886 |
| concepts[2].level | 4 |
| concepts[2].score | 0.6876025199890137 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q5121689 |
| concepts[2].display_name | Circulating tumor cell |
| concepts[3].id | https://openalex.org/C152110520 |
| concepts[3].level | 5 |
| concepts[3].score | 0.5613117814064026 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q14225730 |
| concepts[3].display_name | Cell-free fetal DNA |
| concepts[4].id | https://openalex.org/C142724271 |
| concepts[4].level | 1 |
| concepts[4].score | 0.5586827993392944 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q7208 |
| concepts[4].display_name | Pathology |
| concepts[5].id | https://openalex.org/C2775934546 |
| concepts[5].level | 2 |
| concepts[5].score | 0.5247523784637451 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q179991 |
| concepts[5].display_name | Biopsy |
| concepts[6].id | https://openalex.org/C181152851 |
| concepts[6].level | 2 |
| concepts[6].score | 0.47885772585868835 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q712604 |
| concepts[6].display_name | Lymphatic system |
| concepts[7].id | https://openalex.org/C2781261824 |
| concepts[7].level | 5 |
| concepts[7].score | 0.4681209623813629 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q903634 |
| concepts[7].display_name | Exosome |
| concepts[8].id | https://openalex.org/C121608353 |
| concepts[8].level | 2 |
| concepts[8].score | 0.4331378638744354 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q12078 |
| concepts[8].display_name | Cancer |
| concepts[9].id | https://openalex.org/C20518536 |
| concepts[9].level | 4 |
| concepts[9].score | 0.41499146819114685 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q6840469 |
| concepts[9].display_name | Microvesicles |
| concepts[10].id | https://openalex.org/C145059251 |
| concepts[10].level | 3 |
| concepts[10].score | 0.38893938064575195 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q310899 |
| concepts[10].display_name | microRNA |
| concepts[11].id | https://openalex.org/C86803240 |
| concepts[11].level | 0 |
| concepts[11].score | 0.14808595180511475 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[11].display_name | Biology |
| concepts[12].id | https://openalex.org/C126322002 |
| concepts[12].level | 1 |
| concepts[12].score | 0.13543078303337097 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q11180 |
| concepts[12].display_name | Internal medicine |
| concepts[13].id | https://openalex.org/C2779013556 |
| concepts[13].level | 3 |
| concepts[13].score | 0.11469092965126038 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q181876 |
| concepts[13].display_name | Metastasis |
| concepts[14].id | https://openalex.org/C104317684 |
| concepts[14].level | 2 |
| concepts[14].score | 0.10608449578285217 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[14].display_name | Gene |
| concepts[15].id | https://openalex.org/C54355233 |
| concepts[15].level | 1 |
| concepts[15].score | 0.0 |
| concepts[15].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[15].display_name | Genetics |
| concepts[16].id | https://openalex.org/C2778258057 |
| concepts[16].level | 4 |
| concepts[16].score | 0.0 |
| concepts[16].wikidata | https://www.wikidata.org/wiki/Q1136569 |
| concepts[16].display_name | Prenatal diagnosis |
| concepts[17].id | https://openalex.org/C172680121 |
| concepts[17].level | 3 |
| concepts[17].score | 0.0 |
| concepts[17].wikidata | https://www.wikidata.org/wiki/Q26513 |
| concepts[17].display_name | Fetus |
| concepts[18].id | https://openalex.org/C2779234561 |
| concepts[18].level | 2 |
| concepts[18].score | 0.0 |
| concepts[18].wikidata | https://www.wikidata.org/wiki/Q11995 |
| concepts[18].display_name | Pregnancy |
| concepts[19].id | https://openalex.org/C55493867 |
| concepts[19].level | 1 |
| concepts[19].score | 0.0 |
| concepts[19].wikidata | https://www.wikidata.org/wiki/Q7094 |
| concepts[19].display_name | Biochemistry |
| keywords[0].id | https://openalex.org/keywords/liquid-biopsy |
| keywords[0].score | 0.857001781463623 |
| keywords[0].display_name | Liquid biopsy |
| keywords[1].id | https://openalex.org/keywords/medicine |
| keywords[1].score | 0.7554597854614258 |
| keywords[1].display_name | Medicine |
| keywords[2].id | https://openalex.org/keywords/circulating-tumor-cell |
| keywords[2].score | 0.6876025199890137 |
| keywords[2].display_name | Circulating tumor cell |
| keywords[3].id | https://openalex.org/keywords/cell-free-fetal-dna |
| keywords[3].score | 0.5613117814064026 |
| keywords[3].display_name | Cell-free fetal DNA |
| keywords[4].id | https://openalex.org/keywords/pathology |
| keywords[4].score | 0.5586827993392944 |
| keywords[4].display_name | Pathology |
| keywords[5].id | https://openalex.org/keywords/biopsy |
| keywords[5].score | 0.5247523784637451 |
| keywords[5].display_name | Biopsy |
| keywords[6].id | https://openalex.org/keywords/lymphatic-system |
| keywords[6].score | 0.47885772585868835 |
| keywords[6].display_name | Lymphatic system |
| keywords[7].id | https://openalex.org/keywords/exosome |
| keywords[7].score | 0.4681209623813629 |
| keywords[7].display_name | Exosome |
| keywords[8].id | https://openalex.org/keywords/cancer |
| keywords[8].score | 0.4331378638744354 |
| keywords[8].display_name | Cancer |
| keywords[9].id | https://openalex.org/keywords/microvesicles |
| keywords[9].score | 0.41499146819114685 |
| keywords[9].display_name | Microvesicles |
| keywords[10].id | https://openalex.org/keywords/microrna |
| keywords[10].score | 0.38893938064575195 |
| keywords[10].display_name | microRNA |
| keywords[11].id | https://openalex.org/keywords/biology |
| keywords[11].score | 0.14808595180511475 |
| keywords[11].display_name | Biology |
| keywords[12].id | https://openalex.org/keywords/internal-medicine |
| keywords[12].score | 0.13543078303337097 |
| keywords[12].display_name | Internal medicine |
| keywords[13].id | https://openalex.org/keywords/metastasis |
| keywords[13].score | 0.11469092965126038 |
| keywords[13].display_name | Metastasis |
| keywords[14].id | https://openalex.org/keywords/gene |
| keywords[14].score | 0.10608449578285217 |
| keywords[14].display_name | Gene |
| language | en |
| locations[0].id | doi:10.4103/jss.jss_23_21 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S2738012530 |
| locations[0].source.issn | 0974-5009, 2278-7127 |
| locations[0].source.type | journal |
| locations[0].source.is_oa | True |
| locations[0].source.issn_l | 0974-5009 |
| locations[0].source.is_core | False |
| locations[0].source.is_in_doaj | True |
| locations[0].source.display_name | Journal of the Scientific Society |
| locations[0].source.host_organization | https://openalex.org/P4310320448 |
| locations[0].source.host_organization_name | Medknow |
| locations[0].source.host_organization_lineage | https://openalex.org/P4310320448, https://openalex.org/P4310318547 |
| locations[0].source.host_organization_lineage_names | Medknow, Wolters Kluwer |
| locations[0].license | cc-by-nc-sa |
| locations[0].pdf_url | |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | https://openalex.org/licenses/cc-by-nc-sa |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | Journal of the Scientific Society |
| locations[0].landing_page_url | https://doi.org/10.4103/jss.jss_23_21 |
| locations[1].id | pmh:oai:doaj.org/article:7b9b223d6e014029a4005ef617420bfe |
| locations[1].is_oa | True |
| locations[1].source.id | https://openalex.org/S4306401280 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | DOAJ (DOAJ: Directory of Open Access Journals) |
| locations[1].source.host_organization | |
| locations[1].source.host_organization_name | |
| locations[1].license | cc-by-sa |
| locations[1].pdf_url | |
| locations[1].version | submittedVersion |
| locations[1].raw_type | article |
| locations[1].license_id | https://openalex.org/licenses/cc-by-sa |
| locations[1].is_accepted | False |
| locations[1].is_published | False |
| locations[1].raw_source_name | Journal of the Scientific Society, Vol 48, Iss 1, Pp 1-2 (2021) |
| locations[1].landing_page_url | https://doaj.org/article/7b9b223d6e014029a4005ef617420bfe |
| indexed_in | crossref, doaj |
| authorships[0].author.id | https://openalex.org/A5078485015 |
| authorships[0].author.orcid | |
| authorships[0].author.display_name | RajendraB Nerli |
| authorships[0].countries | GB, IN, US |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I4210120668 |
| authorships[0].affiliations[0].raw_affiliation_string | This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms. |
| authorships[0].affiliations[1].institution_ids | https://openalex.org/I4210125823 |
| authorships[0].affiliations[1].raw_affiliation_string | 1Department of Urology, JN Medical College, KLE Academy of Higher Education and Research, JNMC Campus, Belagavi, Karnataka, India |
| authorships[0].affiliations[2].institution_ids | https://openalex.org/I2799525281 |
| authorships[0].affiliations[2].raw_affiliation_string | Received in revised form March 29, 2021 |
| authorships[0].affiliations[3].raw_affiliation_string | Accepted March 30, 2021 |
| authorships[0].affiliations[4].raw_affiliation_string | Received March 25, 2021 |
| authorships[0].affiliations[5].institution_ids | https://openalex.org/I4210135707 |
| authorships[0].affiliations[5].raw_affiliation_string | Department of Urology, KLES Kidney Foundation, KLES Dr. Prabhakar Kore Hospital and Medical Research Centre, Belagavi, Karnataka, India |
| authorships[0].institutions[0].id | https://openalex.org/I2799525281 |
| authorships[0].institutions[0].ror | https://ror.org/04m4td435 |
| authorships[0].institutions[0].type | other |
| authorships[0].institutions[0].lineage | https://openalex.org/I2799525281 |
| authorships[0].institutions[0].country_code | GB |
| authorships[0].institutions[0].display_name | Christie's |
| authorships[0].institutions[1].id | https://openalex.org/I4210135707 |
| authorships[0].institutions[1].ror | https://ror.org/02sv2ex42 |
| authorships[0].institutions[1].type | healthcare |
| authorships[0].institutions[1].lineage | https://openalex.org/I4210135707 |
| authorships[0].institutions[1].country_code | IN |
| authorships[0].institutions[1].display_name | KLE Society Hospital |
| authorships[0].institutions[2].id | https://openalex.org/I4210125823 |
| authorships[0].institutions[2].ror | https://ror.org/03aam9155 |
| authorships[0].institutions[2].type | education |
| authorships[0].institutions[2].lineage | https://openalex.org/I4210125823 |
| authorships[0].institutions[2].country_code | IN |
| authorships[0].institutions[2].display_name | KLE University |
| authorships[0].institutions[3].id | https://openalex.org/I4210120668 |
| authorships[0].institutions[3].ror | https://ror.org/02ed4cj64 |
| authorships[0].institutions[3].type | nonprofit |
| authorships[0].institutions[3].lineage | https://openalex.org/I4210120668 |
| authorships[0].institutions[3].country_code | US |
| authorships[0].institutions[3].display_name | Creative Commons |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | RajendraB Nerli |
| authorships[0].is_corresponding | True |
| authorships[0].raw_affiliation_strings | 1Department of Urology, JN Medical College, KLE Academy of Higher Education and Research, JNMC Campus, Belagavi, Karnataka, India, Accepted March 30, 2021, Department of Urology, KLES Kidney Foundation, KLES Dr. Prabhakar Kore Hospital and Medical Research Centre, Belagavi, Karnataka, India, Received March 25, 2021, Received in revised form March 29, 2021, This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms. |
| authorships[1].author.id | https://openalex.org/A5034308055 |
| authorships[1].author.orcid | |
| authorships[1].author.display_name | Shreya Chandra |
| authorships[1].countries | GB, IN, US |
| authorships[1].affiliations[0].raw_affiliation_string | Accepted March 30, 2021 |
| authorships[1].affiliations[1].institution_ids | https://openalex.org/I4210125823 |
| authorships[1].affiliations[1].raw_affiliation_string | 1Department of Urology, JN Medical College, KLE Academy of Higher Education and Research, JNMC Campus, Belagavi, Karnataka, India |
| authorships[1].affiliations[2].raw_affiliation_string | Received March 25, 2021 |
| authorships[1].affiliations[3].institution_ids | https://openalex.org/I4210120668 |
| authorships[1].affiliations[3].raw_affiliation_string | This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms. |
| authorships[1].affiliations[4].institution_ids | https://openalex.org/I2799525281 |
| authorships[1].affiliations[4].raw_affiliation_string | Received in revised form March 29, 2021 |
| authorships[1].affiliations[5].institution_ids | https://openalex.org/I4210135707 |
| authorships[1].affiliations[5].raw_affiliation_string | Department of Urology, KLES Kidney Foundation, KLES Dr. Prabhakar Kore Hospital and Medical Research Centre, Belagavi, Karnataka, India |
| authorships[1].institutions[0].id | https://openalex.org/I2799525281 |
| authorships[1].institutions[0].ror | https://ror.org/04m4td435 |
| authorships[1].institutions[0].type | other |
| authorships[1].institutions[0].lineage | https://openalex.org/I2799525281 |
| authorships[1].institutions[0].country_code | GB |
| authorships[1].institutions[0].display_name | Christie's |
| authorships[1].institutions[1].id | https://openalex.org/I4210135707 |
| authorships[1].institutions[1].ror | https://ror.org/02sv2ex42 |
| authorships[1].institutions[1].type | healthcare |
| authorships[1].institutions[1].lineage | https://openalex.org/I4210135707 |
| authorships[1].institutions[1].country_code | IN |
| authorships[1].institutions[1].display_name | KLE Society Hospital |
| authorships[1].institutions[2].id | https://openalex.org/I4210125823 |
| authorships[1].institutions[2].ror | https://ror.org/03aam9155 |
| authorships[1].institutions[2].type | education |
| authorships[1].institutions[2].lineage | https://openalex.org/I4210125823 |
| authorships[1].institutions[2].country_code | IN |
| authorships[1].institutions[2].display_name | KLE University |
| authorships[1].institutions[3].id | https://openalex.org/I4210120668 |
| authorships[1].institutions[3].ror | https://ror.org/02ed4cj64 |
| authorships[1].institutions[3].type | nonprofit |
| authorships[1].institutions[3].lineage | https://openalex.org/I4210120668 |
| authorships[1].institutions[3].country_code | US |
| authorships[1].institutions[3].display_name | Creative Commons |
| authorships[1].author_position | last |
| authorships[1].raw_author_name | Shreya Chandra |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | 1Department of Urology, JN Medical College, KLE Academy of Higher Education and Research, JNMC Campus, Belagavi, Karnataka, India, Accepted March 30, 2021, Department of Urology, KLES Kidney Foundation, KLES Dr. Prabhakar Kore Hospital and Medical Research Centre, Belagavi, Karnataka, India, Received March 25, 2021, Received in revised form March 29, 2021, This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms. |
| has_content.pdf | False |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://doi.org/10.4103/jss.jss_23_21 |
| open_access.oa_status | diamond |
| open_access.any_repository_has_fulltext | False |
| created_date | 2021-06-22T00:00:00 |
| display_name | Scope of Liquid Biopsy in Urological Malignancies |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T11287 |
| primary_topic.field.id | https://openalex.org/fields/13 |
| primary_topic.field.display_name | Biochemistry, Genetics and Molecular Biology |
| primary_topic.score | 0.9993000030517578 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1306 |
| primary_topic.subfield.display_name | Cancer Research |
| primary_topic.display_name | Cancer Genomics and Diagnostics |
| related_works | https://openalex.org/W3008588181, https://openalex.org/W4386033707, https://openalex.org/W1970650594, https://openalex.org/W4387637728, https://openalex.org/W4378085118, https://openalex.org/W2111545764, https://openalex.org/W2896759930, https://openalex.org/W2067275497, https://openalex.org/W2529223554, https://openalex.org/W1851952311 |
| cited_by_count | 0 |
| locations_count | 2 |
| best_oa_location.id | doi:10.4103/jss.jss_23_21 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S2738012530 |
| best_oa_location.source.issn | 0974-5009, 2278-7127 |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | True |
| best_oa_location.source.issn_l | 0974-5009 |
| best_oa_location.source.is_core | False |
| best_oa_location.source.is_in_doaj | True |
| best_oa_location.source.display_name | Journal of the Scientific Society |
| best_oa_location.source.host_organization | https://openalex.org/P4310320448 |
| best_oa_location.source.host_organization_name | Medknow |
| best_oa_location.source.host_organization_lineage | https://openalex.org/P4310320448, https://openalex.org/P4310318547 |
| best_oa_location.source.host_organization_lineage_names | Medknow, Wolters Kluwer |
| best_oa_location.license | cc-by-nc-sa |
| best_oa_location.pdf_url | |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | journal-article |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by-nc-sa |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | Journal of the Scientific Society |
| best_oa_location.landing_page_url | https://doi.org/10.4103/jss.jss_23_21 |
| primary_location.id | doi:10.4103/jss.jss_23_21 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S2738012530 |
| primary_location.source.issn | 0974-5009, 2278-7127 |
| primary_location.source.type | journal |
| primary_location.source.is_oa | True |
| primary_location.source.issn_l | 0974-5009 |
| primary_location.source.is_core | False |
| primary_location.source.is_in_doaj | True |
| primary_location.source.display_name | Journal of the Scientific Society |
| primary_location.source.host_organization | https://openalex.org/P4310320448 |
| primary_location.source.host_organization_name | Medknow |
| primary_location.source.host_organization_lineage | https://openalex.org/P4310320448, https://openalex.org/P4310318547 |
| primary_location.source.host_organization_lineage_names | Medknow, Wolters Kluwer |
| primary_location.license | cc-by-nc-sa |
| primary_location.pdf_url | |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | https://openalex.org/licenses/cc-by-nc-sa |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | Journal of the Scientific Society |
| primary_location.landing_page_url | https://doi.org/10.4103/jss.jss_23_21 |
| publication_date | 2021-01-01 |
| publication_year | 2021 |
| referenced_works | https://openalex.org/W2143483118, https://openalex.org/W2999395828, https://openalex.org/W6777013225, https://openalex.org/W2025993990, https://openalex.org/W2037179980, https://openalex.org/W2094618536, https://openalex.org/W2402146695, https://openalex.org/W2061398002, https://openalex.org/W3023827600 |
| referenced_works_count | 9 |
| abstract_inverted_index.a | 39, 58, 450, 615, 649, 674, 687, 709, 777, 791 |
| abstract_inverted_index.8, | 572 |
| abstract_inverted_index.In | 672 |
| abstract_inverted_index.It | 689, 737 |
| abstract_inverted_index.So | 813 |
| abstract_inverted_index.an | 692, 745 |
| abstract_inverted_index.as | 57, 109, 163, 426, 449, 480, 614 |
| abstract_inverted_index.at | 588, 722 |
| abstract_inverted_index.be | 824 |
| abstract_inverted_index.by | 159, 223, 264, 362 |
| abstract_inverted_index.do | 641 |
| abstract_inverted_index.et | 414 |
| abstract_inverted_index.in | 5, 31, 150, 185, 218, 280, 326, 352, 392, 421, 459, 475, 488, 506, 576, 597, 629, 686, 806 |
| abstract_inverted_index.is | 77, 114, 143, 309, 329, 338, 389, 662, 717, 738, 752, 790 |
| abstract_inverted_index.it | 703 |
| abstract_inverted_index.of | 8, 26, 34, 41, 52, 60, 73, 92, 120, 124, 190, 321, 344, 456, 512, 519, 541, 557, 565, 632, 681, 719, 727, 747, 770, 784, 800, 819, 829 |
| abstract_inverted_index.on | 755 |
| abstract_inverted_index.or | 199, 222, 365, 496, 721 |
| abstract_inverted_index.to | 1, 49, 78, 131, 145, 312, 340, 471, 554, 635, 694, 698, 707, 712, 763, 823 |
| abstract_inverted_index.we | 741 |
| abstract_inverted_index.(70 | 334 |
| abstract_inverted_index.79% | 427 |
| abstract_inverted_index.84% | 430 |
| abstract_inverted_index.CTC | 499, 507, 532, 552 |
| abstract_inverted_index.DNA | 165, 314, 350, 355, 397, 411 |
| abstract_inverted_index.Due | 0 |
| abstract_inverted_index.PCR | 446 |
| abstract_inverted_index.The | 44, 66, 188, 306, 348, 543 |
| abstract_inverted_index.Via | 227 |
| abstract_inverted_index.all | 682 |
| abstract_inverted_index.and | 17, 28, 177, 183, 239, 246, 275, 297, 300, 316, 387, 410, 429, 462, 526, 539, 545, 570, 603, 651, 655, 667, 734, 802, 826 |
| abstract_inverted_index.are | 14, 102, 262, 284, 467, 486, 580, 742 |
| abstract_inverted_index.but | 106 |
| abstract_inverted_index.can | 204 |
| abstract_inverted_index.era | 631, 746 |
| abstract_inverted_index.for | 19, 63, 68, 117, 434, 453, 619, 669, 780, 795, 810 |
| abstract_inverted_index.get | 595 |
| abstract_inverted_index.has | 37, 382, 447, 548, 731 |
| abstract_inverted_index.may | 232, 478 |
| abstract_inverted_index.not | 103, 642, 664 |
| abstract_inverted_index.one | 481 |
| abstract_inverted_index.the | 2, 6, 24, 32, 50, 85, 90, 99, 118, 121, 140, 147, 206, 219, 228, 234, 256, 265, 269, 318, 322, 327, 342, 358, 363, 373, 375, 454, 489, 493, 510, 550, 630, 670, 678, 705, 713, 725, 760, 767, 781, 815 |
| abstract_inverted_index.too | 303 |
| abstract_inverted_index.use | 51, 726 |
| abstract_inverted_index.18+, | 573 |
| abstract_inverted_index.19+) | 575 |
| abstract_inverted_index.>200 | 581 |
| abstract_inverted_index.CTCs | 238, 474, 564 |
| abstract_inverted_index.Cell | 258 |
| abstract_inverted_index.DNA. | 347 |
| abstract_inverted_index.Drug | 546 |
| abstract_inverted_index.Food | 544 |
| abstract_inverted_index.More | 443 |
| abstract_inverted_index.RCCs | 299 |
| abstract_inverted_index.RNAs | 171, 175 |
| abstract_inverted_index.Test | 553 |
| abstract_inverted_index.This | 336, 561 |
| abstract_inverted_index.UCA1 | 609 |
| abstract_inverted_index.With | 89 |
| abstract_inverted_index.also | 107, 247, 690 |
| abstract_inverted_index.are, | 625 |
| abstract_inverted_index.base | 332 |
| abstract_inverted_index.been | 612 |
| abstract_inverted_index.body | 53, 186, 191 |
| abstract_inverted_index.both | 647 |
| abstract_inverted_index.cell | 521 |
| abstract_inverted_index.far, | 814 |
| abstract_inverted_index.form | 33 |
| abstract_inverted_index.free | 346 |
| abstract_inverted_index.from | 98, 139, 208, 286, 292, 357, 378, 492 |
| abstract_inverted_index.gene | 586 |
| abstract_inverted_index.have | 611 |
| abstract_inverted_index.into | 255, 268, 372, 399, 744 |
| abstract_inverted_index.like | 288 |
| abstract_inverted_index.long | 173, 583 |
| abstract_inverted_index.many | 20 |
| abstract_inverted_index.mean | 319 |
| abstract_inverted_index.most | 816 |
| abstract_inverted_index.node | 537 |
| abstract_inverted_index.only | 104, 311 |
| abstract_inverted_index.rise | 4 |
| abstract_inverted_index.same | 86 |
| abstract_inverted_index.seem | 822 |
| abstract_inverted_index.shed | 263, 285, 304 |
| abstract_inverted_index.size | 320 |
| abstract_inverted_index.such | 162 |
| abstract_inverted_index.take | 695 |
| abstract_inverted_index.term | 45 |
| abstract_inverted_index.test | 562 |
| abstract_inverted_index.than | 759 |
| abstract_inverted_index.that | 381, 401, 485, 584, 740 |
| abstract_inverted_index.tool | 452, 794 |
| abstract_inverted_index.used | 18 |
| abstract_inverted_index.when | 83, 127, 282 |
| abstract_inverted_index.with | 423, 523, 535, 607 |
| abstract_inverted_index.KDa). | 335 |
| abstract_inverted_index.Since | 368 |
| abstract_inverted_index.These | 594 |
| abstract_inverted_index.Thus, | 23, 112 |
| abstract_inverted_index.Tumor | 202, 242, 271 |
| abstract_inverted_index.acids | 277 |
| abstract_inverted_index.acini | 388 |
| abstract_inverted_index.avoid | 79 |
| abstract_inverted_index.based | 754 |
| abstract_inverted_index.being | 15, 469 |
| abstract_inverted_index.blood | 56, 213, 477, 490, 531 |
| abstract_inverted_index.cells | 169, 203, 231, 253, 360, 484 |
| abstract_inverted_index.cfDNA | 240, 323, 419, 435, 472, 801 |
| abstract_inverted_index.chain | 439 |
| abstract_inverted_index.clear | 739 |
| abstract_inverted_index.ducts | 386 |
| abstract_inverted_index.early | 827 |
| abstract_inverted_index.foci. | 498 |
| abstract_inverted_index.forms | 248 |
| abstract_inverted_index.found | 351 |
| abstract_inverted_index.gives | 691 |
| abstract_inverted_index.gland | 370 |
| abstract_inverted_index.hence | 236 |
| abstract_inverted_index.least | 723 |
| abstract_inverted_index.lymph | 536 |
| abstract_inverted_index.major | 42 |
| abstract_inverted_index.occur | 479 |
| abstract_inverted_index.often | 663 |
| abstract_inverted_index.other | 803 |
| abstract_inverted_index.pairs | 333 |
| abstract_inverted_index.point | 40, 406, 457 |
| abstract_inverted_index.reach | 205, 233 |
| abstract_inverted_index.renal | 293, 366, 520 |
| abstract_inverted_index.serve | 108 |
| abstract_inverted_index.stage | 525 |
| abstract_inverted_index.their | 209, 273 |
| abstract_inverted_index.there | 113 |
| abstract_inverted_index.these | 283 |
| abstract_inverted_index.trend | 7 |
| abstract_inverted_index.truly | 643 |
| abstract_inverted_index.tumor | 12, 168, 220, 224, 230, 252, 266, 495, 600, 645, 650, 654, 683, 756 |
| abstract_inverted_index.urine | 281, 328, 353, 418 |
| abstract_inverted_index.using | 69 |
| abstract_inverted_index.where | 251, 750 |
| abstract_inverted_index.which | 730 |
| abstract_inverted_index.whole | 359, 463, 577 |
| abstract_inverted_index.Liquid | 154, 715, 788 |
| abstract_inverted_index.RNAs), | 179 |
| abstract_inverted_index.Tumors | 395 |
| abstract_inverted_index.actual | 74 |
| abstract_inverted_index.al.[5] | 415 |
| abstract_inverted_index.allows | 704 |
| abstract_inverted_index.and/or | 55, 214, 574, 591, 773 |
| abstract_inverted_index.assess | 341 |
| abstract_inverted_index.become | 38, 278 |
| abstract_inverted_index.biopsy | 36, 47, 155, 661, 676, 716, 729, 789 |
| abstract_inverted_index.blood, | 195 |
| abstract_inverted_index.blood. | 578 |
| abstract_inverted_index.cancer | 94, 157, 380 |
| abstract_inverted_index.cells, | 272 |
| abstract_inverted_index.cfDNA. | 460 |
| abstract_inverted_index.clone, | 142 |
| abstract_inverted_index.counts | 500, 508, 563 |
| abstract_inverted_index.course | 511 |
| abstract_inverted_index.ctDNA, | 261 |
| abstract_inverted_index.demand | 116 |
| abstract_inverted_index.drains | 371 |
| abstract_inverted_index.during | 509, 658 |
| abstract_inverted_index.easily | 390 |
| abstract_inverted_index.fluid, | 198 |
| abstract_inverted_index.fluids | 54, 192, 809 |
| abstract_inverted_index.focus. | 43 |
| abstract_inverted_index.genome | 464 |
| abstract_inverted_index.global | 768 |
| abstract_inverted_index.hence, | 626 |
| abstract_inverted_index.itself | 243, 267 |
| abstract_inverted_index.kidney | 307 |
| abstract_inverted_index.levels | 533, 610 |
| abstract_inverted_index.liquid | 35, 46, 70, 675, 820 |
| abstract_inverted_index.lumen. | 257 |
| abstract_inverted_index.marker | 618 |
| abstract_inverted_index.moving | 743 |
| abstract_inverted_index.myriad | 189 |
| abstract_inverted_index.needed | 628 |
| abstract_inverted_index.origin | 567 |
| abstract_inverted_index.rather | 758 |
| abstract_inverted_index.recent | 3 |
| abstract_inverted_index.refers | 48 |
| abstract_inverted_index.risky, | 666 |
| abstract_inverted_index.serial | 696 |
| abstract_inverted_index.showed | 416, 529 |
| abstract_inverted_index.source | 343 |
| abstract_inverted_index.spaces | 250 |
| abstract_inverted_index.spread | 383 |
| abstract_inverted_index.status | 538 |
| abstract_inverted_index.stroma | 221 |
| abstract_inverted_index.tissue | 61, 75 |
| abstract_inverted_index.tumors | 287 |
| abstract_inverted_index.urine, | 196 |
| abstract_inverted_index.urine. | 394 |
| abstract_inverted_index.voided | 393 |
| abstract_inverted_index.within | 384, 648 |
| abstract_inverted_index.(CD45-, | 568 |
| abstract_inverted_index.(CTCs), | 170 |
| abstract_inverted_index.Casadio | 413 |
| abstract_inverted_index.Classic | 432 |
| abstract_inverted_index.EPCAM+, | 569 |
| abstract_inverted_index.LncRNAs | 579 |
| abstract_inverted_index.Precise | 622 |
| abstract_inverted_index.Urinary | 605 |
| abstract_inverted_index.altered | 596 |
| abstract_inverted_index.applied | 470 |
| abstract_inverted_index.arising | 291 |
| abstract_inverted_index.average | 761 |
| abstract_inverted_index.barrier | 308 |
| abstract_inverted_index.between | 652 |
| abstract_inverted_index.bladder | 296, 620 |
| abstract_inverted_index.burden, | 503 |
| abstract_inverted_index.cancer. | 560 |
| abstract_inverted_index.cancers | 598 |
| abstract_inverted_index.capable | 718 |
| abstract_inverted_index.changes | 505 |
| abstract_inverted_index.contain | 402 |
| abstract_inverted_index.derived | 356 |
| abstract_inverted_index.develop | 129 |
| abstract_inverted_index.digital | 445 |
| abstract_inverted_index.disease | 502 |
| abstract_inverted_index.emerged | 448 |
| abstract_inverted_index.fluids. | 187, 201 |
| abstract_inverted_index.genetic | 376, 679 |
| abstract_inverted_index.guiding | 151 |
| abstract_inverted_index.include | 193, 437 |
| abstract_inverted_index.induces | 244 |
| abstract_inverted_index.instead | 72 |
| abstract_inverted_index.levels. | 593 |
| abstract_inverted_index.limited | 732 |
| abstract_inverted_index.located | 217 |
| abstract_inverted_index.methods | 433 |
| abstract_inverted_index.million | 483 |
| abstract_inverted_index.monitor | 555, 699 |
| abstract_inverted_index.nucleic | 276 |
| abstract_inverted_index.painful | 668 |
| abstract_inverted_index.pelvis, | 294 |
| abstract_inverted_index.predict | 501 |
| abstract_inverted_index.present | 685 |
| abstract_inverted_index.primary | 494, 653 |
| abstract_inverted_index.profile | 680 |
| abstract_inverted_index.release | 396 |
| abstract_inverted_index.samples | 62, 76, 96, 697 |
| abstract_inverted_index.success | 733 |
| abstract_inverted_index.support | 636 |
| abstract_inverted_index.therapy | 514, 711 |
| abstract_inverted_index.through | 212 |
| abstract_inverted_index.tissues | 101 |
| abstract_inverted_index.tumoral | 700 |
| abstract_inverted_index.tumors. | 621 |
| abstract_inverted_index.ureter, | 295 |
| abstract_inverted_index.urinary | 345 |
| abstract_inverted_index.various | 807 |
| abstract_inverted_index.vessels | 216 |
| abstract_inverted_index.whereas | 504 |
| abstract_inverted_index.(cfDNA), | 166 |
| abstract_inverted_index.50–100 | 331 |
| abstract_inverted_index.Patients | 518 |
| abstract_inverted_index.Targeted | 461 |
| abstract_inverted_index.advanced | 524 |
| abstract_inverted_index.analysis | 769 |
| abstract_inverted_index.approved | 549 |
| abstract_inverted_index.behavior | 757 |
| abstract_inverted_index.biopsies | 71, 640, 821 |
| abstract_inverted_index.cell/100 | 482 |
| abstract_inverted_index.changes. | 701 |
| abstract_inverted_index.clinical | 21, 637 |
| abstract_inverted_index.critical | 144, 778 |
| abstract_inverted_index.detected | 391 |
| abstract_inverted_index.effusion | 200 |
| abstract_inverted_index.elevated | 417, 530, 608 |
| abstract_inverted_index.enabling | 798 |
| abstract_inverted_index.exciting | 817 |
| abstract_inverted_index.exosomes | 184 |
| abstract_inverted_index.failure. | 831 |
| abstract_inverted_index.genomic, | 771 |
| abstract_inverted_index.indicate | 515 |
| abstract_inverted_index.invasive | 81, 728 |
| abstract_inverted_index.involves | 156 |
| abstract_inverted_index.location | 211 |
| abstract_inverted_index.material | 377 |
| abstract_inverted_index.medicine | 634 |
| abstract_inverted_index.neoplasm | 290 |
| abstract_inverted_index.obtained | 97 |
| abstract_inverted_index.original | 210 |
| abstract_inverted_index.patient. | 671 |
| abstract_inverted_index.patients | 128, 422, 556 |
| abstract_inverted_index.presence | 540 |
| abstract_inverted_index.prostate | 301, 369, 379, 424, 559 |
| abstract_inverted_index.proteins | 274 |
| abstract_inverted_index.protrude | 254 |
| abstract_inverted_index.reaction | 440 |
| abstract_inverted_index.reflects | 677 |
| abstract_inverted_index.regulate | 585 |
| abstract_inverted_index.released | 325, 361 |
| abstract_inverted_index.relevant | 339 |
| abstract_inverted_index.repeated | 660 |
| abstract_inverted_index.response | 762 |
| abstract_inverted_index.samples, | 137 |
| abstract_inverted_index.specific | 27 |
| abstract_inverted_index.systemic | 513 |
| abstract_inverted_index.tailored | 753 |
| abstract_inverted_index.targeted | 132 |
| abstract_inverted_index.therapy, | 95, 133 |
| abstract_inverted_index.therapy. | 764 |
| abstract_inverted_index.urethra, | 374 |
| abstract_inverted_index.urethra. | 298 |
| abstract_inverted_index.urgently | 627 |
| abstract_inverted_index.vascular | 225 |
| abstract_inverted_index.Molecular | 765 |
| abstract_inverted_index.Moreover, | 657 |
| abstract_inverted_index.analysis. | 473 |
| abstract_inverted_index.biomarker | 811 |
| abstract_inverted_index.cancerous | 100 |
| abstract_inverted_index.carcinoma | 522 |
| abstract_inverted_index.cell-free | 164, 180 |
| abstract_inverted_index.clinician | 706 |
| abstract_inverted_index.contrast, | 673 |
| abstract_inverted_index.delineate | 146 |
| abstract_inverted_index.detection | 119, 455 |
| abstract_inverted_index.effective | 29 |
| abstract_inverted_index.feasible, | 665 |
| abstract_inverted_index.fragments | 315, 324, 398 |
| abstract_inverted_index.including | 260, 405 |
| abstract_inverted_index.integrity | 420 |
| abstract_inverted_index.invasion. | 226 |
| abstract_inverted_index.lymphatic | 215 |
| abstract_inverted_index.mechanism | 149 |
| abstract_inverted_index.medicine, | 749 |
| abstract_inverted_index.messenger | 178 |
| abstract_inverted_index.molecular | 623, 796 |
| abstract_inverted_index.molecules | 805 |
| abstract_inverted_index.mutations | 458 |
| abstract_inverted_index.noncoding | 174 |
| abstract_inverted_index.obtaining | 134, 160 |
| abstract_inverted_index.peptides, | 182 |
| abstract_inverted_index.permeable | 310 |
| abstract_inverted_index.potential | 616 |
| abstract_inverted_index.precision | 748 |
| abstract_inverted_index.profiles, | 775 |
| abstract_inverted_index.prognosis | 825 |
| abstract_inverted_index.promising | 792 |
| abstract_inverted_index.promoting | 599 |
| abstract_inverted_index.proteins, | 181 |
| abstract_inverted_index.proteomic | 774 |
| abstract_inverted_index.providing | 84 |
| abstract_inverted_index.purposes. | 65 |
| abstract_inverted_index.rationale | 67 |
| abstract_inverted_index.recently, | 444 |
| abstract_inverted_index.redundant | 80 |
| abstract_inverted_index.rendering | 237 |
| abstract_inverted_index.replacing | 720 |
| abstract_inverted_index.resistant | 141 |
| abstract_inverted_index.response. | 517 |
| abstract_inverted_index.sediments | 606 |
| abstract_inverted_index.sensitive | 428, 451 |
| abstract_inverted_index.settings. | 22 |
| abstract_inverted_index.specific. | 431 |
| abstract_inverted_index.subclones | 684 |
| abstract_inverted_index.surrogate | 59 |
| abstract_inverted_index.therefore | 317 |
| abstract_inverted_index.these.[3] | 305 |
| abstract_inverted_index.treatment | 516, 751, 786, 830 |
| abstract_inverted_index.Excisional | 639 |
| abstract_inverted_index.Meanwhile, | 702 |
| abstract_inverted_index.[lncRNAs], | 176 |
| abstract_inverted_index.additional | 135 |
| abstract_inverted_index.administer | 708 |
| abstract_inverted_index.aggressive | 527 |
| abstract_inverted_index.assessment | 436, 799, 828 |
| abstract_inverted_index.associated | 735 |
| abstract_inverted_index.augmenting | 724 |
| abstract_inverted_index.biological | 808 |
| abstract_inverted_index.biomarkers | 13, 30, 624 |
| abstract_inverted_index.carcinomas | 302 |
| abstract_inverted_index.detectable | 279 |
| abstract_inverted_index.diagnostic | 64, 87, 105, 136, 617 |
| abstract_inverted_index.discovery. | 812 |
| abstract_inverted_index.epigenetic | 592 |
| abstract_inverted_index.epithelial | 566 |
| abstract_inverted_index.expression | 587 |
| abstract_inverted_index.formation, | 601 |
| abstract_inverted_index.identified | 613 |
| abstract_inverted_index.increasing | 115 |
| abstract_inverted_index.metastatic | 497, 558 |
| abstract_inverted_index.mutations, | 407 |
| abstract_inverted_index.peripheral | 194, 476 |
| abstract_inverted_index.phenotypes | 528 |
| abstract_inverted_index.polymerase | 438 |
| abstract_inverted_index.predictive | 110 |
| abstract_inverted_index.preferably | 138 |
| abstract_inverted_index.procedures | 82 |
| abstract_inverted_index.profiling, | 766, 797 |
| abstract_inverted_index.repertoire | 123 |
| abstract_inverted_index.represents | 776 |
| abstract_inverted_index.researched | 16 |
| abstract_inverted_index.resistance | 130, 148 |
| abstract_inverted_index.sequencing | 465 |
| abstract_inverted_index.sinusoidal | 249 |
| abstract_inverted_index.speculates | 354 |
| abstract_inverted_index.successful | 782 |
| abstract_inverted_index.treatment. | 153 |
| abstract_inverted_index.urological | 9 |
| abstract_inverted_index.urothelial | 289 |
| abstract_inverted_index.urothelium | 364 |
| abstract_inverted_index.variation, | 409 |
| abstract_inverted_index.(PCR)-based | 441 |
| abstract_inverted_index.(microRNAs, | 172 |
| abstract_inverted_index.advancement | 91 |
| abstract_inverted_index.alterations | 404 |
| abstract_inverted_index.approaches. | 442 |
| abstract_inverted_index.appropriate | 152 |
| abstract_inverted_index.biomarkers. | 111, 125 |
| abstract_inverted_index.bloodstream | 207 |
| abstract_inverted_index.circulating | 167, 487, 804 |
| abstract_inverted_index.circulation | 400 |
| abstract_inverted_index.components, | 259 |
| abstract_inverted_index.copy-number | 408 |
| abstract_inverted_index.correlating | 534 |
| abstract_inverted_index.demonstrate | 644 |
| abstract_inverted_index.development | 783 |
| abstract_inverted_index.information | 337 |
| abstract_inverted_index.lymphatics, | 229 |
| abstract_inverted_index.metastases. | 656 |
| abstract_inverted_index.metastasis. | 604 |
| abstract_inverted_index.noninvasive | 11, 793 |
| abstract_inverted_index.nucleotides | 582 |
| abstract_inverted_index.opportunity | 693 |
| abstract_inverted_index.originating | 491 |
| abstract_inverted_index.patient.[7] | 688 |
| abstract_inverted_index.preexisting | 385 |
| abstract_inverted_index.strategies. | 787 |
| abstract_inverted_index.tubules.[4] | 367 |
| abstract_inverted_index.CELLSEARCH® | 551 |
| abstract_inverted_index.Furthermore, | 126 |
| abstract_inverted_index.angiogenesis | 245 |
| abstract_inverted_index.applications | 818 |
| abstract_inverted_index.biomolecules | 161 |
| abstract_inverted_index.bloodstream, | 235 |
| abstract_inverted_index.bloodstream. | 270 |
| abstract_inverted_index.cytokeratins | 571 |
| abstract_inverted_index.increasingly | 468 |
| abstract_inverted_index.methylation. | 412 |
| abstract_inverted_index.patients.[8] | 714 |
| abstract_inverted_index.personalized | 93, 633, 710 |
| abstract_inverted_index.prerequisite | 779 |
| abstract_inverted_index.progression, | 602 |
| abstract_inverted_index.technologies | 466 |
| abstract_inverted_index.approximately | 330 |
| abstract_inverted_index.cerebrospinal | 197 |
| abstract_inverted_index.malignancies, | 10 |
| abstract_inverted_index.Administration | 547 |
| abstract_inverted_index.adenocarcinoma | 425 |
| abstract_inverted_index.categorization | 158 |
| abstract_inverted_index.complications. | 736 |
| abstract_inverted_index.detectable.[2] | 241 |
| abstract_inverted_index.ever-expanding | 122 |
| abstract_inverted_index.heterogeneity, | 646 |
| abstract_inverted_index.identification | 25 |
| abstract_inverted_index.individualized | 785 |
| abstract_inverted_index.metastasis.[6] | 542 |
| abstract_inverted_index.tumor-specific | 403 |
| abstract_inverted_index.information.[1] | 88 |
| abstract_inverted_index.transcriptomic, | 772 |
| abstract_inverted_index.decision-making. | 638 |
| abstract_inverted_index.transcriptional, | 589 |
| abstract_inverted_index.monitoring/therapy, | 659 |
| abstract_inverted_index.low-molecular-weight | 313 |
| abstract_inverted_index.posttranscriptional, | 590 |
| abstract_inverted_index.high-molecular-weight | 349 |
| cited_by_percentile_year | |
| corresponding_author_ids | https://openalex.org/A5078485015 |
| countries_distinct_count | 3 |
| institutions_distinct_count | 2 |
| corresponding_institution_ids | https://openalex.org/I2799525281, https://openalex.org/I4210120668, https://openalex.org/I4210125823, https://openalex.org/I4210135707 |
| sustainable_development_goals[0].id | https://metadata.un.org/sdg/3 |
| sustainable_development_goals[0].score | 0.7799999713897705 |
| sustainable_development_goals[0].display_name | Good health and well-being |
| citation_normalized_percentile.value | 0.06163615 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |