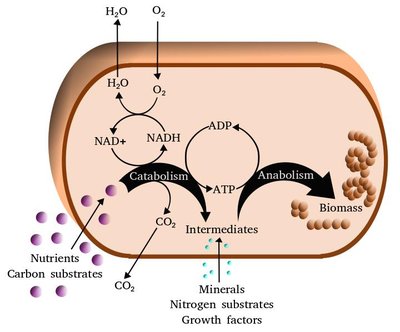
Staphylococcus aureus Transcriptome Data and Metabolic Modelling Investigate the Interplay of Ser/Thr Kinase PknB, Its Phosphatase Stp, the glmR/yvcK Regulon and the cdaA Operon for Metabolic Adaptation Article Swipe
 YOU?
·
· 2021
· Open Access
·
· DOI: https://doi.org/10.3390/microorganisms9102148
YOU?
·
· 2021
· Open Access
·
· DOI: https://doi.org/10.3390/microorganisms9102148
Serine/threonine kinase PknB and its corresponding phosphatase Stp are important regulators of many cell functions in the pathogen S. aureus. Genome-scale gene expression data of S. aureus strain NewHG (sigB+) elucidated their effect on physiological functions. Moreover, metabolic modelling from these data inferred metabolic adaptations. We compared wild-type to deletion strains lacking pknB, stp or both. Ser/Thr phosphorylation of target proteins by PknB switched amino acid catabolism off and gluconeogenesis on to provide the cell with sufficient components. We revealed a significant impact of PknB and Stp on peptidoglycan, nucleotide and aromatic amino acid synthesis, as well as catabolism involving aspartate transaminase. Moreover, pyrimidine synthesis was dramatically impaired by stp deletion but only slightly by functional loss of PknB. In double knockouts, higher activity concerned genes involved in peptidoglycan, purine and aromatic amino acid synthesis from glucose but lower activity of pyrimidine synthesis from glucose compared to the wild type. A second transcriptome dataset from S. aureus NCTC 8325 (sigB−) validated the predictions. For this metabolic adaptation, PknB was found to interact with CdaA and the yvcK/glmR regulon. The involved GlmR structure and the GlmS riboswitch were modelled. Furthermore, PknB phosphorylation lowered the expression of many virulence factors, and the study shed light on S. aureus infection processes.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.3390/microorganisms9102148
- https://www.mdpi.com/2076-2607/9/10/2148/pdf?version=1634292897
- OA Status
- gold
- Cited By
- 10
- References
- 54
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W3207666385
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W3207666385Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.3390/microorganisms9102148Digital Object Identifier
- Title
-
Staphylococcus aureus Transcriptome Data and Metabolic Modelling Investigate the Interplay of Ser/Thr Kinase PknB, Its Phosphatase Stp, the glmR/yvcK Regulon and the cdaA Operon for Metabolic AdaptationWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2021Year of publication
- Publication date
-
2021-10-14Full publication date if available
- Authors
-
Chunguang Liang, Ana B. Rios‐Miguel, Marcel Jarick, Priya Neurgaonkar, Myriam Girard, Patrice François, Jacques Schrenzel, Eslam S. Ibrahim, Knut Ohlsen, Thomas DandekarList of authors in order
- Landing page
-
https://doi.org/10.3390/microorganisms9102148Publisher landing page
- PDF URL
-
https://www.mdpi.com/2076-2607/9/10/2148/pdf?version=1634292897Direct link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
goldOpen access status per OpenAlex
- OA URL
-
https://www.mdpi.com/2076-2607/9/10/2148/pdf?version=1634292897Direct OA link when available
- Concepts
-
Regulon, Staphylococcus aureus, Operon, Transcriptome, Metabolic activity, Adaptation (eye), Biology, Phosphatase, Microbiology, Enzyme, Biochemistry, Genetics, Biological system, Bacterial protein, Neuroscience, Bacteria, Gene, Gene expression, Escherichia coliTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
10Total citation count in OpenAlex
- Citations by year (recent)
-
2025: 4, 2024: 1, 2023: 4, 2022: 1Per-year citation counts (last 5 years)
- References (count)
-
54Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W3207666385 |
|---|---|
| doi | https://doi.org/10.3390/microorganisms9102148 |
| ids.doi | https://doi.org/10.3390/microorganisms9102148 |
| ids.mag | 3207666385 |
| ids.pmid | https://pubmed.ncbi.nlm.nih.gov/34683468 |
| ids.openalex | https://openalex.org/W3207666385 |
| fwci | 0.58519456 |
| type | article |
| title | Staphylococcus aureus Transcriptome Data and Metabolic Modelling Investigate the Interplay of Ser/Thr Kinase PknB, Its Phosphatase Stp, the glmR/yvcK Regulon and the cdaA Operon for Metabolic Adaptation |
| biblio.issue | 10 |
| biblio.volume | 9 |
| biblio.last_page | 2148 |
| biblio.first_page | 2148 |
| grants[0].funder | https://openalex.org/F4320320879 |
| grants[0].award_id | GRK 2157, 270563345; |
| grants[0].funder_display_name | Deutsche Forschungsgemeinschaft |
| grants[1].funder | https://openalex.org/F4320323237 |
| grants[1].award_id | contribution to DFG projectNo 324392634/TR221 |
| grants[1].funder_display_name | Bayerisches Staatsministerium für Wissenschaft, Forschung und Kunst |
| topics[0].id | https://openalex.org/T10932 |
| topics[0].field.id | https://openalex.org/fields/13 |
| topics[0].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[0].score | 0.9986000061035156 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1312 |
| topics[0].subfield.display_name | Molecular Biology |
| topics[0].display_name | Microbial Metabolic Engineering and Bioproduction |
| topics[1].id | https://openalex.org/T10120 |
| topics[1].field.id | https://openalex.org/fields/13 |
| topics[1].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[1].score | 0.9980000257492065 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1311 |
| topics[1].subfield.display_name | Genetics |
| topics[1].display_name | Bacterial Genetics and Biotechnology |
| topics[2].id | https://openalex.org/T10521 |
| topics[2].field.id | https://openalex.org/fields/13 |
| topics[2].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[2].score | 0.9977999925613403 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1312 |
| topics[2].subfield.display_name | Molecular Biology |
| topics[2].display_name | RNA and protein synthesis mechanisms |
| funders[0].id | https://openalex.org/F4320320879 |
| funders[0].ror | https://ror.org/018mejw64 |
| funders[0].display_name | Deutsche Forschungsgemeinschaft |
| funders[1].id | https://openalex.org/F4320323237 |
| funders[1].ror | https://ror.org/01a44gd51 |
| funders[1].display_name | Bayerisches Staatsministerium für Wissenschaft, Forschung und Kunst |
| is_xpac | False |
| apc_list.value | 2200 |
| apc_list.currency | CHF |
| apc_list.value_usd | 2382 |
| apc_paid.value | 2200 |
| apc_paid.currency | CHF |
| apc_paid.value_usd | 2382 |
| concepts[0].id | https://openalex.org/C62361671 |
| concepts[0].level | 4 |
| concepts[0].score | 0.8609878420829773 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q1754612 |
| concepts[0].display_name | Regulon |
| concepts[1].id | https://openalex.org/C2779489039 |
| concepts[1].level | 3 |
| concepts[1].score | 0.5623898506164551 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q188121 |
| concepts[1].display_name | Staphylococcus aureus |
| concepts[2].id | https://openalex.org/C203075996 |
| concepts[2].level | 4 |
| concepts[2].score | 0.5371703505516052 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q139677 |
| concepts[2].display_name | Operon |
| concepts[3].id | https://openalex.org/C162317418 |
| concepts[3].level | 4 |
| concepts[3].score | 0.5059027075767517 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q252857 |
| concepts[3].display_name | Transcriptome |
| concepts[4].id | https://openalex.org/C2993927895 |
| concepts[4].level | 2 |
| concepts[4].score | 0.4793477952480316 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q1057 |
| concepts[4].display_name | Metabolic activity |
| concepts[5].id | https://openalex.org/C139807058 |
| concepts[5].level | 2 |
| concepts[5].score | 0.4491193890571594 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q352374 |
| concepts[5].display_name | Adaptation (eye) |
| concepts[6].id | https://openalex.org/C86803240 |
| concepts[6].level | 0 |
| concepts[6].score | 0.4487171471118927 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[6].display_name | Biology |
| concepts[7].id | https://openalex.org/C178666793 |
| concepts[7].level | 3 |
| concepts[7].score | 0.425687313079834 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q422476 |
| concepts[7].display_name | Phosphatase |
| concepts[8].id | https://openalex.org/C89423630 |
| concepts[8].level | 1 |
| concepts[8].score | 0.4025386869907379 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q7193 |
| concepts[8].display_name | Microbiology |
| concepts[9].id | https://openalex.org/C181199279 |
| concepts[9].level | 2 |
| concepts[9].score | 0.27793365716934204 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q8047 |
| concepts[9].display_name | Enzyme |
| concepts[10].id | https://openalex.org/C55493867 |
| concepts[10].level | 1 |
| concepts[10].score | 0.27664434909820557 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q7094 |
| concepts[10].display_name | Biochemistry |
| concepts[11].id | https://openalex.org/C54355233 |
| concepts[11].level | 1 |
| concepts[11].score | 0.19339331984519958 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[11].display_name | Genetics |
| concepts[12].id | https://openalex.org/C186060115 |
| concepts[12].level | 1 |
| concepts[12].score | 0.12603449821472168 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q30336093 |
| concepts[12].display_name | Biological system |
| concepts[13].id | https://openalex.org/C2993175405 |
| concepts[13].level | 3 |
| concepts[13].score | 0.1109054684638977 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q8054 |
| concepts[13].display_name | Bacterial protein |
| concepts[14].id | https://openalex.org/C169760540 |
| concepts[14].level | 1 |
| concepts[14].score | 0.10248368978500366 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q207011 |
| concepts[14].display_name | Neuroscience |
| concepts[15].id | https://openalex.org/C523546767 |
| concepts[15].level | 2 |
| concepts[15].score | 0.09940958023071289 |
| concepts[15].wikidata | https://www.wikidata.org/wiki/Q10876 |
| concepts[15].display_name | Bacteria |
| concepts[16].id | https://openalex.org/C104317684 |
| concepts[16].level | 2 |
| concepts[16].score | 0.08101692795753479 |
| concepts[16].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[16].display_name | Gene |
| concepts[17].id | https://openalex.org/C150194340 |
| concepts[17].level | 3 |
| concepts[17].score | 0.06253144145011902 |
| concepts[17].wikidata | https://www.wikidata.org/wiki/Q26972 |
| concepts[17].display_name | Gene expression |
| concepts[18].id | https://openalex.org/C547475151 |
| concepts[18].level | 3 |
| concepts[18].score | 0.0 |
| concepts[18].wikidata | https://www.wikidata.org/wiki/Q25419 |
| concepts[18].display_name | Escherichia coli |
| keywords[0].id | https://openalex.org/keywords/regulon |
| keywords[0].score | 0.8609878420829773 |
| keywords[0].display_name | Regulon |
| keywords[1].id | https://openalex.org/keywords/staphylococcus-aureus |
| keywords[1].score | 0.5623898506164551 |
| keywords[1].display_name | Staphylococcus aureus |
| keywords[2].id | https://openalex.org/keywords/operon |
| keywords[2].score | 0.5371703505516052 |
| keywords[2].display_name | Operon |
| keywords[3].id | https://openalex.org/keywords/transcriptome |
| keywords[3].score | 0.5059027075767517 |
| keywords[3].display_name | Transcriptome |
| keywords[4].id | https://openalex.org/keywords/metabolic-activity |
| keywords[4].score | 0.4793477952480316 |
| keywords[4].display_name | Metabolic activity |
| keywords[5].id | https://openalex.org/keywords/adaptation |
| keywords[5].score | 0.4491193890571594 |
| keywords[5].display_name | Adaptation (eye) |
| keywords[6].id | https://openalex.org/keywords/biology |
| keywords[6].score | 0.4487171471118927 |
| keywords[6].display_name | Biology |
| keywords[7].id | https://openalex.org/keywords/phosphatase |
| keywords[7].score | 0.425687313079834 |
| keywords[7].display_name | Phosphatase |
| keywords[8].id | https://openalex.org/keywords/microbiology |
| keywords[8].score | 0.4025386869907379 |
| keywords[8].display_name | Microbiology |
| keywords[9].id | https://openalex.org/keywords/enzyme |
| keywords[9].score | 0.27793365716934204 |
| keywords[9].display_name | Enzyme |
| keywords[10].id | https://openalex.org/keywords/biochemistry |
| keywords[10].score | 0.27664434909820557 |
| keywords[10].display_name | Biochemistry |
| keywords[11].id | https://openalex.org/keywords/genetics |
| keywords[11].score | 0.19339331984519958 |
| keywords[11].display_name | Genetics |
| keywords[12].id | https://openalex.org/keywords/biological-system |
| keywords[12].score | 0.12603449821472168 |
| keywords[12].display_name | Biological system |
| keywords[13].id | https://openalex.org/keywords/bacterial-protein |
| keywords[13].score | 0.1109054684638977 |
| keywords[13].display_name | Bacterial protein |
| keywords[14].id | https://openalex.org/keywords/neuroscience |
| keywords[14].score | 0.10248368978500366 |
| keywords[14].display_name | Neuroscience |
| keywords[15].id | https://openalex.org/keywords/bacteria |
| keywords[15].score | 0.09940958023071289 |
| keywords[15].display_name | Bacteria |
| keywords[16].id | https://openalex.org/keywords/gene |
| keywords[16].score | 0.08101692795753479 |
| keywords[16].display_name | Gene |
| keywords[17].id | https://openalex.org/keywords/gene-expression |
| keywords[17].score | 0.06253144145011902 |
| keywords[17].display_name | Gene expression |
| language | en |
| locations[0].id | doi:10.3390/microorganisms9102148 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4210222090 |
| locations[0].source.issn | 2076-2607 |
| locations[0].source.type | journal |
| locations[0].source.is_oa | True |
| locations[0].source.issn_l | 2076-2607 |
| locations[0].source.is_core | True |
| locations[0].source.is_in_doaj | True |
| locations[0].source.display_name | Microorganisms |
| locations[0].source.host_organization | https://openalex.org/P4310310987 |
| locations[0].source.host_organization_name | Multidisciplinary Digital Publishing Institute |
| locations[0].source.host_organization_lineage | https://openalex.org/P4310310987 |
| locations[0].license | cc-by |
| locations[0].pdf_url | https://www.mdpi.com/2076-2607/9/10/2148/pdf?version=1634292897 |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | Microorganisms |
| locations[0].landing_page_url | https://doi.org/10.3390/microorganisms9102148 |
| locations[1].id | pmid:34683468 |
| locations[1].is_oa | False |
| locations[1].source.id | https://openalex.org/S4306525036 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | PubMed |
| locations[1].source.host_organization | https://openalex.org/I1299303238 |
| locations[1].source.host_organization_name | National Institutes of Health |
| locations[1].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[1].license | |
| locations[1].pdf_url | |
| locations[1].version | publishedVersion |
| locations[1].raw_type | |
| locations[1].license_id | |
| locations[1].is_accepted | True |
| locations[1].is_published | True |
| locations[1].raw_source_name | Microorganisms |
| locations[1].landing_page_url | https://pubmed.ncbi.nlm.nih.gov/34683468 |
| locations[2].id | pmh:oai:unige.ch:aou:unige:162618 |
| locations[2].is_oa | True |
| locations[2].source.id | https://openalex.org/S4306402259 |
| locations[2].source.issn | |
| locations[2].source.type | repository |
| locations[2].source.is_oa | False |
| locations[2].source.issn_l | |
| locations[2].source.is_core | False |
| locations[2].source.is_in_doaj | False |
| locations[2].source.display_name | Archive ouverte UNIGE (University of Geneva) |
| locations[2].source.host_organization | https://openalex.org/I114457229 |
| locations[2].source.host_organization_name | University of Geneva |
| locations[2].source.host_organization_lineage | https://openalex.org/I114457229 |
| locations[2].license | other-oa |
| locations[2].pdf_url | |
| locations[2].version | publishedVersion |
| locations[2].raw_type | info:eu-repo/semantics/article |
| locations[2].license_id | https://openalex.org/licenses/other-oa |
| locations[2].is_accepted | True |
| locations[2].is_published | True |
| locations[2].raw_source_name | ISSN: 2076-2607 |
| locations[2].landing_page_url | https://archive-ouverte.unige.ch/unige:162618 |
| locations[3].id | pmh:oai:repository.ubn.ru.nl:2066/247225 |
| locations[3].is_oa | False |
| locations[3].source.id | https://openalex.org/S4306401067 |
| locations[3].source.issn | |
| locations[3].source.type | repository |
| locations[3].source.is_oa | False |
| locations[3].source.issn_l | |
| locations[3].source.is_core | False |
| locations[3].source.is_in_doaj | False |
| locations[3].source.display_name | Radboud Repository (Radboud University) |
| locations[3].source.host_organization | https://openalex.org/I145872427 |
| locations[3].source.host_organization_name | Radboud University Nijmegen |
| locations[3].source.host_organization_lineage | https://openalex.org/I145872427 |
| locations[3].license | |
| locations[3].pdf_url | |
| locations[3].version | submittedVersion |
| locations[3].raw_type | Article / Letter to editor |
| locations[3].license_id | |
| locations[3].is_accepted | False |
| locations[3].is_published | False |
| locations[3].raw_source_name | Microorganisms, 9, 10, pp. 1-30 |
| locations[3].landing_page_url | https://repository.ubn.ru.nl/handle/2066/247225 |
| locations[4].id | pmh:oai:mdpi.com:/2076-2607/9/10/2148/ |
| locations[4].is_oa | True |
| locations[4].source.id | https://openalex.org/S4306400947 |
| locations[4].source.issn | |
| locations[4].source.type | repository |
| locations[4].source.is_oa | True |
| locations[4].source.issn_l | |
| locations[4].source.is_core | False |
| locations[4].source.is_in_doaj | False |
| locations[4].source.display_name | MDPI (MDPI AG) |
| locations[4].source.host_organization | https://openalex.org/I4210097602 |
| locations[4].source.host_organization_name | Multidisciplinary Digital Publishing Institute (Switzerland) |
| locations[4].source.host_organization_lineage | https://openalex.org/I4210097602 |
| locations[4].license | cc-by |
| locations[4].pdf_url | |
| locations[4].version | submittedVersion |
| locations[4].raw_type | Text |
| locations[4].license_id | https://openalex.org/licenses/cc-by |
| locations[4].is_accepted | False |
| locations[4].is_published | False |
| locations[4].raw_source_name | Microorganisms; Volume 9; Issue 10; Pages: 2148 |
| locations[4].landing_page_url | https://dx.doi.org/10.3390/microorganisms9102148 |
| locations[5].id | pmh:oai:pubmedcentral.nih.gov:8537086 |
| locations[5].is_oa | True |
| locations[5].source.id | https://openalex.org/S2764455111 |
| locations[5].source.issn | |
| locations[5].source.type | repository |
| locations[5].source.is_oa | False |
| locations[5].source.issn_l | |
| locations[5].source.is_core | False |
| locations[5].source.is_in_doaj | False |
| locations[5].source.display_name | PubMed Central |
| locations[5].source.host_organization | https://openalex.org/I1299303238 |
| locations[5].source.host_organization_name | National Institutes of Health |
| locations[5].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[5].license | other-oa |
| locations[5].pdf_url | |
| locations[5].version | submittedVersion |
| locations[5].raw_type | Text |
| locations[5].license_id | https://openalex.org/licenses/other-oa |
| locations[5].is_accepted | False |
| locations[5].is_published | False |
| locations[5].raw_source_name | Microorganisms |
| locations[5].landing_page_url | https://www.ncbi.nlm.nih.gov/pmc/articles/8537086 |
| locations[6].id | pmh:ru:oai:repository.ubn.ru.nl:2066/247225 |
| locations[6].is_oa | True |
| locations[6].source.id | https://openalex.org/S4306401843 |
| locations[6].source.issn | |
| locations[6].source.type | repository |
| locations[6].source.is_oa | False |
| locations[6].source.issn_l | |
| locations[6].source.is_core | False |
| locations[6].source.is_in_doaj | False |
| locations[6].source.display_name | Data Archiving and Networked Services (DANS) |
| locations[6].source.host_organization | https://openalex.org/I1322597698 |
| locations[6].source.host_organization_name | Royal Netherlands Academy of Arts and Sciences |
| locations[6].source.host_organization_lineage | https://openalex.org/I1322597698 |
| locations[6].license | other-oa |
| locations[6].pdf_url | |
| locations[6].version | submittedVersion |
| locations[6].raw_type | info:eu-repo/semantics/article |
| locations[6].license_id | https://openalex.org/licenses/other-oa |
| locations[6].is_accepted | False |
| locations[6].is_published | False |
| locations[6].raw_source_name | Microorganisms, 9, 1 - 30 |
| locations[6].landing_page_url | http://hdl.handle.net/2066/247225 |
| locations[7].id | pmh:oai:doaj.org/article:65b0974dbaee4046a3a6639425446db3 |
| locations[7].is_oa | True |
| locations[7].source.id | https://openalex.org/S4306401280 |
| locations[7].source.issn | |
| locations[7].source.type | repository |
| locations[7].source.is_oa | False |
| locations[7].source.issn_l | |
| locations[7].source.is_core | False |
| locations[7].source.is_in_doaj | False |
| locations[7].source.display_name | DOAJ (DOAJ: Directory of Open Access Journals) |
| locations[7].source.host_organization | |
| locations[7].source.host_organization_name | |
| locations[7].source.host_organization_lineage | |
| locations[7].license | cc-by-sa |
| locations[7].pdf_url | |
| locations[7].version | submittedVersion |
| locations[7].raw_type | article |
| locations[7].license_id | https://openalex.org/licenses/cc-by-sa |
| locations[7].is_accepted | False |
| locations[7].is_published | False |
| locations[7].raw_source_name | Microorganisms, Vol 9, Iss 10, p 2148 (2021) |
| locations[7].landing_page_url | https://doaj.org/article/65b0974dbaee4046a3a6639425446db3 |
| locations[8].id | pmh:oai:opus.bibliothek.uni-wuerzburg.de:24845 |
| locations[8].is_oa | True |
| locations[8].source.id | https://openalex.org/S4306401635 |
| locations[8].source.issn | |
| locations[8].source.type | repository |
| locations[8].source.is_oa | False |
| locations[8].source.issn_l | |
| locations[8].source.is_core | False |
| locations[8].source.is_in_doaj | False |
| locations[8].source.display_name | Online Publication Service of Würzburg University (Würzburg University) |
| locations[8].source.host_organization | https://openalex.org/I25974101 |
| locations[8].source.host_organization_name | University of Würzburg |
| locations[8].source.host_organization_lineage | https://openalex.org/I25974101 |
| locations[8].license | cc-by |
| locations[8].pdf_url | |
| locations[8].version | submittedVersion |
| locations[8].raw_type | article |
| locations[8].license_id | https://openalex.org/licenses/cc-by |
| locations[8].is_accepted | False |
| locations[8].is_published | False |
| locations[8].raw_source_name | |
| locations[8].landing_page_url | https://nbn-resolving.org/urn:nbn:de:bvb:20-opus-248459 |
| indexed_in | crossref, doaj, pubmed |
| authorships[0].author.id | https://openalex.org/A5008078406 |
| authorships[0].author.orcid | https://orcid.org/0000-0002-0305-2522 |
| authorships[0].author.display_name | Chunguang Liang |
| authorships[0].countries | DE |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I25974101 |
| authorships[0].affiliations[0].raw_affiliation_string | Department of Bioinformatics, Biocenter, Am Hubland, University of Würzburg, 97074 Würzburg, Germany |
| authorships[0].institutions[0].id | https://openalex.org/I25974101 |
| authorships[0].institutions[0].ror | https://ror.org/00fbnyb24 |
| authorships[0].institutions[0].type | education |
| authorships[0].institutions[0].lineage | https://openalex.org/I25974101 |
| authorships[0].institutions[0].country_code | DE |
| authorships[0].institutions[0].display_name | University of Würzburg |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Chunguang Liang |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Department of Bioinformatics, Biocenter, Am Hubland, University of Würzburg, 97074 Würzburg, Germany |
| authorships[1].author.id | https://openalex.org/A5015491144 |
| authorships[1].author.orcid | https://orcid.org/0000-0002-9399-044X |
| authorships[1].author.display_name | Ana B. Rios‐Miguel |
| authorships[1].countries | NL |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I145872427 |
| authorships[1].affiliations[0].raw_affiliation_string | Department of Environmental Microbiology, Institute of Water and Wetland Research, Radboud University, 6525 AJ Nijmegen, The Netherlands |
| authorships[1].institutions[0].id | https://openalex.org/I145872427 |
| authorships[1].institutions[0].ror | https://ror.org/016xsfp80 |
| authorships[1].institutions[0].type | education |
| authorships[1].institutions[0].lineage | https://openalex.org/I145872427 |
| authorships[1].institutions[0].country_code | NL |
| authorships[1].institutions[0].display_name | Radboud University Nijmegen |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Ana B. Rios-Miguel |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | Department of Environmental Microbiology, Institute of Water and Wetland Research, Radboud University, 6525 AJ Nijmegen, The Netherlands |
| authorships[2].author.id | https://openalex.org/A5061129172 |
| authorships[2].author.orcid | |
| authorships[2].author.display_name | Marcel Jarick |
| authorships[2].countries | DE |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I25974101 |
| authorships[2].affiliations[0].raw_affiliation_string | Institute for Molecular Infection Biology, Josef-Schneider-Straße 2/D15, University of Würzburg, 97080 Würzburg, Germany |
| authorships[2].institutions[0].id | https://openalex.org/I25974101 |
| authorships[2].institutions[0].ror | https://ror.org/00fbnyb24 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I25974101 |
| authorships[2].institutions[0].country_code | DE |
| authorships[2].institutions[0].display_name | University of Würzburg |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Marcel Jarick |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | Institute for Molecular Infection Biology, Josef-Schneider-Straße 2/D15, University of Würzburg, 97080 Würzburg, Germany |
| authorships[3].author.id | https://openalex.org/A5056925664 |
| authorships[3].author.orcid | |
| authorships[3].author.display_name | Priya Neurgaonkar |
| authorships[3].countries | DE |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I25974101 |
| authorships[3].affiliations[0].raw_affiliation_string | Department of Bioinformatics, Biocenter, Am Hubland, University of Würzburg, 97074 Würzburg, Germany |
| authorships[3].institutions[0].id | https://openalex.org/I25974101 |
| authorships[3].institutions[0].ror | https://ror.org/00fbnyb24 |
| authorships[3].institutions[0].type | education |
| authorships[3].institutions[0].lineage | https://openalex.org/I25974101 |
| authorships[3].institutions[0].country_code | DE |
| authorships[3].institutions[0].display_name | University of Würzburg |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Priya Neurgaonkar |
| authorships[3].is_corresponding | False |
| authorships[3].raw_affiliation_strings | Department of Bioinformatics, Biocenter, Am Hubland, University of Würzburg, 97074 Würzburg, Germany |
| authorships[4].author.id | https://openalex.org/A5004068707 |
| authorships[4].author.orcid | |
| authorships[4].author.display_name | Myriam Girard |
| authorships[4].countries | CH |
| authorships[4].affiliations[0].institution_ids | https://openalex.org/I114457229 |
| authorships[4].affiliations[0].raw_affiliation_string | Genomic Research Laboratory, Service of Infectious Diseases, University of Geneva Hospitals, CH-1211 Geneva 14, Switzerland |
| authorships[4].institutions[0].id | https://openalex.org/I114457229 |
| authorships[4].institutions[0].ror | https://ror.org/01swzsf04 |
| authorships[4].institutions[0].type | education |
| authorships[4].institutions[0].lineage | https://openalex.org/I114457229 |
| authorships[4].institutions[0].country_code | CH |
| authorships[4].institutions[0].display_name | University of Geneva |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Myriam Girard |
| authorships[4].is_corresponding | False |
| authorships[4].raw_affiliation_strings | Genomic Research Laboratory, Service of Infectious Diseases, University of Geneva Hospitals, CH-1211 Geneva 14, Switzerland |
| authorships[5].author.id | https://openalex.org/A5010055797 |
| authorships[5].author.orcid | https://orcid.org/0000-0002-0540-750X |
| authorships[5].author.display_name | Patrice François |
| authorships[5].countries | CH |
| authorships[5].affiliations[0].institution_ids | https://openalex.org/I114457229 |
| authorships[5].affiliations[0].raw_affiliation_string | Genomic Research Laboratory, Service of Infectious Diseases, University of Geneva Hospitals, CH-1211 Geneva 14, Switzerland |
| authorships[5].institutions[0].id | https://openalex.org/I114457229 |
| authorships[5].institutions[0].ror | https://ror.org/01swzsf04 |
| authorships[5].institutions[0].type | education |
| authorships[5].institutions[0].lineage | https://openalex.org/I114457229 |
| authorships[5].institutions[0].country_code | CH |
| authorships[5].institutions[0].display_name | University of Geneva |
| authorships[5].author_position | middle |
| authorships[5].raw_author_name | Patrice François |
| authorships[5].is_corresponding | False |
| authorships[5].raw_affiliation_strings | Genomic Research Laboratory, Service of Infectious Diseases, University of Geneva Hospitals, CH-1211 Geneva 14, Switzerland |
| authorships[6].author.id | https://openalex.org/A5088214706 |
| authorships[6].author.orcid | https://orcid.org/0000-0001-5464-7764 |
| authorships[6].author.display_name | Jacques Schrenzel |
| authorships[6].countries | CH |
| authorships[6].affiliations[0].institution_ids | https://openalex.org/I114457229 |
| authorships[6].affiliations[0].raw_affiliation_string | Genomic Research Laboratory, Service of Infectious Diseases, University of Geneva Hospitals, CH-1211 Geneva 14, Switzerland |
| authorships[6].institutions[0].id | https://openalex.org/I114457229 |
| authorships[6].institutions[0].ror | https://ror.org/01swzsf04 |
| authorships[6].institutions[0].type | education |
| authorships[6].institutions[0].lineage | https://openalex.org/I114457229 |
| authorships[6].institutions[0].country_code | CH |
| authorships[6].institutions[0].display_name | University of Geneva |
| authorships[6].author_position | middle |
| authorships[6].raw_author_name | Jacques Schrenzel |
| authorships[6].is_corresponding | False |
| authorships[6].raw_affiliation_strings | Genomic Research Laboratory, Service of Infectious Diseases, University of Geneva Hospitals, CH-1211 Geneva 14, Switzerland |
| authorships[7].author.id | https://openalex.org/A5029089879 |
| authorships[7].author.orcid | https://orcid.org/0000-0002-0206-5321 |
| authorships[7].author.display_name | Eslam S. Ibrahim |
| authorships[7].countries | DE, EG |
| authorships[7].affiliations[0].institution_ids | https://openalex.org/I25974101 |
| authorships[7].affiliations[0].raw_affiliation_string | Institute for Molecular Infection Biology, Josef-Schneider-Straße 2/D15, University of Würzburg, 97080 Würzburg, Germany |
| authorships[7].affiliations[1].institution_ids | https://openalex.org/I145487455 |
| authorships[7].affiliations[1].raw_affiliation_string | Department of Microbiology and Immunology, Faculty of Pharmacy, Cairo University, Cairo 11562, Egypt |
| authorships[7].institutions[0].id | https://openalex.org/I25974101 |
| authorships[7].institutions[0].ror | https://ror.org/00fbnyb24 |
| authorships[7].institutions[0].type | education |
| authorships[7].institutions[0].lineage | https://openalex.org/I25974101 |
| authorships[7].institutions[0].country_code | DE |
| authorships[7].institutions[0].display_name | University of Würzburg |
| authorships[7].institutions[1].id | https://openalex.org/I145487455 |
| authorships[7].institutions[1].ror | https://ror.org/03q21mh05 |
| authorships[7].institutions[1].type | education |
| authorships[7].institutions[1].lineage | https://openalex.org/I145487455 |
| authorships[7].institutions[1].country_code | EG |
| authorships[7].institutions[1].display_name | Cairo University |
| authorships[7].author_position | middle |
| authorships[7].raw_author_name | Eslam S. Ibrahim |
| authorships[7].is_corresponding | False |
| authorships[7].raw_affiliation_strings | Department of Microbiology and Immunology, Faculty of Pharmacy, Cairo University, Cairo 11562, Egypt, Institute for Molecular Infection Biology, Josef-Schneider-Straße 2/D15, University of Würzburg, 97080 Würzburg, Germany |
| authorships[8].author.id | https://openalex.org/A5107393601 |
| authorships[8].author.orcid | |
| authorships[8].author.display_name | Knut Ohlsen |
| authorships[8].countries | DE |
| authorships[8].affiliations[0].institution_ids | https://openalex.org/I25974101 |
| authorships[8].affiliations[0].raw_affiliation_string | Institute for Molecular Infection Biology, Josef-Schneider-Straße 2/D15, University of Würzburg, 97080 Würzburg, Germany |
| authorships[8].institutions[0].id | https://openalex.org/I25974101 |
| authorships[8].institutions[0].ror | https://ror.org/00fbnyb24 |
| authorships[8].institutions[0].type | education |
| authorships[8].institutions[0].lineage | https://openalex.org/I25974101 |
| authorships[8].institutions[0].country_code | DE |
| authorships[8].institutions[0].display_name | University of Würzburg |
| authorships[8].author_position | middle |
| authorships[8].raw_author_name | Knut Ohlsen |
| authorships[8].is_corresponding | True |
| authorships[8].raw_affiliation_strings | Institute for Molecular Infection Biology, Josef-Schneider-Straße 2/D15, University of Würzburg, 97080 Würzburg, Germany |
| authorships[9].author.id | https://openalex.org/A5086202312 |
| authorships[9].author.orcid | https://orcid.org/0000-0003-1886-7625 |
| authorships[9].author.display_name | Thomas Dandekar |
| authorships[9].countries | DE |
| authorships[9].affiliations[0].institution_ids | https://openalex.org/I25974101 |
| authorships[9].affiliations[0].raw_affiliation_string | Department of Bioinformatics, Biocenter, Am Hubland, University of Würzburg, 97074 Würzburg, Germany |
| authorships[9].institutions[0].id | https://openalex.org/I25974101 |
| authorships[9].institutions[0].ror | https://ror.org/00fbnyb24 |
| authorships[9].institutions[0].type | education |
| authorships[9].institutions[0].lineage | https://openalex.org/I25974101 |
| authorships[9].institutions[0].country_code | DE |
| authorships[9].institutions[0].display_name | University of Würzburg |
| authorships[9].author_position | last |
| authorships[9].raw_author_name | Thomas Dandekar |
| authorships[9].is_corresponding | True |
| authorships[9].raw_affiliation_strings | Department of Bioinformatics, Biocenter, Am Hubland, University of Würzburg, 97074 Würzburg, Germany |
| has_content.pdf | True |
| has_content.grobid_xml | True |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://www.mdpi.com/2076-2607/9/10/2148/pdf?version=1634292897 |
| open_access.oa_status | gold |
| open_access.any_repository_has_fulltext | False |
| created_date | 2021-10-25T00:00:00 |
| display_name | Staphylococcus aureus Transcriptome Data and Metabolic Modelling Investigate the Interplay of Ser/Thr Kinase PknB, Its Phosphatase Stp, the glmR/yvcK Regulon and the cdaA Operon for Metabolic Adaptation |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T10932 |
| primary_topic.field.id | https://openalex.org/fields/13 |
| primary_topic.field.display_name | Biochemistry, Genetics and Molecular Biology |
| primary_topic.score | 0.9986000061035156 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1312 |
| primary_topic.subfield.display_name | Molecular Biology |
| primary_topic.display_name | Microbial Metabolic Engineering and Bioproduction |
| related_works | https://openalex.org/W61127607, https://openalex.org/W1984136937, https://openalex.org/W1786268878, https://openalex.org/W2794893700, https://openalex.org/W4281651837, https://openalex.org/W4313622135, https://openalex.org/W4282031271, https://openalex.org/W2171972144, https://openalex.org/W2132790938, https://openalex.org/W1796329565 |
| cited_by_count | 10 |
| counts_by_year[0].year | 2025 |
| counts_by_year[0].cited_by_count | 4 |
| counts_by_year[1].year | 2024 |
| counts_by_year[1].cited_by_count | 1 |
| counts_by_year[2].year | 2023 |
| counts_by_year[2].cited_by_count | 4 |
| counts_by_year[3].year | 2022 |
| counts_by_year[3].cited_by_count | 1 |
| locations_count | 9 |
| best_oa_location.id | doi:10.3390/microorganisms9102148 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4210222090 |
| best_oa_location.source.issn | 2076-2607 |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | True |
| best_oa_location.source.issn_l | 2076-2607 |
| best_oa_location.source.is_core | True |
| best_oa_location.source.is_in_doaj | True |
| best_oa_location.source.display_name | Microorganisms |
| best_oa_location.source.host_organization | https://openalex.org/P4310310987 |
| best_oa_location.source.host_organization_name | Multidisciplinary Digital Publishing Institute |
| best_oa_location.source.host_organization_lineage | https://openalex.org/P4310310987 |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | https://www.mdpi.com/2076-2607/9/10/2148/pdf?version=1634292897 |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | journal-article |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | Microorganisms |
| best_oa_location.landing_page_url | https://doi.org/10.3390/microorganisms9102148 |
| primary_location.id | doi:10.3390/microorganisms9102148 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4210222090 |
| primary_location.source.issn | 2076-2607 |
| primary_location.source.type | journal |
| primary_location.source.is_oa | True |
| primary_location.source.issn_l | 2076-2607 |
| primary_location.source.is_core | True |
| primary_location.source.is_in_doaj | True |
| primary_location.source.display_name | Microorganisms |
| primary_location.source.host_organization | https://openalex.org/P4310310987 |
| primary_location.source.host_organization_name | Multidisciplinary Digital Publishing Institute |
| primary_location.source.host_organization_lineage | https://openalex.org/P4310310987 |
| primary_location.license | cc-by |
| primary_location.pdf_url | https://www.mdpi.com/2076-2607/9/10/2148/pdf?version=1634292897 |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | Microorganisms |
| primary_location.landing_page_url | https://doi.org/10.3390/microorganisms9102148 |
| publication_date | 2021-10-14 |
| publication_year | 2021 |
| referenced_works | https://openalex.org/W2109613442, https://openalex.org/W2131178894, https://openalex.org/W2000072052, https://openalex.org/W2083178365, https://openalex.org/W2055149442, https://openalex.org/W2020799925, https://openalex.org/W2113452278, https://openalex.org/W2073829378, https://openalex.org/W2890773099, https://openalex.org/W2134354446, https://openalex.org/W2028067981, https://openalex.org/W2072608250, https://openalex.org/W1994538322, https://openalex.org/W2059444426, https://openalex.org/W2135673226, https://openalex.org/W2037450561, https://openalex.org/W2122282339, https://openalex.org/W2654770002, https://openalex.org/W2547260309, https://openalex.org/W2111587697, https://openalex.org/W3114019208, https://openalex.org/W2028222221, https://openalex.org/W2892540384, https://openalex.org/W2108244474, https://openalex.org/W1986355672, https://openalex.org/W2116857869, https://openalex.org/W2800763431, https://openalex.org/W2124036689, https://openalex.org/W1998881688, https://openalex.org/W1975289975, https://openalex.org/W2044935306, https://openalex.org/W2007477611, https://openalex.org/W2175946349, https://openalex.org/W1988419460, https://openalex.org/W2013381224, https://openalex.org/W2605275799, https://openalex.org/W2588283941, https://openalex.org/W2164191772, https://openalex.org/W2156126133, https://openalex.org/W4234320099, https://openalex.org/W1920924307, https://openalex.org/W2147674484, https://openalex.org/W2113247851, https://openalex.org/W108176572, https://openalex.org/W2140040278, https://openalex.org/W2105924489, https://openalex.org/W2132926880, https://openalex.org/W2799524357, https://openalex.org/W2149525061, https://openalex.org/W1990046762, https://openalex.org/W2108086845, https://openalex.org/W2782327081, https://openalex.org/W2027730892, https://openalex.org/W2580767461 |
| referenced_works_count | 54 |
| abstract_inverted_index.A | 150 |
| abstract_inverted_index.a | 80 |
| abstract_inverted_index.In | 119 |
| abstract_inverted_index.S. | 18, 25, 155, 204 |
| abstract_inverted_index.We | 45, 78 |
| abstract_inverted_index.as | 95, 97 |
| abstract_inverted_index.by | 61, 108, 114 |
| abstract_inverted_index.in | 15, 127 |
| abstract_inverted_index.of | 11, 24, 58, 83, 117, 140, 194 |
| abstract_inverted_index.on | 33, 70, 87, 203 |
| abstract_inverted_index.or | 54 |
| abstract_inverted_index.to | 48, 71, 146, 170 |
| abstract_inverted_index.For | 163 |
| abstract_inverted_index.Stp | 7, 86 |
| abstract_inverted_index.The | 178 |
| abstract_inverted_index.and | 3, 68, 85, 90, 130, 174, 182, 198 |
| abstract_inverted_index.are | 8 |
| abstract_inverted_index.but | 111, 137 |
| abstract_inverted_index.its | 4 |
| abstract_inverted_index.off | 67 |
| abstract_inverted_index.stp | 53, 109 |
| abstract_inverted_index.the | 16, 73, 147, 161, 175, 183, 192, 199 |
| abstract_inverted_index.was | 105, 168 |
| abstract_inverted_index.8325 | 158 |
| abstract_inverted_index.CdaA | 173 |
| abstract_inverted_index.GlmR | 180 |
| abstract_inverted_index.GlmS | 184 |
| abstract_inverted_index.NCTC | 157 |
| abstract_inverted_index.PknB | 2, 62, 84, 167, 189 |
| abstract_inverted_index.acid | 65, 93, 133 |
| abstract_inverted_index.cell | 13, 74 |
| abstract_inverted_index.data | 23, 41 |
| abstract_inverted_index.from | 39, 135, 143, 154 |
| abstract_inverted_index.gene | 21 |
| abstract_inverted_index.loss | 116 |
| abstract_inverted_index.many | 12, 195 |
| abstract_inverted_index.only | 112 |
| abstract_inverted_index.shed | 201 |
| abstract_inverted_index.this | 164 |
| abstract_inverted_index.well | 96 |
| abstract_inverted_index.were | 186 |
| abstract_inverted_index.wild | 148 |
| abstract_inverted_index.with | 75, 172 |
| abstract_inverted_index.NewHG | 28 |
| abstract_inverted_index.PknB. | 118 |
| abstract_inverted_index.amino | 64, 92, 132 |
| abstract_inverted_index.both. | 55 |
| abstract_inverted_index.found | 169 |
| abstract_inverted_index.genes | 125 |
| abstract_inverted_index.light | 202 |
| abstract_inverted_index.lower | 138 |
| abstract_inverted_index.pknB, | 52 |
| abstract_inverted_index.study | 200 |
| abstract_inverted_index.their | 31 |
| abstract_inverted_index.these | 40 |
| abstract_inverted_index.type. | 149 |
| abstract_inverted_index.aureus | 26, 156, 205 |
| abstract_inverted_index.double | 120 |
| abstract_inverted_index.effect | 32 |
| abstract_inverted_index.higher | 122 |
| abstract_inverted_index.impact | 82 |
| abstract_inverted_index.kinase | 1 |
| abstract_inverted_index.purine | 129 |
| abstract_inverted_index.second | 151 |
| abstract_inverted_index.strain | 27 |
| abstract_inverted_index.target | 59 |
| abstract_inverted_index.(sigB+) | 29 |
| abstract_inverted_index.Ser/Thr | 56 |
| abstract_inverted_index.aureus. | 19 |
| abstract_inverted_index.dataset | 153 |
| abstract_inverted_index.glucose | 136, 144 |
| abstract_inverted_index.lacking | 51 |
| abstract_inverted_index.lowered | 191 |
| abstract_inverted_index.provide | 72 |
| abstract_inverted_index.strains | 50 |
| abstract_inverted_index.activity | 123, 139 |
| abstract_inverted_index.aromatic | 91, 131 |
| abstract_inverted_index.compared | 46, 145 |
| abstract_inverted_index.deletion | 49, 110 |
| abstract_inverted_index.factors, | 197 |
| abstract_inverted_index.impaired | 107 |
| abstract_inverted_index.inferred | 42 |
| abstract_inverted_index.interact | 171 |
| abstract_inverted_index.involved | 126, 179 |
| abstract_inverted_index.pathogen | 17 |
| abstract_inverted_index.proteins | 60 |
| abstract_inverted_index.regulon. | 177 |
| abstract_inverted_index.revealed | 79 |
| abstract_inverted_index.slightly | 113 |
| abstract_inverted_index.switched | 63 |
| abstract_inverted_index.(sigB−) | 159 |
| abstract_inverted_index.Moreover, | 36, 102 |
| abstract_inverted_index.aspartate | 100 |
| abstract_inverted_index.concerned | 124 |
| abstract_inverted_index.functions | 14 |
| abstract_inverted_index.important | 9 |
| abstract_inverted_index.infection | 206 |
| abstract_inverted_index.involving | 99 |
| abstract_inverted_index.metabolic | 37, 43, 165 |
| abstract_inverted_index.modelled. | 187 |
| abstract_inverted_index.modelling | 38 |
| abstract_inverted_index.structure | 181 |
| abstract_inverted_index.synthesis | 104, 134, 142 |
| abstract_inverted_index.validated | 160 |
| abstract_inverted_index.virulence | 196 |
| abstract_inverted_index.wild-type | 47 |
| abstract_inverted_index.yvcK/glmR | 176 |
| abstract_inverted_index.catabolism | 66, 98 |
| abstract_inverted_index.elucidated | 30 |
| abstract_inverted_index.expression | 22, 193 |
| abstract_inverted_index.functional | 115 |
| abstract_inverted_index.functions. | 35 |
| abstract_inverted_index.knockouts, | 121 |
| abstract_inverted_index.nucleotide | 89 |
| abstract_inverted_index.processes. | 207 |
| abstract_inverted_index.pyrimidine | 103, 141 |
| abstract_inverted_index.regulators | 10 |
| abstract_inverted_index.riboswitch | 185 |
| abstract_inverted_index.sufficient | 76 |
| abstract_inverted_index.synthesis, | 94 |
| abstract_inverted_index.adaptation, | 166 |
| abstract_inverted_index.components. | 77 |
| abstract_inverted_index.phosphatase | 6 |
| abstract_inverted_index.significant | 81 |
| abstract_inverted_index.Furthermore, | 188 |
| abstract_inverted_index.Genome-scale | 20 |
| abstract_inverted_index.adaptations. | 44 |
| abstract_inverted_index.dramatically | 106 |
| abstract_inverted_index.predictions. | 162 |
| abstract_inverted_index.corresponding | 5 |
| abstract_inverted_index.physiological | 34 |
| abstract_inverted_index.transaminase. | 101 |
| abstract_inverted_index.transcriptome | 152 |
| abstract_inverted_index.peptidoglycan, | 88, 128 |
| abstract_inverted_index.gluconeogenesis | 69 |
| abstract_inverted_index.phosphorylation | 57, 190 |
| abstract_inverted_index.Serine/threonine | 0 |
| cited_by_percentile_year.max | 98 |
| cited_by_percentile_year.min | 89 |
| corresponding_author_ids | https://openalex.org/A5086202312, https://openalex.org/A5107393601 |
| countries_distinct_count | 4 |
| institutions_distinct_count | 10 |
| corresponding_institution_ids | https://openalex.org/I25974101 |
| citation_normalized_percentile.value | 0.64223858 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |