Spatially resolved whole transcriptome profiling in human and mouse tissue using Digital Spatial Profiling Article Swipe
 YOU?
·
· 2021
· Open Access
·
· DOI: https://doi.org/10.1101/2021.09.29.462442
YOU?
·
· 2021
· Open Access
·
· DOI: https://doi.org/10.1101/2021.09.29.462442
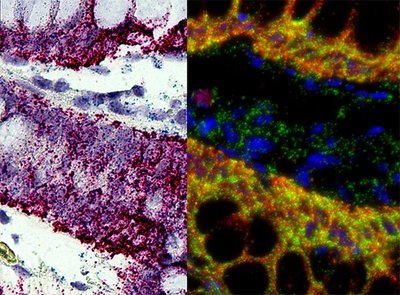
Emerging spatial profiling technology has enabled high-plex molecular profiling in biological tissues, preserving the spatial and morphological context of gene expression. Here we describe expanding the chemistry for the Digital Spatial Profiling platform to quantify whole transcriptomes in human and mouse tissues using a wide range of spatial profiling strategies and sample types. We designed multiplexed in situ hybridization probe pools targeting the protein-coding genes in the human and mouse transcriptomes, hereafter referred to as the human or mouse Whole Transcriptome Atlas (WTA). We validated the human and mouse WTA using cell lines to demonstrate concordance with orthogonal gene expression profiling methods in profiled region sizes ranging from ~10-500 cells. By benchmarking against bulk RNAseq and fluorescence in situ hybridization, we demonstrate robust transcript detection possible down to ~100 transcripts per region. To assess the performance of WTA across tissue and sample types, we applied WTA to biological questions in cancer, molecular pathology, and developmental biology. We show that spatial profiling with WTA can detect expected spatial gene expression differences between tumor and tumor microenvironment, identify spatial disease-specific heterogeneity in gene expression in histological structures of the human kidney, and comprehensively map transcriptional programs in anatomical substructures of nine organs in the developing mouse embryo. Digital Spatial Profiling technology with the WTA assays provides a flexible method for spatial whole transcriptome profiling applicable to diverse tissue types and biological contexts.
Related Topics
- Type
- preprint
- Language
- en
- Landing Page
- https://doi.org/10.1101/2021.09.29.462442
- https://www.biorxiv.org/content/biorxiv/early/2022/04/08/2021.09.29.462442.full.pdf
- OA Status
- green
- Cited By
- 32
- References
- 59
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W3202978190
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W3202978190Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1101/2021.09.29.462442Digital Object Identifier
- Title
-
Spatially resolved whole transcriptome profiling in human and mouse tissue using Digital Spatial ProfilingWork title
- Type
-
preprintOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2021Year of publication
- Publication date
-
2021-10-01Full publication date if available
- Authors
-
Stephanie M. Zimmerman, Robin Fropf, Bridget R. Kulasekara, Maddy Griswold, Oliver K. Appelbe, Arya Bahrami, Rich Boykin, Derek L. Buhr, Kit Fuhrman, Margaret L. Hoang, Quoc Huynh, Lesley Isgur, Andrew Klock, Alecksandr Kutchma, Alexa E. Lasley, Yan Liang, Jill McKay-Fleisch, Jeffrey S. Nelson, Karen Nguyen, Erin Piazza, Aric B.E. Rininger, Daniel R. Zollinger, Michael Rhodes, Joseph BeechemList of authors in order
- Landing page
-
https://doi.org/10.1101/2021.09.29.462442Publisher landing page
- PDF URL
-
https://www.biorxiv.org/content/biorxiv/early/2022/04/08/2021.09.29.462442.full.pdfDirect link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
greenOpen access status per OpenAlex
- OA URL
-
https://www.biorxiv.org/content/biorxiv/early/2022/04/08/2021.09.29.462442.full.pdfDirect OA link when available
- Concepts
-
Transcriptome, Gene expression profiling, Biology, Computational biology, Profiling (computer programming), In situ hybridization, Gene expression, Gene, Genetics, Computer science, Operating systemTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
32Total citation count in OpenAlex
- Citations by year (recent)
-
2025: 10, 2024: 8, 2023: 9, 2022: 5Per-year citation counts (last 5 years)
- References (count)
-
59Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W3202978190 |
|---|---|
| doi | https://doi.org/10.1101/2021.09.29.462442 |
| ids.doi | https://doi.org/10.1101/2021.09.29.462442 |
| ids.mag | 3202978190 |
| ids.openalex | https://openalex.org/W3202978190 |
| fwci | 2.1457134 |
| type | preprint |
| title | Spatially resolved whole transcriptome profiling in human and mouse tissue using Digital Spatial Profiling |
| biblio.issue | |
| biblio.volume | |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T11289 |
| topics[0].field.id | https://openalex.org/fields/13 |
| topics[0].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[0].score | 0.9998999834060669 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1312 |
| topics[0].subfield.display_name | Molecular Biology |
| topics[0].display_name | Single-cell and spatial transcriptomics |
| topics[1].id | https://openalex.org/T10207 |
| topics[1].field.id | https://openalex.org/fields/13 |
| topics[1].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[1].score | 0.9779999852180481 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1312 |
| topics[1].subfield.display_name | Molecular Biology |
| topics[1].display_name | Advanced biosensing and bioanalysis techniques |
| topics[2].id | https://openalex.org/T11970 |
| topics[2].field.id | https://openalex.org/fields/13 |
| topics[2].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[2].score | 0.9726999998092651 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1312 |
| topics[2].subfield.display_name | Molecular Biology |
| topics[2].display_name | Molecular Biology Techniques and Applications |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C162317418 |
| concepts[0].level | 4 |
| concepts[0].score | 0.7099893689155579 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q252857 |
| concepts[0].display_name | Transcriptome |
| concepts[1].id | https://openalex.org/C18431079 |
| concepts[1].level | 4 |
| concepts[1].score | 0.6579415798187256 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q1502169 |
| concepts[1].display_name | Gene expression profiling |
| concepts[2].id | https://openalex.org/C86803240 |
| concepts[2].level | 0 |
| concepts[2].score | 0.6487178206443787 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[2].display_name | Biology |
| concepts[3].id | https://openalex.org/C70721500 |
| concepts[3].level | 1 |
| concepts[3].score | 0.6077204346656799 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q177005 |
| concepts[3].display_name | Computational biology |
| concepts[4].id | https://openalex.org/C187191949 |
| concepts[4].level | 2 |
| concepts[4].score | 0.5423427820205688 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q1138496 |
| concepts[4].display_name | Profiling (computer programming) |
| concepts[5].id | https://openalex.org/C81439078 |
| concepts[5].level | 4 |
| concepts[5].score | 0.42638924717903137 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q2304142 |
| concepts[5].display_name | In situ hybridization |
| concepts[6].id | https://openalex.org/C150194340 |
| concepts[6].level | 3 |
| concepts[6].score | 0.4137257933616638 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q26972 |
| concepts[6].display_name | Gene expression |
| concepts[7].id | https://openalex.org/C104317684 |
| concepts[7].level | 2 |
| concepts[7].score | 0.37725159525871277 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[7].display_name | Gene |
| concepts[8].id | https://openalex.org/C54355233 |
| concepts[8].level | 1 |
| concepts[8].score | 0.316737562417984 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[8].display_name | Genetics |
| concepts[9].id | https://openalex.org/C41008148 |
| concepts[9].level | 0 |
| concepts[9].score | 0.15366709232330322 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q21198 |
| concepts[9].display_name | Computer science |
| concepts[10].id | https://openalex.org/C111919701 |
| concepts[10].level | 1 |
| concepts[10].score | 0.0 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q9135 |
| concepts[10].display_name | Operating system |
| keywords[0].id | https://openalex.org/keywords/transcriptome |
| keywords[0].score | 0.7099893689155579 |
| keywords[0].display_name | Transcriptome |
| keywords[1].id | https://openalex.org/keywords/gene-expression-profiling |
| keywords[1].score | 0.6579415798187256 |
| keywords[1].display_name | Gene expression profiling |
| keywords[2].id | https://openalex.org/keywords/biology |
| keywords[2].score | 0.6487178206443787 |
| keywords[2].display_name | Biology |
| keywords[3].id | https://openalex.org/keywords/computational-biology |
| keywords[3].score | 0.6077204346656799 |
| keywords[3].display_name | Computational biology |
| keywords[4].id | https://openalex.org/keywords/profiling |
| keywords[4].score | 0.5423427820205688 |
| keywords[4].display_name | Profiling (computer programming) |
| keywords[5].id | https://openalex.org/keywords/in-situ-hybridization |
| keywords[5].score | 0.42638924717903137 |
| keywords[5].display_name | In situ hybridization |
| keywords[6].id | https://openalex.org/keywords/gene-expression |
| keywords[6].score | 0.4137257933616638 |
| keywords[6].display_name | Gene expression |
| keywords[7].id | https://openalex.org/keywords/gene |
| keywords[7].score | 0.37725159525871277 |
| keywords[7].display_name | Gene |
| keywords[8].id | https://openalex.org/keywords/genetics |
| keywords[8].score | 0.316737562417984 |
| keywords[8].display_name | Genetics |
| keywords[9].id | https://openalex.org/keywords/computer-science |
| keywords[9].score | 0.15366709232330322 |
| keywords[9].display_name | Computer science |
| language | en |
| locations[0].id | doi:10.1101/2021.09.29.462442 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4306402567 |
| locations[0].source.issn | |
| locations[0].source.type | repository |
| locations[0].source.is_oa | False |
| locations[0].source.issn_l | |
| locations[0].source.is_core | False |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| locations[0].source.host_organization | https://openalex.org/I2750212522 |
| locations[0].source.host_organization_name | Cold Spring Harbor Laboratory |
| locations[0].source.host_organization_lineage | https://openalex.org/I2750212522 |
| locations[0].license | cc-by-nc-nd |
| locations[0].pdf_url | https://www.biorxiv.org/content/biorxiv/early/2022/04/08/2021.09.29.462442.full.pdf |
| locations[0].version | acceptedVersion |
| locations[0].raw_type | posted-content |
| locations[0].license_id | https://openalex.org/licenses/cc-by-nc-nd |
| locations[0].is_accepted | True |
| locations[0].is_published | False |
| locations[0].raw_source_name | |
| locations[0].landing_page_url | https://doi.org/10.1101/2021.09.29.462442 |
| indexed_in | crossref |
| authorships[0].author.id | https://openalex.org/A5102751302 |
| authorships[0].author.orcid | https://orcid.org/0000-0003-1646-120X |
| authorships[0].author.display_name | Stephanie M. Zimmerman |
| authorships[0].affiliations[0].raw_affiliation_string | ®, Seattle, WA 9809, USA |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Stephanie M. Zimmerman |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | ®, Seattle, WA 9809, USA |
| authorships[1].author.id | https://openalex.org/A5050509398 |
| authorships[1].author.orcid | https://orcid.org/0000-0002-4095-6504 |
| authorships[1].author.display_name | Robin Fropf |
| authorships[1].affiliations[0].raw_affiliation_string | ®, Seattle, WA 9809, USA |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Robin Fropf |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | ®, Seattle, WA 9809, USA |
| authorships[2].author.id | https://openalex.org/A5030147092 |
| authorships[2].author.orcid | |
| authorships[2].author.display_name | Bridget R. Kulasekara |
| authorships[2].affiliations[0].raw_affiliation_string | ®, Seattle, WA 9809, USA |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Bridget R. Kulasekara |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | ®, Seattle, WA 9809, USA |
| authorships[3].author.id | https://openalex.org/A5034128279 |
| authorships[3].author.orcid | |
| authorships[3].author.display_name | Maddy Griswold |
| authorships[3].affiliations[0].raw_affiliation_string | ®, Seattle, WA 9809, USA |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Maddy Griswold |
| authorships[3].is_corresponding | False |
| authorships[3].raw_affiliation_strings | ®, Seattle, WA 9809, USA |
| authorships[4].author.id | https://openalex.org/A5081162981 |
| authorships[4].author.orcid | https://orcid.org/0000-0003-3504-3396 |
| authorships[4].author.display_name | Oliver K. Appelbe |
| authorships[4].affiliations[0].raw_affiliation_string | ®, Seattle, WA 9809, USA |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Oliver Appelbe |
| authorships[4].is_corresponding | False |
| authorships[4].raw_affiliation_strings | ®, Seattle, WA 9809, USA |
| authorships[5].author.id | https://openalex.org/A5089368745 |
| authorships[5].author.orcid | |
| authorships[5].author.display_name | Arya Bahrami |
| authorships[5].affiliations[0].raw_affiliation_string | ®, Seattle, WA 9809, USA |
| authorships[5].author_position | middle |
| authorships[5].raw_author_name | Arya Bahrami |
| authorships[5].is_corresponding | False |
| authorships[5].raw_affiliation_strings | ®, Seattle, WA 9809, USA |
| authorships[6].author.id | https://openalex.org/A5068915700 |
| authorships[6].author.orcid | |
| authorships[6].author.display_name | Rich Boykin |
| authorships[6].affiliations[0].raw_affiliation_string | ®, Seattle, WA 9809, USA |
| authorships[6].author_position | middle |
| authorships[6].raw_author_name | Rich Boykin |
| authorships[6].is_corresponding | False |
| authorships[6].raw_affiliation_strings | ®, Seattle, WA 9809, USA |
| authorships[7].author.id | https://openalex.org/A5044078490 |
| authorships[7].author.orcid | |
| authorships[7].author.display_name | Derek L. Buhr |
| authorships[7].affiliations[0].raw_affiliation_string | ®, Seattle, WA 9809, USA |
| authorships[7].author_position | middle |
| authorships[7].raw_author_name | Derek L. Buhr |
| authorships[7].is_corresponding | False |
| authorships[7].raw_affiliation_strings | ®, Seattle, WA 9809, USA |
| authorships[8].author.id | https://openalex.org/A5070396842 |
| authorships[8].author.orcid | |
| authorships[8].author.display_name | Kit Fuhrman |
| authorships[8].affiliations[0].raw_affiliation_string | ®, Seattle, WA 9809, USA |
| authorships[8].author_position | middle |
| authorships[8].raw_author_name | Kit Fuhrman |
| authorships[8].is_corresponding | False |
| authorships[8].raw_affiliation_strings | ®, Seattle, WA 9809, USA |
| authorships[9].author.id | https://openalex.org/A5057611393 |
| authorships[9].author.orcid | https://orcid.org/0000-0003-0442-855X |
| authorships[9].author.display_name | Margaret L. Hoang |
| authorships[9].affiliations[0].raw_affiliation_string | ®, Seattle, WA 9809, USA |
| authorships[9].author_position | middle |
| authorships[9].raw_author_name | Margaret L. Hoang |
| authorships[9].is_corresponding | False |
| authorships[9].raw_affiliation_strings | ®, Seattle, WA 9809, USA |
| authorships[10].author.id | https://openalex.org/A5112105437 |
| authorships[10].author.orcid | |
| authorships[10].author.display_name | Quoc Huynh |
| authorships[10].affiliations[0].raw_affiliation_string | ®, Seattle, WA 9809, USA |
| authorships[10].author_position | middle |
| authorships[10].raw_author_name | Quoc Huynh |
| authorships[10].is_corresponding | False |
| authorships[10].raw_affiliation_strings | ®, Seattle, WA 9809, USA |
| authorships[11].author.id | https://openalex.org/A5029139576 |
| authorships[11].author.orcid | |
| authorships[11].author.display_name | Lesley Isgur |
| authorships[11].affiliations[0].raw_affiliation_string | ®, Seattle, WA 9809, USA |
| authorships[11].author_position | middle |
| authorships[11].raw_author_name | Lesley Isgur |
| authorships[11].is_corresponding | False |
| authorships[11].raw_affiliation_strings | ®, Seattle, WA 9809, USA |
| authorships[12].author.id | https://openalex.org/A5071731452 |
| authorships[12].author.orcid | |
| authorships[12].author.display_name | Andrew Klock |
| authorships[12].affiliations[0].raw_affiliation_string | ®, Seattle, WA 9809, USA |
| authorships[12].author_position | middle |
| authorships[12].raw_author_name | Andrew Klock |
| authorships[12].is_corresponding | False |
| authorships[12].raw_affiliation_strings | ®, Seattle, WA 9809, USA |
| authorships[13].author.id | https://openalex.org/A5036882000 |
| authorships[13].author.orcid | |
| authorships[13].author.display_name | Alecksandr Kutchma |
| authorships[13].affiliations[0].raw_affiliation_string | ®, Seattle, WA 9809, USA |
| authorships[13].author_position | middle |
| authorships[13].raw_author_name | Alecksandr Kutchma |
| authorships[13].is_corresponding | False |
| authorships[13].raw_affiliation_strings | ®, Seattle, WA 9809, USA |
| authorships[14].author.id | https://openalex.org/A5069710732 |
| authorships[14].author.orcid | |
| authorships[14].author.display_name | Alexa E. Lasley |
| authorships[14].affiliations[0].raw_affiliation_string | ®, Seattle, WA 9809, USA |
| authorships[14].author_position | middle |
| authorships[14].raw_author_name | Alexa E. Lasley |
| authorships[14].is_corresponding | False |
| authorships[14].raw_affiliation_strings | ®, Seattle, WA 9809, USA |
| authorships[15].author.id | https://openalex.org/A5012138398 |
| authorships[15].author.orcid | https://orcid.org/0000-0002-8536-5951 |
| authorships[15].author.display_name | Yan Liang |
| authorships[15].affiliations[0].raw_affiliation_string | ®, Seattle, WA 9809, USA |
| authorships[15].author_position | middle |
| authorships[15].raw_author_name | Yan Liang |
| authorships[15].is_corresponding | False |
| authorships[15].raw_affiliation_strings | ®, Seattle, WA 9809, USA |
| authorships[16].author.id | https://openalex.org/A5004637786 |
| authorships[16].author.orcid | |
| authorships[16].author.display_name | Jill McKay-Fleisch |
| authorships[16].affiliations[0].raw_affiliation_string | ®, Seattle, WA 9809, USA |
| authorships[16].author_position | middle |
| authorships[16].raw_author_name | Jill McKay-Fleisch |
| authorships[16].is_corresponding | False |
| authorships[16].raw_affiliation_strings | ®, Seattle, WA 9809, USA |
| authorships[17].author.id | https://openalex.org/A5112392302 |
| authorships[17].author.orcid | |
| authorships[17].author.display_name | Jeffrey S. Nelson |
| authorships[17].affiliations[0].raw_affiliation_string | ®, Seattle, WA 9809, USA |
| authorships[17].author_position | middle |
| authorships[17].raw_author_name | Jeffrey S. Nelson |
| authorships[17].is_corresponding | False |
| authorships[17].raw_affiliation_strings | ®, Seattle, WA 9809, USA |
| authorships[18].author.id | https://openalex.org/A5111061699 |
| authorships[18].author.orcid | https://orcid.org/0009-0004-7898-1969 |
| authorships[18].author.display_name | Karen Nguyen |
| authorships[18].affiliations[0].raw_affiliation_string | ®, Seattle, WA 9809, USA |
| authorships[18].author_position | middle |
| authorships[18].raw_author_name | Karen Nguyen |
| authorships[18].is_corresponding | False |
| authorships[18].raw_affiliation_strings | ®, Seattle, WA 9809, USA |
| authorships[19].author.id | https://openalex.org/A5077463542 |
| authorships[19].author.orcid | https://orcid.org/0000-0002-3263-5946 |
| authorships[19].author.display_name | Erin Piazza |
| authorships[19].affiliations[0].raw_affiliation_string | ®, Seattle, WA 9809, USA |
| authorships[19].author_position | middle |
| authorships[19].raw_author_name | Erin Piazza |
| authorships[19].is_corresponding | False |
| authorships[19].raw_affiliation_strings | ®, Seattle, WA 9809, USA |
| authorships[20].author.id | https://openalex.org/A5011039391 |
| authorships[20].author.orcid | https://orcid.org/0000-0002-9856-8811 |
| authorships[20].author.display_name | Aric B.E. Rininger |
| authorships[20].affiliations[0].raw_affiliation_string | ®, Seattle, WA 9809, USA |
| authorships[20].author_position | middle |
| authorships[20].raw_author_name | Aric Rininger |
| authorships[20].is_corresponding | False |
| authorships[20].raw_affiliation_strings | ®, Seattle, WA 9809, USA |
| authorships[21].author.id | https://openalex.org/A5080291812 |
| authorships[21].author.orcid | https://orcid.org/0000-0003-0181-4228 |
| authorships[21].author.display_name | Daniel R. Zollinger |
| authorships[21].affiliations[0].raw_affiliation_string | ®, Seattle, WA 9809, USA |
| authorships[21].author_position | middle |
| authorships[21].raw_author_name | Daniel R. Zollinger |
| authorships[21].is_corresponding | False |
| authorships[21].raw_affiliation_strings | ®, Seattle, WA 9809, USA |
| authorships[22].author.id | https://openalex.org/A5091661224 |
| authorships[22].author.orcid | https://orcid.org/0009-0000-3718-813X |
| authorships[22].author.display_name | Michael Rhodes |
| authorships[22].affiliations[0].raw_affiliation_string | ®, Seattle, WA 9809, USA |
| authorships[22].author_position | middle |
| authorships[22].raw_author_name | Michael Rhodes |
| authorships[22].is_corresponding | False |
| authorships[22].raw_affiliation_strings | ®, Seattle, WA 9809, USA |
| authorships[23].author.id | https://openalex.org/A5070065201 |
| authorships[23].author.orcid | https://orcid.org/0000-0001-8372-5899 |
| authorships[23].author.display_name | Joseph Beechem |
| authorships[23].affiliations[0].raw_affiliation_string | ®, Seattle, WA 9809, USA |
| authorships[23].author_position | last |
| authorships[23].raw_author_name | Joseph M. Beechem |
| authorships[23].is_corresponding | True |
| authorships[23].raw_affiliation_strings | ®, Seattle, WA 9809, USA |
| has_content.pdf | True |
| has_content.grobid_xml | True |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://www.biorxiv.org/content/biorxiv/early/2022/04/08/2021.09.29.462442.full.pdf |
| open_access.oa_status | green |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Spatially resolved whole transcriptome profiling in human and mouse tissue using Digital Spatial Profiling |
| has_fulltext | True |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T11289 |
| primary_topic.field.id | https://openalex.org/fields/13 |
| primary_topic.field.display_name | Biochemistry, Genetics and Molecular Biology |
| primary_topic.score | 0.9998999834060669 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1312 |
| primary_topic.subfield.display_name | Molecular Biology |
| primary_topic.display_name | Single-cell and spatial transcriptomics |
| related_works | https://openalex.org/W3012091774, https://openalex.org/W3170099453, https://openalex.org/W4294954873, https://openalex.org/W4229335045, https://openalex.org/W1971124177, https://openalex.org/W1985562003, https://openalex.org/W2590201838, https://openalex.org/W2563598809, https://openalex.org/W2734403094, https://openalex.org/W2111172999 |
| cited_by_count | 32 |
| counts_by_year[0].year | 2025 |
| counts_by_year[0].cited_by_count | 10 |
| counts_by_year[1].year | 2024 |
| counts_by_year[1].cited_by_count | 8 |
| counts_by_year[2].year | 2023 |
| counts_by_year[2].cited_by_count | 9 |
| counts_by_year[3].year | 2022 |
| counts_by_year[3].cited_by_count | 5 |
| locations_count | 1 |
| best_oa_location.id | doi:10.1101/2021.09.29.462442 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4306402567 |
| best_oa_location.source.issn | |
| best_oa_location.source.type | repository |
| best_oa_location.source.is_oa | False |
| best_oa_location.source.issn_l | |
| best_oa_location.source.is_core | False |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| best_oa_location.source.host_organization | https://openalex.org/I2750212522 |
| best_oa_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| best_oa_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| best_oa_location.license | cc-by-nc-nd |
| best_oa_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2022/04/08/2021.09.29.462442.full.pdf |
| best_oa_location.version | acceptedVersion |
| best_oa_location.raw_type | posted-content |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by-nc-nd |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | False |
| best_oa_location.raw_source_name | |
| best_oa_location.landing_page_url | https://doi.org/10.1101/2021.09.29.462442 |
| primary_location.id | doi:10.1101/2021.09.29.462442 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4306402567 |
| primary_location.source.issn | |
| primary_location.source.type | repository |
| primary_location.source.is_oa | False |
| primary_location.source.issn_l | |
| primary_location.source.is_core | False |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| primary_location.source.host_organization | https://openalex.org/I2750212522 |
| primary_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| primary_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| primary_location.license | cc-by-nc-nd |
| primary_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2022/04/08/2021.09.29.462442.full.pdf |
| primary_location.version | acceptedVersion |
| primary_location.raw_type | posted-content |
| primary_location.license_id | https://openalex.org/licenses/cc-by-nc-nd |
| primary_location.is_accepted | True |
| primary_location.is_published | False |
| primary_location.raw_source_name | |
| primary_location.landing_page_url | https://doi.org/10.1101/2021.09.29.462442 |
| publication_date | 2021-10-01 |
| publication_year | 2021 |
| referenced_works | https://openalex.org/W3035892631, https://openalex.org/W1987470886, https://openalex.org/W3198493858, https://openalex.org/W1982379359, https://openalex.org/W3134382826, https://openalex.org/W2612724210, https://openalex.org/W3104282543, https://openalex.org/W2042789810, https://openalex.org/W4206199829, https://openalex.org/W3157263169, https://openalex.org/W3112860625, https://openalex.org/W2126867710, https://openalex.org/W3216834392, https://openalex.org/W2109847775, https://openalex.org/W2923300855, https://openalex.org/W2029549744, https://openalex.org/W2113284270, https://openalex.org/W2944727022, https://openalex.org/W2159707944, https://openalex.org/W3080363208, https://openalex.org/W3121990740, https://openalex.org/W4200416514, https://openalex.org/W2140733306, https://openalex.org/W2097383747, https://openalex.org/W3206922912, https://openalex.org/W2031190177, https://openalex.org/W2952452404, https://openalex.org/W2112138633, https://openalex.org/W2807661081, https://openalex.org/W3023701177, https://openalex.org/W2139828607, https://openalex.org/W2012858641, https://openalex.org/W2086108787, https://openalex.org/W2592811885, https://openalex.org/W3193419934, https://openalex.org/W2125305686, https://openalex.org/W3190202966, https://openalex.org/W2043873569, https://openalex.org/W2046976185, https://openalex.org/W3145026833, https://openalex.org/W2122082071, https://openalex.org/W3201406237, https://openalex.org/W2773852483, https://openalex.org/W2197124664, https://openalex.org/W2471536144, https://openalex.org/W3112563877, https://openalex.org/W2394782024, https://openalex.org/W2012034410, https://openalex.org/W2972983657, https://openalex.org/W1985901090, https://openalex.org/W1960895270, https://openalex.org/W2158485828, https://openalex.org/W2397467012, https://openalex.org/W2972445800, https://openalex.org/W2887326710, https://openalex.org/W3016981080, https://openalex.org/W2946054009, https://openalex.org/W3047419512, https://openalex.org/W3133845106 |
| referenced_works_count | 59 |
| abstract_inverted_index.a | 44, 215 |
| abstract_inverted_index.By | 111 |
| abstract_inverted_index.To | 133 |
| abstract_inverted_index.We | 54, 84, 157 |
| abstract_inverted_index.as | 75 |
| abstract_inverted_index.in | 10, 38, 57, 66, 103, 118, 150, 180, 183, 195, 201 |
| abstract_inverted_index.of | 19, 47, 137, 186, 198 |
| abstract_inverted_index.or | 78 |
| abstract_inverted_index.to | 34, 74, 94, 128, 147, 224 |
| abstract_inverted_index.we | 23, 121, 144 |
| abstract_inverted_index.WTA | 90, 138, 146, 163, 212 |
| abstract_inverted_index.and | 16, 40, 51, 69, 88, 116, 141, 154, 173, 190, 228 |
| abstract_inverted_index.can | 164 |
| abstract_inverted_index.for | 28, 218 |
| abstract_inverted_index.has | 5 |
| abstract_inverted_index.map | 192 |
| abstract_inverted_index.per | 131 |
| abstract_inverted_index.the | 14, 26, 29, 63, 67, 76, 86, 135, 187, 202, 211 |
| abstract_inverted_index.Here | 22 |
| abstract_inverted_index.bulk | 114 |
| abstract_inverted_index.cell | 92 |
| abstract_inverted_index.down | 127 |
| abstract_inverted_index.from | 108 |
| abstract_inverted_index.gene | 20, 99, 168, 181 |
| abstract_inverted_index.nine | 199 |
| abstract_inverted_index.show | 158 |
| abstract_inverted_index.situ | 58, 119 |
| abstract_inverted_index.that | 159 |
| abstract_inverted_index.wide | 45 |
| abstract_inverted_index.with | 97, 162, 210 |
| abstract_inverted_index.~100 | 129 |
| abstract_inverted_index.Atlas | 82 |
| abstract_inverted_index.Whole | 80 |
| abstract_inverted_index.genes | 65 |
| abstract_inverted_index.human | 39, 68, 77, 87, 188 |
| abstract_inverted_index.lines | 93 |
| abstract_inverted_index.mouse | 41, 70, 79, 89, 204 |
| abstract_inverted_index.pools | 61 |
| abstract_inverted_index.probe | 60 |
| abstract_inverted_index.range | 46 |
| abstract_inverted_index.sizes | 106 |
| abstract_inverted_index.tumor | 172, 174 |
| abstract_inverted_index.types | 227 |
| abstract_inverted_index.using | 43, 91 |
| abstract_inverted_index.whole | 36, 220 |
| abstract_inverted_index.(WTA). | 83 |
| abstract_inverted_index.RNAseq | 115 |
| abstract_inverted_index.across | 139 |
| abstract_inverted_index.assays | 213 |
| abstract_inverted_index.assess | 134 |
| abstract_inverted_index.cells. | 110 |
| abstract_inverted_index.detect | 165 |
| abstract_inverted_index.method | 217 |
| abstract_inverted_index.organs | 200 |
| abstract_inverted_index.region | 105 |
| abstract_inverted_index.robust | 123 |
| abstract_inverted_index.sample | 52, 142 |
| abstract_inverted_index.tissue | 140, 226 |
| abstract_inverted_index.types, | 143 |
| abstract_inverted_index.types. | 53 |
| abstract_inverted_index.Digital | 30, 206 |
| abstract_inverted_index.Spatial | 31, 207 |
| abstract_inverted_index.against | 113 |
| abstract_inverted_index.applied | 145 |
| abstract_inverted_index.between | 171 |
| abstract_inverted_index.cancer, | 151 |
| abstract_inverted_index.context | 18 |
| abstract_inverted_index.diverse | 225 |
| abstract_inverted_index.embryo. | 205 |
| abstract_inverted_index.enabled | 6 |
| abstract_inverted_index.kidney, | 189 |
| abstract_inverted_index.methods | 102 |
| abstract_inverted_index.ranging | 107 |
| abstract_inverted_index.region. | 132 |
| abstract_inverted_index.spatial | 2, 15, 48, 160, 167, 177, 219 |
| abstract_inverted_index.tissues | 42 |
| abstract_inverted_index.~10-500 | 109 |
| abstract_inverted_index.Abstract | 0 |
| abstract_inverted_index.Emerging | 1 |
| abstract_inverted_index.biology. | 156 |
| abstract_inverted_index.describe | 24 |
| abstract_inverted_index.designed | 55 |
| abstract_inverted_index.expected | 166 |
| abstract_inverted_index.flexible | 216 |
| abstract_inverted_index.identify | 176 |
| abstract_inverted_index.platform | 33 |
| abstract_inverted_index.possible | 126 |
| abstract_inverted_index.profiled | 104 |
| abstract_inverted_index.programs | 194 |
| abstract_inverted_index.provides | 214 |
| abstract_inverted_index.quantify | 35 |
| abstract_inverted_index.referred | 73 |
| abstract_inverted_index.tissues, | 12 |
| abstract_inverted_index.Profiling | 32, 208 |
| abstract_inverted_index.chemistry | 27 |
| abstract_inverted_index.contexts. | 230 |
| abstract_inverted_index.detection | 125 |
| abstract_inverted_index.expanding | 25 |
| abstract_inverted_index.hereafter | 72 |
| abstract_inverted_index.high-plex | 7 |
| abstract_inverted_index.molecular | 8, 152 |
| abstract_inverted_index.profiling | 3, 9, 49, 101, 161, 222 |
| abstract_inverted_index.questions | 149 |
| abstract_inverted_index.targeting | 62 |
| abstract_inverted_index.validated | 85 |
| abstract_inverted_index.anatomical | 196 |
| abstract_inverted_index.applicable | 223 |
| abstract_inverted_index.biological | 11, 148, 229 |
| abstract_inverted_index.developing | 203 |
| abstract_inverted_index.expression | 100, 169, 182 |
| abstract_inverted_index.orthogonal | 98 |
| abstract_inverted_index.pathology, | 153 |
| abstract_inverted_index.preserving | 13 |
| abstract_inverted_index.strategies | 50 |
| abstract_inverted_index.structures | 185 |
| abstract_inverted_index.technology | 4, 209 |
| abstract_inverted_index.transcript | 124 |
| abstract_inverted_index.concordance | 96 |
| abstract_inverted_index.demonstrate | 95, 122 |
| abstract_inverted_index.differences | 170 |
| abstract_inverted_index.expression. | 21 |
| abstract_inverted_index.multiplexed | 56 |
| abstract_inverted_index.performance | 136 |
| abstract_inverted_index.transcripts | 130 |
| abstract_inverted_index.benchmarking | 112 |
| abstract_inverted_index.fluorescence | 117 |
| abstract_inverted_index.histological | 184 |
| abstract_inverted_index.Transcriptome | 81 |
| abstract_inverted_index.developmental | 155 |
| abstract_inverted_index.heterogeneity | 179 |
| abstract_inverted_index.hybridization | 59 |
| abstract_inverted_index.morphological | 17 |
| abstract_inverted_index.substructures | 197 |
| abstract_inverted_index.transcriptome | 221 |
| abstract_inverted_index.hybridization, | 120 |
| abstract_inverted_index.protein-coding | 64 |
| abstract_inverted_index.transcriptomes | 37 |
| abstract_inverted_index.comprehensively | 191 |
| abstract_inverted_index.transcriptional | 193 |
| abstract_inverted_index.transcriptomes, | 71 |
| abstract_inverted_index.disease-specific | 178 |
| abstract_inverted_index.microenvironment, | 175 |
| cited_by_percentile_year.max | 99 |
| cited_by_percentile_year.min | 97 |
| corresponding_author_ids | https://openalex.org/A5070065201 |
| countries_distinct_count | 0 |
| institutions_distinct_count | 24 |
| sustainable_development_goals[0].id | https://metadata.un.org/sdg/9 |
| sustainable_development_goals[0].score | 0.44999998807907104 |
| sustainable_development_goals[0].display_name | Industry, innovation and infrastructure |
| citation_normalized_percentile.value | 0.86653956 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |