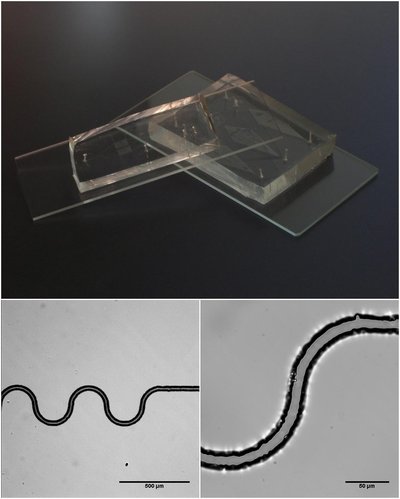
spinDrop: a droplet microfluidic platform to maximise single-cell sequencing information content Article Swipe
 YOU?
·
· 2023
· Open Access
·
· DOI: https://doi.org/10.1101/2023.01.12.523500
YOU?
·
· 2023
· Open Access
·
· DOI: https://doi.org/10.1101/2023.01.12.523500
Droplet microfluidic methods have massively increased the throughput of single-cell sequencing campaigns. The benefit of scale-up is, however, accompanied by increased background noise when processing challenging samples and the overall RNA capture efficiency is lower. These drawbacks stem from the lack of strategies to enrich for high-quality material or specific cell types at the moment of cell encapsulation and the absence of implementable multi-step enzymatic processes that increase capture. Here we alleviate both bottlenecks using fluorescence-activated droplet sorting to enrich for droplets that contain single viable cells, intact nuclei, fixed cells or target cell types and use reagent addition to droplets by picoinjection to perform multi-step lysis and reverse transcription. Our methodology increases gene detection rates fivefold, while reducing background noise by up to half. We harness these unique properties to deliver a high-quality molecular atlas of mouse brain development, despite starting with highly damaged input material, and provide an atlas of nascent RNA transcription during mouse organogenesis. Our method is broadly applicable to other droplet-based workflows to deliver sensitive and accurate single-cell profiling at a reduced cost.
Related Topics
- Type
- preprint
- Language
- en
- Landing Page
- https://doi.org/10.1101/2023.01.12.523500
- https://www.biorxiv.org/content/biorxiv/early/2023/01/13/2023.01.12.523500.full.pdf
- OA Status
- green
- Cited By
- 2
- References
- 68
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4316020640
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4316020640Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1101/2023.01.12.523500Digital Object Identifier
- Title
-
spinDrop: a droplet microfluidic platform to maximise single-cell sequencing information contentWork title
- Type
-
preprintOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2023Year of publication
- Publication date
-
2023-01-13Full publication date if available
- Authors
-
Joachim De Jonghe, Tomasz S. Kamiński, David B. Morse, Marcin Tabaka, Anna L. Ellermann, Timo N. Kohler, Gianluca Amadei, Charlotte E. Handford, Gregory M. Findlay, Magdalena Zernicka‐Goetz, Sarah A. Teichmann, Florian HollfelderList of authors in order
- Landing page
-
https://doi.org/10.1101/2023.01.12.523500Publisher landing page
- PDF URL
-
https://www.biorxiv.org/content/biorxiv/early/2023/01/13/2023.01.12.523500.full.pdfDirect link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
greenOpen access status per OpenAlex
- OA URL
-
https://www.biorxiv.org/content/biorxiv/early/2023/01/13/2023.01.12.523500.full.pdfDirect OA link when available
- Concepts
-
Microfluidics, Workflow, RNA, Nanotechnology, Cell, Single-cell analysis, Computational biology, Computer science, Massively parallel, Lysis, Chemistry, Cell biology, Biology, Biological system, Materials science, Gene, Molecular biology, Biochemistry, Parallel computing, DatabaseTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
2Total citation count in OpenAlex
- Citations by year (recent)
-
2023: 2Per-year citation counts (last 5 years)
- References (count)
-
68Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4316020640 |
|---|---|
| doi | https://doi.org/10.1101/2023.01.12.523500 |
| ids.doi | https://doi.org/10.25418/crick.23943333.v1 |
| ids.openalex | https://openalex.org/W4316020640 |
| fwci | 0.37134082 |
| type | preprint |
| title | spinDrop: a droplet microfluidic platform to maximise single-cell sequencing information content |
| biblio.issue | |
| biblio.volume | |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T11289 |
| topics[0].field.id | https://openalex.org/fields/13 |
| topics[0].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[0].score | 0.9998999834060669 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1312 |
| topics[0].subfield.display_name | Molecular Biology |
| topics[0].display_name | Single-cell and spatial transcriptomics |
| topics[1].id | https://openalex.org/T11407 |
| topics[1].field.id | https://openalex.org/fields/22 |
| topics[1].field.display_name | Engineering |
| topics[1].score | 0.9944999814033508 |
| topics[1].domain.id | https://openalex.org/domains/3 |
| topics[1].domain.display_name | Physical Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/2204 |
| topics[1].subfield.display_name | Biomedical Engineering |
| topics[1].display_name | Innovative Microfluidic and Catalytic Techniques Innovation |
| topics[2].id | https://openalex.org/T12859 |
| topics[2].field.id | https://openalex.org/fields/13 |
| topics[2].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[2].score | 0.9564999938011169 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1304 |
| topics[2].subfield.display_name | Biophysics |
| topics[2].display_name | Cell Image Analysis Techniques |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C8673954 |
| concepts[0].level | 2 |
| concepts[0].score | 0.7240808010101318 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q138845 |
| concepts[0].display_name | Microfluidics |
| concepts[1].id | https://openalex.org/C177212765 |
| concepts[1].level | 2 |
| concepts[1].score | 0.6552172899246216 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q627335 |
| concepts[1].display_name | Workflow |
| concepts[2].id | https://openalex.org/C67705224 |
| concepts[2].level | 3 |
| concepts[2].score | 0.5111389756202698 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q11053 |
| concepts[2].display_name | RNA |
| concepts[3].id | https://openalex.org/C171250308 |
| concepts[3].level | 1 |
| concepts[3].score | 0.5041605234146118 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q11468 |
| concepts[3].display_name | Nanotechnology |
| concepts[4].id | https://openalex.org/C1491633281 |
| concepts[4].level | 2 |
| concepts[4].score | 0.48720425367355347 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q7868 |
| concepts[4].display_name | Cell |
| concepts[5].id | https://openalex.org/C2776950831 |
| concepts[5].level | 3 |
| concepts[5].score | 0.4829115569591522 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q15635432 |
| concepts[5].display_name | Single-cell analysis |
| concepts[6].id | https://openalex.org/C70721500 |
| concepts[6].level | 1 |
| concepts[6].score | 0.4665018916130066 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q177005 |
| concepts[6].display_name | Computational biology |
| concepts[7].id | https://openalex.org/C41008148 |
| concepts[7].level | 0 |
| concepts[7].score | 0.45543915033340454 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q21198 |
| concepts[7].display_name | Computer science |
| concepts[8].id | https://openalex.org/C190475519 |
| concepts[8].level | 2 |
| concepts[8].score | 0.43378937244415283 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q544384 |
| concepts[8].display_name | Massively parallel |
| concepts[9].id | https://openalex.org/C57409179 |
| concepts[9].level | 2 |
| concepts[9].score | 0.42249977588653564 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q1218293 |
| concepts[9].display_name | Lysis |
| concepts[10].id | https://openalex.org/C185592680 |
| concepts[10].level | 0 |
| concepts[10].score | 0.39178627729415894 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q2329 |
| concepts[10].display_name | Chemistry |
| concepts[11].id | https://openalex.org/C95444343 |
| concepts[11].level | 1 |
| concepts[11].score | 0.38374483585357666 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q7141 |
| concepts[11].display_name | Cell biology |
| concepts[12].id | https://openalex.org/C86803240 |
| concepts[12].level | 0 |
| concepts[12].score | 0.363767147064209 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[12].display_name | Biology |
| concepts[13].id | https://openalex.org/C186060115 |
| concepts[13].level | 1 |
| concepts[13].score | 0.3417586088180542 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q30336093 |
| concepts[13].display_name | Biological system |
| concepts[14].id | https://openalex.org/C192562407 |
| concepts[14].level | 0 |
| concepts[14].score | 0.23371508717536926 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q228736 |
| concepts[14].display_name | Materials science |
| concepts[15].id | https://openalex.org/C104317684 |
| concepts[15].level | 2 |
| concepts[15].score | 0.21574968099594116 |
| concepts[15].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[15].display_name | Gene |
| concepts[16].id | https://openalex.org/C153911025 |
| concepts[16].level | 1 |
| concepts[16].score | 0.18166208267211914 |
| concepts[16].wikidata | https://www.wikidata.org/wiki/Q7202 |
| concepts[16].display_name | Molecular biology |
| concepts[17].id | https://openalex.org/C55493867 |
| concepts[17].level | 1 |
| concepts[17].score | 0.13853245973587036 |
| concepts[17].wikidata | https://www.wikidata.org/wiki/Q7094 |
| concepts[17].display_name | Biochemistry |
| concepts[18].id | https://openalex.org/C173608175 |
| concepts[18].level | 1 |
| concepts[18].score | 0.11622953414916992 |
| concepts[18].wikidata | https://www.wikidata.org/wiki/Q232661 |
| concepts[18].display_name | Parallel computing |
| concepts[19].id | https://openalex.org/C77088390 |
| concepts[19].level | 1 |
| concepts[19].score | 0.0 |
| concepts[19].wikidata | https://www.wikidata.org/wiki/Q8513 |
| concepts[19].display_name | Database |
| keywords[0].id | https://openalex.org/keywords/microfluidics |
| keywords[0].score | 0.7240808010101318 |
| keywords[0].display_name | Microfluidics |
| keywords[1].id | https://openalex.org/keywords/workflow |
| keywords[1].score | 0.6552172899246216 |
| keywords[1].display_name | Workflow |
| keywords[2].id | https://openalex.org/keywords/rna |
| keywords[2].score | 0.5111389756202698 |
| keywords[2].display_name | RNA |
| keywords[3].id | https://openalex.org/keywords/nanotechnology |
| keywords[3].score | 0.5041605234146118 |
| keywords[3].display_name | Nanotechnology |
| keywords[4].id | https://openalex.org/keywords/cell |
| keywords[4].score | 0.48720425367355347 |
| keywords[4].display_name | Cell |
| keywords[5].id | https://openalex.org/keywords/single-cell-analysis |
| keywords[5].score | 0.4829115569591522 |
| keywords[5].display_name | Single-cell analysis |
| keywords[6].id | https://openalex.org/keywords/computational-biology |
| keywords[6].score | 0.4665018916130066 |
| keywords[6].display_name | Computational biology |
| keywords[7].id | https://openalex.org/keywords/computer-science |
| keywords[7].score | 0.45543915033340454 |
| keywords[7].display_name | Computer science |
| keywords[8].id | https://openalex.org/keywords/massively-parallel |
| keywords[8].score | 0.43378937244415283 |
| keywords[8].display_name | Massively parallel |
| keywords[9].id | https://openalex.org/keywords/lysis |
| keywords[9].score | 0.42249977588653564 |
| keywords[9].display_name | Lysis |
| keywords[10].id | https://openalex.org/keywords/chemistry |
| keywords[10].score | 0.39178627729415894 |
| keywords[10].display_name | Chemistry |
| keywords[11].id | https://openalex.org/keywords/cell-biology |
| keywords[11].score | 0.38374483585357666 |
| keywords[11].display_name | Cell biology |
| keywords[12].id | https://openalex.org/keywords/biology |
| keywords[12].score | 0.363767147064209 |
| keywords[12].display_name | Biology |
| keywords[13].id | https://openalex.org/keywords/biological-system |
| keywords[13].score | 0.3417586088180542 |
| keywords[13].display_name | Biological system |
| keywords[14].id | https://openalex.org/keywords/materials-science |
| keywords[14].score | 0.23371508717536926 |
| keywords[14].display_name | Materials science |
| keywords[15].id | https://openalex.org/keywords/gene |
| keywords[15].score | 0.21574968099594116 |
| keywords[15].display_name | Gene |
| keywords[16].id | https://openalex.org/keywords/molecular-biology |
| keywords[16].score | 0.18166208267211914 |
| keywords[16].display_name | Molecular biology |
| keywords[17].id | https://openalex.org/keywords/biochemistry |
| keywords[17].score | 0.13853245973587036 |
| keywords[17].display_name | Biochemistry |
| keywords[18].id | https://openalex.org/keywords/parallel-computing |
| keywords[18].score | 0.11622953414916992 |
| keywords[18].display_name | Parallel computing |
| language | en |
| locations[0].id | doi:10.1101/2023.01.12.523500 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4306402567 |
| locations[0].source.issn | |
| locations[0].source.type | repository |
| locations[0].source.is_oa | False |
| locations[0].source.issn_l | |
| locations[0].source.is_core | False |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| locations[0].source.host_organization | https://openalex.org/I2750212522 |
| locations[0].source.host_organization_name | Cold Spring Harbor Laboratory |
| locations[0].source.host_organization_lineage | https://openalex.org/I2750212522 |
| locations[0].license | |
| locations[0].pdf_url | https://www.biorxiv.org/content/biorxiv/early/2023/01/13/2023.01.12.523500.full.pdf |
| locations[0].version | acceptedVersion |
| locations[0].raw_type | posted-content |
| locations[0].license_id | |
| locations[0].is_accepted | True |
| locations[0].is_published | False |
| locations[0].raw_source_name | |
| locations[0].landing_page_url | https://doi.org/10.1101/2023.01.12.523500 |
| locations[1].id | pmh:oai:figshare.com:article/23943333 |
| locations[1].is_oa | True |
| locations[1].source.id | https://openalex.org/S4306400572 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | OPAL (Open@LaTrobe) (La Trobe University) |
| locations[1].source.host_organization | https://openalex.org/I196829312 |
| locations[1].source.host_organization_name | La Trobe University |
| locations[1].source.host_organization_lineage | https://openalex.org/I196829312 |
| locations[1].license | cc-by |
| locations[1].pdf_url | |
| locations[1].version | submittedVersion |
| locations[1].raw_type | Text |
| locations[1].license_id | https://openalex.org/licenses/cc-by |
| locations[1].is_accepted | False |
| locations[1].is_published | False |
| locations[1].raw_source_name | |
| locations[1].landing_page_url | https://figshare.com/articles/journal_contribution/spinDrop_a_droplet_microfluidic_platform_to_maximise_single-cell_sequencing_information_content_/23943333 |
| locations[2].id | pmh:oai:www.repository.cam.ac.uk:1810/350854 |
| locations[2].is_oa | True |
| locations[2].source.id | https://openalex.org/S4306401776 |
| locations[2].source.issn | |
| locations[2].source.type | repository |
| locations[2].source.is_oa | False |
| locations[2].source.issn_l | |
| locations[2].source.is_core | False |
| locations[2].source.is_in_doaj | False |
| locations[2].source.display_name | Apollo (University of Cambridge) |
| locations[2].source.host_organization | https://openalex.org/I241749 |
| locations[2].source.host_organization_name | University of Cambridge |
| locations[2].source.host_organization_lineage | https://openalex.org/I241749 |
| locations[2].license | cc-by |
| locations[2].pdf_url | |
| locations[2].version | submittedVersion |
| locations[2].raw_type | Article |
| locations[2].license_id | https://openalex.org/licenses/cc-by |
| locations[2].is_accepted | False |
| locations[2].is_published | False |
| locations[2].raw_source_name | |
| locations[2].landing_page_url | https://www.repository.cam.ac.uk/handle/1810/350854 |
| locations[3].id | doi:10.17863/cam.97135 |
| locations[3].is_oa | True |
| locations[3].source.id | https://openalex.org/S7407050737 |
| locations[3].source.type | repository |
| locations[3].source.is_oa | False |
| locations[3].source.issn_l | |
| locations[3].source.is_core | False |
| locations[3].source.is_in_doaj | False |
| locations[3].source.display_name | Apollo |
| locations[3].source.host_organization | |
| locations[3].source.host_organization_name | |
| locations[3].license | cc-by |
| locations[3].pdf_url | |
| locations[3].version | |
| locations[3].raw_type | article-journal |
| locations[3].license_id | https://openalex.org/licenses/cc-by |
| locations[3].is_accepted | False |
| locations[3].is_published | |
| locations[3].raw_source_name | |
| locations[3].landing_page_url | https://doi.org/10.17863/cam.97135 |
| locations[4].id | doi:10.25418/crick.23943333.v1 |
| locations[4].is_oa | True |
| locations[4].source.id | https://openalex.org/S7407051354 |
| locations[4].source.type | repository |
| locations[4].source.is_oa | False |
| locations[4].source.issn_l | |
| locations[4].source.is_core | False |
| locations[4].source.is_in_doaj | False |
| locations[4].source.display_name | The Francis Crick Institute |
| locations[4].source.host_organization | |
| locations[4].source.host_organization_name | |
| locations[4].license | cc-by |
| locations[4].pdf_url | |
| locations[4].version | |
| locations[4].raw_type | article-journal |
| locations[4].license_id | https://openalex.org/licenses/cc-by |
| locations[4].is_accepted | False |
| locations[4].is_published | |
| locations[4].raw_source_name | |
| locations[4].landing_page_url | https://doi.org/10.25418/crick.23943333.v1 |
| indexed_in | crossref, datacite |
| authorships[0].author.id | https://openalex.org/A5033311273 |
| authorships[0].author.orcid | https://orcid.org/0000-0003-0584-8265 |
| authorships[0].author.display_name | Joachim De Jonghe |
| authorships[0].countries | GB |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I241749 |
| authorships[0].affiliations[0].raw_affiliation_string | Department of Biochemistry, University of Cambridge, Cambridge, United Kingdom |
| authorships[0].affiliations[1].institution_ids | https://openalex.org/I2801081054 |
| authorships[0].affiliations[1].raw_affiliation_string | Francis Crick Institute, London, United Kingdom |
| authorships[0].institutions[0].id | https://openalex.org/I2801081054 |
| authorships[0].institutions[0].ror | https://ror.org/04tnbqb63 |
| authorships[0].institutions[0].type | facility |
| authorships[0].institutions[0].lineage | https://openalex.org/I2801081054 |
| authorships[0].institutions[0].country_code | GB |
| authorships[0].institutions[0].display_name | The Francis Crick Institute |
| authorships[0].institutions[1].id | https://openalex.org/I241749 |
| authorships[0].institutions[1].ror | https://ror.org/013meh722 |
| authorships[0].institutions[1].type | education |
| authorships[0].institutions[1].lineage | https://openalex.org/I241749 |
| authorships[0].institutions[1].country_code | GB |
| authorships[0].institutions[1].display_name | University of Cambridge |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Joachim De Jonghe |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Department of Biochemistry, University of Cambridge, Cambridge, United Kingdom, Francis Crick Institute, London, United Kingdom |
| authorships[1].author.id | https://openalex.org/A5072510306 |
| authorships[1].author.orcid | https://orcid.org/0000-0001-5124-4548 |
| authorships[1].author.display_name | Tomasz S. Kamiński |
| authorships[1].countries | GB, PL |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I4654613 |
| authorships[1].affiliations[0].raw_affiliation_string | Department of Environmental Microbiology and Biotechnology, Institute of Microbiology, Faculty of Biology, University of Warsaw, Warsaw, Poland |
| authorships[1].affiliations[1].institution_ids | https://openalex.org/I241749 |
| authorships[1].affiliations[1].raw_affiliation_string | Department of Biochemistry, University of Cambridge, Cambridge, United Kingdom |
| authorships[1].institutions[0].id | https://openalex.org/I241749 |
| authorships[1].institutions[0].ror | https://ror.org/013meh722 |
| authorships[1].institutions[0].type | education |
| authorships[1].institutions[0].lineage | https://openalex.org/I241749 |
| authorships[1].institutions[0].country_code | GB |
| authorships[1].institutions[0].display_name | University of Cambridge |
| authorships[1].institutions[1].id | https://openalex.org/I4654613 |
| authorships[1].institutions[1].ror | https://ror.org/039bjqg32 |
| authorships[1].institutions[1].type | education |
| authorships[1].institutions[1].lineage | https://openalex.org/I4654613 |
| authorships[1].institutions[1].country_code | PL |
| authorships[1].institutions[1].display_name | University of Warsaw |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Tomasz S. Kaminski |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | Department of Biochemistry, University of Cambridge, Cambridge, United Kingdom, Department of Environmental Microbiology and Biotechnology, Institute of Microbiology, Faculty of Biology, University of Warsaw, Warsaw, Poland |
| authorships[2].author.id | https://openalex.org/A5051262526 |
| authorships[2].author.orcid | https://orcid.org/0000-0002-5863-7253 |
| authorships[2].author.display_name | David B. Morse |
| authorships[2].countries | GB |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I241749 |
| authorships[2].affiliations[0].raw_affiliation_string | Department of Chemistry, University of Cambridge, Cambridge, United Kingdom |
| authorships[2].institutions[0].id | https://openalex.org/I241749 |
| authorships[2].institutions[0].ror | https://ror.org/013meh722 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I241749 |
| authorships[2].institutions[0].country_code | GB |
| authorships[2].institutions[0].display_name | University of Cambridge |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | David B. Morse |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | Department of Chemistry, University of Cambridge, Cambridge, United Kingdom |
| authorships[3].author.id | https://openalex.org/A5011787284 |
| authorships[3].author.orcid | https://orcid.org/0000-0002-6166-2820 |
| authorships[3].author.display_name | Marcin Tabaka |
| authorships[3].countries | PL |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I4210157029, https://openalex.org/I99542240 |
| authorships[3].affiliations[0].raw_affiliation_string | International Centre for Translational Eye Research & Institute of Physical Chemistry, Polish Academy of Sciences, Warsaw, Poland |
| authorships[3].institutions[0].id | https://openalex.org/I4210157029 |
| authorships[3].institutions[0].ror | https://ror.org/05e830h62 |
| authorships[3].institutions[0].type | facility |
| authorships[3].institutions[0].lineage | https://openalex.org/I4210157029, https://openalex.org/I99542240 |
| authorships[3].institutions[0].country_code | PL |
| authorships[3].institutions[0].display_name | Institute of Physical Chemistry |
| authorships[3].institutions[1].id | https://openalex.org/I99542240 |
| authorships[3].institutions[1].ror | https://ror.org/01dr6c206 |
| authorships[3].institutions[1].type | government |
| authorships[3].institutions[1].lineage | https://openalex.org/I99542240 |
| authorships[3].institutions[1].country_code | PL |
| authorships[3].institutions[1].display_name | Polish Academy of Sciences |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Marcin Tabaka |
| authorships[3].is_corresponding | False |
| authorships[3].raw_affiliation_strings | International Centre for Translational Eye Research & Institute of Physical Chemistry, Polish Academy of Sciences, Warsaw, Poland |
| authorships[4].author.id | https://openalex.org/A5002967555 |
| authorships[4].author.orcid | https://orcid.org/0000-0002-3713-2870 |
| authorships[4].author.display_name | Anna L. Ellermann |
| authorships[4].countries | GB |
| authorships[4].affiliations[0].institution_ids | https://openalex.org/I241749 |
| authorships[4].affiliations[0].raw_affiliation_string | Department of Biochemistry, University of Cambridge, Cambridge, United Kingdom |
| authorships[4].institutions[0].id | https://openalex.org/I241749 |
| authorships[4].institutions[0].ror | https://ror.org/013meh722 |
| authorships[4].institutions[0].type | education |
| authorships[4].institutions[0].lineage | https://openalex.org/I241749 |
| authorships[4].institutions[0].country_code | GB |
| authorships[4].institutions[0].display_name | University of Cambridge |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Anna L. Ellermann |
| authorships[4].is_corresponding | False |
| authorships[4].raw_affiliation_strings | Department of Biochemistry, University of Cambridge, Cambridge, United Kingdom |
| authorships[5].author.id | https://openalex.org/A5012356388 |
| authorships[5].author.orcid | https://orcid.org/0000-0003-1949-0655 |
| authorships[5].author.display_name | Timo N. Kohler |
| authorships[5].countries | GB |
| authorships[5].affiliations[0].institution_ids | https://openalex.org/I241749 |
| authorships[5].affiliations[0].raw_affiliation_string | Department of Biochemistry, University of Cambridge, Cambridge, United Kingdom |
| authorships[5].institutions[0].id | https://openalex.org/I241749 |
| authorships[5].institutions[0].ror | https://ror.org/013meh722 |
| authorships[5].institutions[0].type | education |
| authorships[5].institutions[0].lineage | https://openalex.org/I241749 |
| authorships[5].institutions[0].country_code | GB |
| authorships[5].institutions[0].display_name | University of Cambridge |
| authorships[5].author_position | middle |
| authorships[5].raw_author_name | Timo N. Kohler |
| authorships[5].is_corresponding | False |
| authorships[5].raw_affiliation_strings | Department of Biochemistry, University of Cambridge, Cambridge, United Kingdom |
| authorships[6].author.id | https://openalex.org/A5023749013 |
| authorships[6].author.orcid | https://orcid.org/0000-0001-5405-968X |
| authorships[6].author.display_name | Gianluca Amadei |
| authorships[6].countries | GB |
| authorships[6].affiliations[0].institution_ids | https://openalex.org/I241749 |
| authorships[6].affiliations[0].raw_affiliation_string | Department of Physiology, Development and Neuroscience, University of Cambridge, Cambridge, United Kingdom |
| authorships[6].institutions[0].id | https://openalex.org/I241749 |
| authorships[6].institutions[0].ror | https://ror.org/013meh722 |
| authorships[6].institutions[0].type | education |
| authorships[6].institutions[0].lineage | https://openalex.org/I241749 |
| authorships[6].institutions[0].country_code | GB |
| authorships[6].institutions[0].display_name | University of Cambridge |
| authorships[6].author_position | middle |
| authorships[6].raw_author_name | Gianluca Amadei |
| authorships[6].is_corresponding | False |
| authorships[6].raw_affiliation_strings | Department of Physiology, Development and Neuroscience, University of Cambridge, Cambridge, United Kingdom |
| authorships[7].author.id | https://openalex.org/A5006260966 |
| authorships[7].author.orcid | https://orcid.org/0000-0002-5245-8027 |
| authorships[7].author.display_name | Charlotte E. Handford |
| authorships[7].countries | GB |
| authorships[7].affiliations[0].institution_ids | https://openalex.org/I241749 |
| authorships[7].affiliations[0].raw_affiliation_string | Department of Physiology, Development and Neuroscience, University of Cambridge, Cambridge, United Kingdom |
| authorships[7].institutions[0].id | https://openalex.org/I241749 |
| authorships[7].institutions[0].ror | https://ror.org/013meh722 |
| authorships[7].institutions[0].type | education |
| authorships[7].institutions[0].lineage | https://openalex.org/I241749 |
| authorships[7].institutions[0].country_code | GB |
| authorships[7].institutions[0].display_name | University of Cambridge |
| authorships[7].author_position | middle |
| authorships[7].raw_author_name | Charlotte Handford |
| authorships[7].is_corresponding | False |
| authorships[7].raw_affiliation_strings | Department of Physiology, Development and Neuroscience, University of Cambridge, Cambridge, United Kingdom |
| authorships[8].author.id | https://openalex.org/A5084597634 |
| authorships[8].author.orcid | https://orcid.org/0000-0002-7767-8608 |
| authorships[8].author.display_name | Gregory M. Findlay |
| authorships[8].countries | GB |
| authorships[8].affiliations[0].institution_ids | https://openalex.org/I2801081054 |
| authorships[8].affiliations[0].raw_affiliation_string | Francis Crick Institute, London, United Kingdom |
| authorships[8].institutions[0].id | https://openalex.org/I2801081054 |
| authorships[8].institutions[0].ror | https://ror.org/04tnbqb63 |
| authorships[8].institutions[0].type | facility |
| authorships[8].institutions[0].lineage | https://openalex.org/I2801081054 |
| authorships[8].institutions[0].country_code | GB |
| authorships[8].institutions[0].display_name | The Francis Crick Institute |
| authorships[8].author_position | middle |
| authorships[8].raw_author_name | Gregory M. Findlay |
| authorships[8].is_corresponding | False |
| authorships[8].raw_affiliation_strings | Francis Crick Institute, London, United Kingdom |
| authorships[9].author.id | https://openalex.org/A5062741380 |
| authorships[9].author.orcid | https://orcid.org/0000-0002-7004-2471 |
| authorships[9].author.display_name | Magdalena Zernicka‐Goetz |
| authorships[9].countries | GB, US |
| authorships[9].affiliations[0].institution_ids | https://openalex.org/I122411786 |
| authorships[9].affiliations[0].raw_affiliation_string | California Institute of Technology, Division of Biology and Biological Engineering, Pasadena, USA |
| authorships[9].affiliations[1].institution_ids | https://openalex.org/I241749 |
| authorships[9].affiliations[1].raw_affiliation_string | Department of Physiology, Development and Neuroscience, University of Cambridge, Cambridge, United Kingdom |
| authorships[9].institutions[0].id | https://openalex.org/I241749 |
| authorships[9].institutions[0].ror | https://ror.org/013meh722 |
| authorships[9].institutions[0].type | education |
| authorships[9].institutions[0].lineage | https://openalex.org/I241749 |
| authorships[9].institutions[0].country_code | GB |
| authorships[9].institutions[0].display_name | University of Cambridge |
| authorships[9].institutions[1].id | https://openalex.org/I122411786 |
| authorships[9].institutions[1].ror | https://ror.org/05dxps055 |
| authorships[9].institutions[1].type | education |
| authorships[9].institutions[1].lineage | https://openalex.org/I122411786 |
| authorships[9].institutions[1].country_code | US |
| authorships[9].institutions[1].display_name | California Institute of Technology |
| authorships[9].author_position | middle |
| authorships[9].raw_author_name | Magdalena Zernicka-Goetz |
| authorships[9].is_corresponding | False |
| authorships[9].raw_affiliation_strings | California Institute of Technology, Division of Biology and Biological Engineering, Pasadena, USA, Department of Physiology, Development and Neuroscience, University of Cambridge, Cambridge, United Kingdom |
| authorships[10].author.id | https://openalex.org/A5028596003 |
| authorships[10].author.orcid | https://orcid.org/0000-0002-6294-6366 |
| authorships[10].author.display_name | Sarah A. Teichmann |
| authorships[10].countries | GB |
| authorships[10].affiliations[0].institution_ids | https://openalex.org/I2802476451 |
| authorships[10].affiliations[0].raw_affiliation_string | Wellcome Trust Sanger Institute, Wellcome Genome Campus, Hinxton, United Kingdom |
| authorships[10].institutions[0].id | https://openalex.org/I2802476451 |
| authorships[10].institutions[0].ror | https://ror.org/05cy4wa09 |
| authorships[10].institutions[0].type | nonprofit |
| authorships[10].institutions[0].lineage | https://openalex.org/I2802476451, https://openalex.org/I87048295 |
| authorships[10].institutions[0].country_code | GB |
| authorships[10].institutions[0].display_name | Wellcome Sanger Institute |
| authorships[10].author_position | middle |
| authorships[10].raw_author_name | Sarah A. Teichmann |
| authorships[10].is_corresponding | False |
| authorships[10].raw_affiliation_strings | Wellcome Trust Sanger Institute, Wellcome Genome Campus, Hinxton, United Kingdom |
| authorships[11].author.id | https://openalex.org/A5051612157 |
| authorships[11].author.orcid | https://orcid.org/0000-0002-1367-6312 |
| authorships[11].author.display_name | Florian Hollfelder |
| authorships[11].countries | GB |
| authorships[11].affiliations[0].institution_ids | https://openalex.org/I241749 |
| authorships[11].affiliations[0].raw_affiliation_string | Department of Biochemistry, University of Cambridge, Cambridge, United Kingdom |
| authorships[11].institutions[0].id | https://openalex.org/I241749 |
| authorships[11].institutions[0].ror | https://ror.org/013meh722 |
| authorships[11].institutions[0].type | education |
| authorships[11].institutions[0].lineage | https://openalex.org/I241749 |
| authorships[11].institutions[0].country_code | GB |
| authorships[11].institutions[0].display_name | University of Cambridge |
| authorships[11].author_position | last |
| authorships[11].raw_author_name | Florian Hollfelder |
| authorships[11].is_corresponding | True |
| authorships[11].raw_affiliation_strings | Department of Biochemistry, University of Cambridge, Cambridge, United Kingdom |
| has_content.pdf | True |
| has_content.grobid_xml | True |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://www.biorxiv.org/content/biorxiv/early/2023/01/13/2023.01.12.523500.full.pdf |
| open_access.oa_status | green |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | spinDrop: a droplet microfluidic platform to maximise single-cell sequencing information content |
| has_fulltext | True |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T11289 |
| primary_topic.field.id | https://openalex.org/fields/13 |
| primary_topic.field.display_name | Biochemistry, Genetics and Molecular Biology |
| primary_topic.score | 0.9998999834060669 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1312 |
| primary_topic.subfield.display_name | Molecular Biology |
| primary_topic.display_name | Single-cell and spatial transcriptomics |
| related_works | https://openalex.org/W2129630532, https://openalex.org/W4402983052, https://openalex.org/W2032078143, https://openalex.org/W2106342149, https://openalex.org/W2970361009, https://openalex.org/W2472280754, https://openalex.org/W2999086771, https://openalex.org/W2909993299, https://openalex.org/W2912241197, https://openalex.org/W2937012602 |
| cited_by_count | 2 |
| counts_by_year[0].year | 2023 |
| counts_by_year[0].cited_by_count | 2 |
| locations_count | 5 |
| best_oa_location.id | doi:10.1101/2023.01.12.523500 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4306402567 |
| best_oa_location.source.issn | |
| best_oa_location.source.type | repository |
| best_oa_location.source.is_oa | False |
| best_oa_location.source.issn_l | |
| best_oa_location.source.is_core | False |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| best_oa_location.source.host_organization | https://openalex.org/I2750212522 |
| best_oa_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| best_oa_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| best_oa_location.license | |
| best_oa_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2023/01/13/2023.01.12.523500.full.pdf |
| best_oa_location.version | acceptedVersion |
| best_oa_location.raw_type | posted-content |
| best_oa_location.license_id | |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | False |
| best_oa_location.raw_source_name | |
| best_oa_location.landing_page_url | https://doi.org/10.1101/2023.01.12.523500 |
| primary_location.id | doi:10.1101/2023.01.12.523500 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4306402567 |
| primary_location.source.issn | |
| primary_location.source.type | repository |
| primary_location.source.is_oa | False |
| primary_location.source.issn_l | |
| primary_location.source.is_core | False |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| primary_location.source.host_organization | https://openalex.org/I2750212522 |
| primary_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| primary_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| primary_location.license | |
| primary_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2023/01/13/2023.01.12.523500.full.pdf |
| primary_location.version | acceptedVersion |
| primary_location.raw_type | posted-content |
| primary_location.license_id | |
| primary_location.is_accepted | True |
| primary_location.is_published | False |
| primary_location.raw_source_name | |
| primary_location.landing_page_url | https://doi.org/10.1101/2023.01.12.523500 |
| publication_date | 2023-01-13 |
| publication_year | 2023 |
| referenced_works | https://openalex.org/W2753908609, https://openalex.org/W3184729641, https://openalex.org/W3103797259, https://openalex.org/W4280578638, https://openalex.org/W3110852430, https://openalex.org/W3088401235, https://openalex.org/W2959566316, https://openalex.org/W2807587036, https://openalex.org/W3215588768, https://openalex.org/W2916020270, https://openalex.org/W2586637063, https://openalex.org/W2149573463, https://openalex.org/W2135937351, https://openalex.org/W2102212449, https://openalex.org/W2952630931, https://openalex.org/W3114078084, https://openalex.org/W2940256315, https://openalex.org/W3217745054, https://openalex.org/W3216579994, https://openalex.org/W3030739213, https://openalex.org/W2900872097, https://openalex.org/W3183469109, https://openalex.org/W2953151710, https://openalex.org/W2817838603, https://openalex.org/W2029886517, https://openalex.org/W2040331257, https://openalex.org/W2564176045, https://openalex.org/W2800028441, https://openalex.org/W1928449592, https://openalex.org/W2110852802, https://openalex.org/W4212780233, https://openalex.org/W3021060601, https://openalex.org/W2884205204, https://openalex.org/W4281928121, https://openalex.org/W4281965038, https://openalex.org/W4283691112, https://openalex.org/W2766437506, https://openalex.org/W2946232537, https://openalex.org/W2009458183, https://openalex.org/W2223794222, https://openalex.org/W2980870655, https://openalex.org/W2707529172, https://openalex.org/W3184208286, https://openalex.org/W2949177718, https://openalex.org/W2129282939, https://openalex.org/W2884148611, https://openalex.org/W1861340857, https://openalex.org/W3009992010, https://openalex.org/W4293247374, https://openalex.org/W3047141442, https://openalex.org/W4205496949, https://openalex.org/W2747841296, https://openalex.org/W3201239023, https://openalex.org/W4293218982, https://openalex.org/W3111222208, https://openalex.org/W3080945506, https://openalex.org/W3091970064, https://openalex.org/W3012809091, https://openalex.org/W4212799764, https://openalex.org/W2979911343, https://openalex.org/W2994871586, https://openalex.org/W2346617579, https://openalex.org/W2044120077, https://openalex.org/W3173187119, https://openalex.org/W2169456326, https://openalex.org/W2153091158, https://openalex.org/W2999403479, https://openalex.org/W3099078529 |
| referenced_works_count | 68 |
| abstract_inverted_index.a | 133, 176 |
| abstract_inverted_index.We | 126 |
| abstract_inverted_index.an | 150 |
| abstract_inverted_index.at | 53, 175 |
| abstract_inverted_index.by | 20, 102, 122 |
| abstract_inverted_index.is | 34, 161 |
| abstract_inverted_index.of | 9, 15, 42, 56, 62, 137, 152 |
| abstract_inverted_index.or | 49, 92 |
| abstract_inverted_index.to | 44, 79, 100, 104, 124, 131, 164, 168 |
| abstract_inverted_index.up | 123 |
| abstract_inverted_index.we | 71 |
| abstract_inverted_index.Our | 111, 159 |
| abstract_inverted_index.RNA | 31, 154 |
| abstract_inverted_index.The | 13 |
| abstract_inverted_index.and | 28, 59, 96, 108, 148, 171 |
| abstract_inverted_index.for | 46, 81 |
| abstract_inverted_index.is, | 17 |
| abstract_inverted_index.the | 7, 29, 40, 54, 60 |
| abstract_inverted_index.use | 97 |
| abstract_inverted_index.Here | 70 |
| abstract_inverted_index.both | 73 |
| abstract_inverted_index.cell | 51, 57, 94 |
| abstract_inverted_index.from | 39 |
| abstract_inverted_index.gene | 114 |
| abstract_inverted_index.have | 4 |
| abstract_inverted_index.lack | 41 |
| abstract_inverted_index.stem | 38 |
| abstract_inverted_index.that | 67, 83 |
| abstract_inverted_index.when | 24 |
| abstract_inverted_index.with | 143 |
| abstract_inverted_index.These | 36 |
| abstract_inverted_index.atlas | 136, 151 |
| abstract_inverted_index.brain | 139 |
| abstract_inverted_index.cells | 91 |
| abstract_inverted_index.cost. | 178 |
| abstract_inverted_index.fixed | 90 |
| abstract_inverted_index.half. | 125 |
| abstract_inverted_index.input | 146 |
| abstract_inverted_index.lysis | 107 |
| abstract_inverted_index.mouse | 138, 157 |
| abstract_inverted_index.noise | 23, 121 |
| abstract_inverted_index.other | 165 |
| abstract_inverted_index.rates | 116 |
| abstract_inverted_index.these | 128 |
| abstract_inverted_index.types | 52, 95 |
| abstract_inverted_index.using | 75 |
| abstract_inverted_index.while | 118 |
| abstract_inverted_index.cells, | 87 |
| abstract_inverted_index.during | 156 |
| abstract_inverted_index.enrich | 45, 80 |
| abstract_inverted_index.highly | 144 |
| abstract_inverted_index.intact | 88 |
| abstract_inverted_index.lower. | 35 |
| abstract_inverted_index.method | 160 |
| abstract_inverted_index.moment | 55 |
| abstract_inverted_index.single | 85 |
| abstract_inverted_index.target | 93 |
| abstract_inverted_index.unique | 129 |
| abstract_inverted_index.viable | 86 |
| abstract_inverted_index.Droplet | 1 |
| abstract_inverted_index.absence | 61 |
| abstract_inverted_index.benefit | 14 |
| abstract_inverted_index.broadly | 162 |
| abstract_inverted_index.capture | 32 |
| abstract_inverted_index.contain | 84 |
| abstract_inverted_index.damaged | 145 |
| abstract_inverted_index.deliver | 132, 169 |
| abstract_inverted_index.despite | 141 |
| abstract_inverted_index.droplet | 77 |
| abstract_inverted_index.harness | 127 |
| abstract_inverted_index.methods | 3 |
| abstract_inverted_index.nascent | 153 |
| abstract_inverted_index.nuclei, | 89 |
| abstract_inverted_index.overall | 30 |
| abstract_inverted_index.perform | 105 |
| abstract_inverted_index.provide | 149 |
| abstract_inverted_index.reagent | 98 |
| abstract_inverted_index.reduced | 177 |
| abstract_inverted_index.reverse | 109 |
| abstract_inverted_index.samples | 27 |
| abstract_inverted_index.sorting | 78 |
| abstract_inverted_index.Abstract | 0 |
| abstract_inverted_index.accurate | 172 |
| abstract_inverted_index.addition | 99 |
| abstract_inverted_index.capture. | 69 |
| abstract_inverted_index.droplets | 82, 101 |
| abstract_inverted_index.however, | 18 |
| abstract_inverted_index.increase | 68 |
| abstract_inverted_index.material | 48 |
| abstract_inverted_index.reducing | 119 |
| abstract_inverted_index.scale-up | 16 |
| abstract_inverted_index.specific | 50 |
| abstract_inverted_index.starting | 142 |
| abstract_inverted_index.alleviate | 72 |
| abstract_inverted_index.detection | 115 |
| abstract_inverted_index.drawbacks | 37 |
| abstract_inverted_index.enzymatic | 65 |
| abstract_inverted_index.fivefold, | 117 |
| abstract_inverted_index.increased | 6, 21 |
| abstract_inverted_index.increases | 113 |
| abstract_inverted_index.massively | 5 |
| abstract_inverted_index.material, | 147 |
| abstract_inverted_index.molecular | 135 |
| abstract_inverted_index.processes | 66 |
| abstract_inverted_index.profiling | 174 |
| abstract_inverted_index.sensitive | 170 |
| abstract_inverted_index.workflows | 167 |
| abstract_inverted_index.applicable | 163 |
| abstract_inverted_index.background | 22, 120 |
| abstract_inverted_index.campaigns. | 12 |
| abstract_inverted_index.efficiency | 33 |
| abstract_inverted_index.multi-step | 64, 106 |
| abstract_inverted_index.processing | 25 |
| abstract_inverted_index.properties | 130 |
| abstract_inverted_index.sequencing | 11 |
| abstract_inverted_index.strategies | 43 |
| abstract_inverted_index.throughput | 8 |
| abstract_inverted_index.accompanied | 19 |
| abstract_inverted_index.bottlenecks | 74 |
| abstract_inverted_index.challenging | 26 |
| abstract_inverted_index.methodology | 112 |
| abstract_inverted_index.single-cell | 10, 173 |
| abstract_inverted_index.development, | 140 |
| abstract_inverted_index.high-quality | 47, 134 |
| abstract_inverted_index.microfluidic | 2 |
| abstract_inverted_index.droplet-based | 166 |
| abstract_inverted_index.encapsulation | 58 |
| abstract_inverted_index.implementable | 63 |
| abstract_inverted_index.picoinjection | 103 |
| abstract_inverted_index.transcription | 155 |
| abstract_inverted_index.organogenesis. | 158 |
| abstract_inverted_index.transcription. | 110 |
| abstract_inverted_index.fluorescence-activated | 76 |
| cited_by_percentile_year.max | 96 |
| cited_by_percentile_year.min | 94 |
| corresponding_author_ids | https://openalex.org/A5051612157 |
| countries_distinct_count | 3 |
| institutions_distinct_count | 12 |
| corresponding_institution_ids | https://openalex.org/I241749 |
| citation_normalized_percentile.value | 0.63175717 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |