Structural conservation of MALAT1 long non-coding RNA in cells and in evolution Article Swipe
 YOU?
·
· 2022
· Open Access
·
· DOI: https://doi.org/10.1101/2022.07.29.502018
YOU?
·
· 2022
· Open Access
·
· DOI: https://doi.org/10.1101/2022.07.29.502018
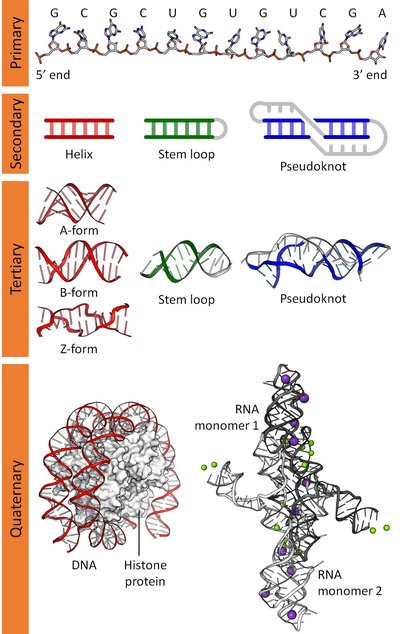
Although not canonically polyadenylated, the long non-coding RNA MALAT1 (Metastasis Associated Lung Adenocarcinoma Transcript 1) is stabilized by a highly conserved 159 nucleotide triple helix structure on its 3’ end. The entire MALAT1 transcript is over 8,000 nucleotides long in humans and is considered one of the most conserved lncRNAs, at both the sequence and structure levels. The strongest structural conservation signal (as measured by co-variation of base-pairs) is in the triple helix structure. Primary sequence analysis of co-variation alone cannot confirm the degree of structural conservation of the entire full-length transcript. Furthermore, RNA structure is often context dependent; RNA binding proteins that are differentially expressed in different cell types may alter structure. We investigate here the in cell and cell free structures of the full-length human and green monkey ( Chlorocebus sabaeus ) MALAT1 transcripts in multiple tissue-derived cell lines using SHAPE chemical probing. Our data reveals surprising levels of uniform structural conservation in different cell lines, in cells and cell free, and even between species, despite significant differences in primary sequence. The uniformity of the structural conservation across the entire transcript suggests that, despite seeing co-variation signals only in the three-helix junction of the lncRNA, the rest of the transcript’s structure is remarkably conserved at least in primates and across multiple cell types and conditions.
Related Topics
- Type
- preprint
- Language
- en
- Landing Page
- https://doi.org/10.1101/2022.07.29.502018
- https://www.biorxiv.org/content/biorxiv/early/2022/07/29/2022.07.29.502018.full.pdf
- OA Status
- green
- Cited By
- 1
- References
- 62
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4288704550
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4288704550Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1101/2022.07.29.502018Digital Object Identifier
- Title
-
Structural conservation of MALAT1 long non-coding RNA in cells and in evolutionWork title
- Type
-
preprintOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2022Year of publication
- Publication date
-
2022-07-29Full publication date if available
- Authors
-
Anaïs Monroy‐Eklund, Colin D. Taylor, Chase A. Weidmann, Christina L. Burch, Alain LaederachList of authors in order
- Landing page
-
https://doi.org/10.1101/2022.07.29.502018Publisher landing page
- PDF URL
-
https://www.biorxiv.org/content/biorxiv/early/2022/07/29/2022.07.29.502018.full.pdfDirect link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
greenOpen access status per OpenAlex
- OA URL
-
https://www.biorxiv.org/content/biorxiv/early/2022/07/29/2022.07.29.502018.full.pdfDirect OA link when available
- Concepts
-
RNA, Biology, Polyadenylation, Conserved sequence, MALAT1, Messenger RNA, Gene, Nucleic acid structure, Coding region, Triple helix, Genetics, Directionality, Peptide sequence, Long non-coding RNATop concepts (fields/topics) attached by OpenAlex
- Cited by
-
1Total citation count in OpenAlex
- Citations by year (recent)
-
2023: 1Per-year citation counts (last 5 years)
- References (count)
-
62Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4288704550 |
|---|---|
| doi | https://doi.org/10.1101/2022.07.29.502018 |
| ids.doi | https://doi.org/10.1101/2022.07.29.502018 |
| ids.openalex | https://openalex.org/W4288704550 |
| fwci | 0.13595652 |
| type | preprint |
| title | Structural conservation of MALAT1 long non-coding RNA in cells and in evolution |
| biblio.issue | |
| biblio.volume | |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T10515 |
| topics[0].field.id | https://openalex.org/fields/13 |
| topics[0].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[0].score | 1.0 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1306 |
| topics[0].subfield.display_name | Cancer Research |
| topics[0].display_name | Cancer-related molecular mechanisms research |
| topics[1].id | https://openalex.org/T10521 |
| topics[1].field.id | https://openalex.org/fields/13 |
| topics[1].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[1].score | 0.9991999864578247 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1312 |
| topics[1].subfield.display_name | Molecular Biology |
| topics[1].display_name | RNA and protein synthesis mechanisms |
| topics[2].id | https://openalex.org/T10604 |
| topics[2].field.id | https://openalex.org/fields/13 |
| topics[2].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[2].score | 0.9984999895095825 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1312 |
| topics[2].subfield.display_name | Molecular Biology |
| topics[2].display_name | RNA Research and Splicing |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C67705224 |
| concepts[0].level | 3 |
| concepts[0].score | 0.7190317511558533 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q11053 |
| concepts[0].display_name | RNA |
| concepts[1].id | https://openalex.org/C86803240 |
| concepts[1].level | 0 |
| concepts[1].score | 0.6908336877822876 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[1].display_name | Biology |
| concepts[2].id | https://openalex.org/C142575336 |
| concepts[2].level | 4 |
| concepts[2].score | 0.6887502074241638 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q412148 |
| concepts[2].display_name | Polyadenylation |
| concepts[3].id | https://openalex.org/C199216141 |
| concepts[3].level | 4 |
| concepts[3].score | 0.6553806066513062 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q4995178 |
| concepts[3].display_name | Conserved sequence |
| concepts[4].id | https://openalex.org/C2779134362 |
| concepts[4].level | 5 |
| concepts[4].score | 0.501326322555542 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q18056223 |
| concepts[4].display_name | MALAT1 |
| concepts[5].id | https://openalex.org/C105580179 |
| concepts[5].level | 3 |
| concepts[5].score | 0.4805110692977905 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q188928 |
| concepts[5].display_name | Messenger RNA |
| concepts[6].id | https://openalex.org/C104317684 |
| concepts[6].level | 2 |
| concepts[6].score | 0.46607327461242676 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[6].display_name | Gene |
| concepts[7].id | https://openalex.org/C126142528 |
| concepts[7].level | 4 |
| concepts[7].score | 0.4644019603729248 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q3771876 |
| concepts[7].display_name | Nucleic acid structure |
| concepts[8].id | https://openalex.org/C91779695 |
| concepts[8].level | 3 |
| concepts[8].score | 0.4454178810119629 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q3780824 |
| concepts[8].display_name | Coding region |
| concepts[9].id | https://openalex.org/C10919887 |
| concepts[9].level | 2 |
| concepts[9].score | 0.4361477792263031 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q7843514 |
| concepts[9].display_name | Triple helix |
| concepts[10].id | https://openalex.org/C54355233 |
| concepts[10].level | 1 |
| concepts[10].score | 0.4338851869106293 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[10].display_name | Genetics |
| concepts[11].id | https://openalex.org/C29648211 |
| concepts[11].level | 2 |
| concepts[11].score | 0.4319894313812256 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q1995607 |
| concepts[11].display_name | Directionality |
| concepts[12].id | https://openalex.org/C167625842 |
| concepts[12].level | 3 |
| concepts[12].score | 0.26507115364074707 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q899763 |
| concepts[12].display_name | Peptide sequence |
| concepts[13].id | https://openalex.org/C62203573 |
| concepts[13].level | 4 |
| concepts[13].score | 0.26385498046875 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q15087973 |
| concepts[13].display_name | Long non-coding RNA |
| keywords[0].id | https://openalex.org/keywords/rna |
| keywords[0].score | 0.7190317511558533 |
| keywords[0].display_name | RNA |
| keywords[1].id | https://openalex.org/keywords/biology |
| keywords[1].score | 0.6908336877822876 |
| keywords[1].display_name | Biology |
| keywords[2].id | https://openalex.org/keywords/polyadenylation |
| keywords[2].score | 0.6887502074241638 |
| keywords[2].display_name | Polyadenylation |
| keywords[3].id | https://openalex.org/keywords/conserved-sequence |
| keywords[3].score | 0.6553806066513062 |
| keywords[3].display_name | Conserved sequence |
| keywords[4].id | https://openalex.org/keywords/malat1 |
| keywords[4].score | 0.501326322555542 |
| keywords[4].display_name | MALAT1 |
| keywords[5].id | https://openalex.org/keywords/messenger-rna |
| keywords[5].score | 0.4805110692977905 |
| keywords[5].display_name | Messenger RNA |
| keywords[6].id | https://openalex.org/keywords/gene |
| keywords[6].score | 0.46607327461242676 |
| keywords[6].display_name | Gene |
| keywords[7].id | https://openalex.org/keywords/nucleic-acid-structure |
| keywords[7].score | 0.4644019603729248 |
| keywords[7].display_name | Nucleic acid structure |
| keywords[8].id | https://openalex.org/keywords/coding-region |
| keywords[8].score | 0.4454178810119629 |
| keywords[8].display_name | Coding region |
| keywords[9].id | https://openalex.org/keywords/triple-helix |
| keywords[9].score | 0.4361477792263031 |
| keywords[9].display_name | Triple helix |
| keywords[10].id | https://openalex.org/keywords/genetics |
| keywords[10].score | 0.4338851869106293 |
| keywords[10].display_name | Genetics |
| keywords[11].id | https://openalex.org/keywords/directionality |
| keywords[11].score | 0.4319894313812256 |
| keywords[11].display_name | Directionality |
| keywords[12].id | https://openalex.org/keywords/peptide-sequence |
| keywords[12].score | 0.26507115364074707 |
| keywords[12].display_name | Peptide sequence |
| keywords[13].id | https://openalex.org/keywords/long-non-coding-rna |
| keywords[13].score | 0.26385498046875 |
| keywords[13].display_name | Long non-coding RNA |
| language | en |
| locations[0].id | doi:10.1101/2022.07.29.502018 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4306402567 |
| locations[0].source.issn | |
| locations[0].source.type | repository |
| locations[0].source.is_oa | False |
| locations[0].source.issn_l | |
| locations[0].source.is_core | False |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| locations[0].source.host_organization | https://openalex.org/I2750212522 |
| locations[0].source.host_organization_name | Cold Spring Harbor Laboratory |
| locations[0].source.host_organization_lineage | https://openalex.org/I2750212522 |
| locations[0].license | cc-by |
| locations[0].pdf_url | https://www.biorxiv.org/content/biorxiv/early/2022/07/29/2022.07.29.502018.full.pdf |
| locations[0].version | acceptedVersion |
| locations[0].raw_type | posted-content |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | True |
| locations[0].is_published | False |
| locations[0].raw_source_name | |
| locations[0].landing_page_url | https://doi.org/10.1101/2022.07.29.502018 |
| indexed_in | crossref |
| authorships[0].author.id | https://openalex.org/A5060696661 |
| authorships[0].author.orcid | |
| authorships[0].author.display_name | Anaïs Monroy‐Eklund |
| authorships[0].affiliations[0].raw_affiliation_string | Deprtment of Biology, University of North Crolin t Chpel Hill, Chpel Hill, NC, 27599 |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Anais Monroy-Eklund |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Deprtment of Biology, University of North Crolin t Chpel Hill, Chpel Hill, NC, 27599 |
| authorships[1].author.id | https://openalex.org/A5024098425 |
| authorships[1].author.orcid | https://orcid.org/0000-0002-1693-5359 |
| authorships[1].author.display_name | Colin D. Taylor |
| authorships[1].affiliations[0].raw_affiliation_string | Deprtment of Biology, University of North Crolin t Chpel Hill, Chpel Hill, NC, 27599 |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Colin Taylor |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | Deprtment of Biology, University of North Crolin t Chpel Hill, Chpel Hill, NC, 27599 |
| authorships[2].author.id | https://openalex.org/A5014076234 |
| authorships[2].author.orcid | https://orcid.org/0000-0002-2941-0500 |
| authorships[2].author.display_name | Chase A. Weidmann |
| authorships[2].countries | US |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I27837315 |
| authorships[2].affiliations[0].raw_affiliation_string | Department of Biological Chemistry, University of Michigan Medical School, Center for RNA Biomedicine, Rogel Cancer Center, Ann Aror Michigan, MI, 48103 |
| authorships[2].institutions[0].id | https://openalex.org/I27837315 |
| authorships[2].institutions[0].ror | https://ror.org/00jmfr291 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I27837315 |
| authorships[2].institutions[0].country_code | US |
| authorships[2].institutions[0].display_name | University of Michigan |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Chase Weidmann |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | Department of Biological Chemistry, University of Michigan Medical School, Center for RNA Biomedicine, Rogel Cancer Center, Ann Aror Michigan, MI, 48103 |
| authorships[3].author.id | https://openalex.org/A5045456930 |
| authorships[3].author.orcid | https://orcid.org/0000-0001-7830-0124 |
| authorships[3].author.display_name | Christina L. Burch |
| authorships[3].affiliations[0].raw_affiliation_string | Deprtment of Biology, University of North Crolin t Chpel Hill, Chpel Hill, NC, 27599 |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Christina Burch |
| authorships[3].is_corresponding | False |
| authorships[3].raw_affiliation_strings | Deprtment of Biology, University of North Crolin t Chpel Hill, Chpel Hill, NC, 27599 |
| authorships[4].author.id | https://openalex.org/A5026069604 |
| authorships[4].author.orcid | https://orcid.org/0000-0002-5088-9907 |
| authorships[4].author.display_name | Alain Laederach |
| authorships[4].affiliations[0].raw_affiliation_string | Deprtment of Biology, University of North Crolin t Chpel Hill, Chpel Hill, NC, 27599 |
| authorships[4].author_position | last |
| authorships[4].raw_author_name | Alain Laederach |
| authorships[4].is_corresponding | True |
| authorships[4].raw_affiliation_strings | Deprtment of Biology, University of North Crolin t Chpel Hill, Chpel Hill, NC, 27599 |
| has_content.pdf | True |
| has_content.grobid_xml | True |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://www.biorxiv.org/content/biorxiv/early/2022/07/29/2022.07.29.502018.full.pdf |
| open_access.oa_status | green |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Structural conservation of MALAT1 long non-coding RNA in cells and in evolution |
| has_fulltext | True |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T10515 |
| primary_topic.field.id | https://openalex.org/fields/13 |
| primary_topic.field.display_name | Biochemistry, Genetics and Molecular Biology |
| primary_topic.score | 1.0 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1306 |
| primary_topic.subfield.display_name | Cancer Research |
| primary_topic.display_name | Cancer-related molecular mechanisms research |
| related_works | https://openalex.org/W2624817431, https://openalex.org/W2950752483, https://openalex.org/W2794987474, https://openalex.org/W4232225082, https://openalex.org/W33032069, https://openalex.org/W1976007281, https://openalex.org/W1599251874, https://openalex.org/W2799337760, https://openalex.org/W3033791775, https://openalex.org/W2029088481 |
| cited_by_count | 1 |
| counts_by_year[0].year | 2023 |
| counts_by_year[0].cited_by_count | 1 |
| locations_count | 1 |
| best_oa_location.id | doi:10.1101/2022.07.29.502018 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4306402567 |
| best_oa_location.source.issn | |
| best_oa_location.source.type | repository |
| best_oa_location.source.is_oa | False |
| best_oa_location.source.issn_l | |
| best_oa_location.source.is_core | False |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| best_oa_location.source.host_organization | https://openalex.org/I2750212522 |
| best_oa_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| best_oa_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2022/07/29/2022.07.29.502018.full.pdf |
| best_oa_location.version | acceptedVersion |
| best_oa_location.raw_type | posted-content |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | False |
| best_oa_location.raw_source_name | |
| best_oa_location.landing_page_url | https://doi.org/10.1101/2022.07.29.502018 |
| primary_location.id | doi:10.1101/2022.07.29.502018 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4306402567 |
| primary_location.source.issn | |
| primary_location.source.type | repository |
| primary_location.source.is_oa | False |
| primary_location.source.issn_l | |
| primary_location.source.is_core | False |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| primary_location.source.host_organization | https://openalex.org/I2750212522 |
| primary_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| primary_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| primary_location.license | cc-by |
| primary_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2022/07/29/2022.07.29.502018.full.pdf |
| primary_location.version | acceptedVersion |
| primary_location.raw_type | posted-content |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | True |
| primary_location.is_published | False |
| primary_location.raw_source_name | |
| primary_location.landing_page_url | https://doi.org/10.1101/2022.07.29.502018 |
| publication_date | 2022-07-29 |
| publication_year | 2022 |
| referenced_works | https://openalex.org/W3033791775, https://openalex.org/W2032741347, https://openalex.org/W1986919130, https://openalex.org/W2606870443, https://openalex.org/W2767747597, https://openalex.org/W2160333540, https://openalex.org/W2767405538, https://openalex.org/W2963600783, https://openalex.org/W2143407915, https://openalex.org/W2071883352, https://openalex.org/W2166810745, https://openalex.org/W2068728873, https://openalex.org/W2148725278, https://openalex.org/W2101497682, https://openalex.org/W1602695373, https://openalex.org/W1879996757, https://openalex.org/W2113224683, https://openalex.org/W2128619556, https://openalex.org/W2740984310, https://openalex.org/W1983000672, https://openalex.org/W2897570487, https://openalex.org/W2031621142, https://openalex.org/W1594381616, https://openalex.org/W2486806490, https://openalex.org/W2789830781, https://openalex.org/W2158069307, https://openalex.org/W2784296343, https://openalex.org/W2098553089, https://openalex.org/W2793097545, https://openalex.org/W2115819912, https://openalex.org/W1969836160, https://openalex.org/W2088069070, https://openalex.org/W2167852161, https://openalex.org/W1964663067, https://openalex.org/W3136689663, https://openalex.org/W2114104545, https://openalex.org/W2003993220, https://openalex.org/W3004711671, https://openalex.org/W1985278607, https://openalex.org/W2911390449, https://openalex.org/W3111995113, https://openalex.org/W2509925948, https://openalex.org/W2212720078, https://openalex.org/W2802573885, https://openalex.org/W2783205930, https://openalex.org/W2810359426, https://openalex.org/W2774855547, https://openalex.org/W2921097287, https://openalex.org/W3014620365, https://openalex.org/W2058970225, https://openalex.org/W2102110811, https://openalex.org/W2153268494, https://openalex.org/W3208185524, https://openalex.org/W1637390375, https://openalex.org/W2132589661, https://openalex.org/W2005636452, https://openalex.org/W2141172277, https://openalex.org/W2626866735, https://openalex.org/W2581660941, https://openalex.org/W2777294334, https://openalex.org/W2055505035, https://openalex.org/W2747089052 |
| referenced_works_count | 62 |
| abstract_inverted_index.( | 131 |
| abstract_inverted_index.) | 134 |
| abstract_inverted_index.a | 19 |
| abstract_inverted_index.1) | 15 |
| abstract_inverted_index.We | 114 |
| abstract_inverted_index.at | 51, 207 |
| abstract_inverted_index.by | 18, 65 |
| abstract_inverted_index.in | 40, 70, 107, 118, 137, 155, 159, 171, 191, 209 |
| abstract_inverted_index.is | 16, 35, 43, 69, 96, 204 |
| abstract_inverted_index.of | 46, 67, 78, 85, 88, 124, 151, 176, 195, 200 |
| abstract_inverted_index.on | 27 |
| abstract_inverted_index.(as | 63 |
| abstract_inverted_index.159 | 22 |
| abstract_inverted_index.Our | 146 |
| abstract_inverted_index.RNA | 8, 94, 100 |
| abstract_inverted_index.The | 31, 58, 174 |
| abstract_inverted_index.and | 42, 55, 120, 128, 161, 164, 211, 216 |
| abstract_inverted_index.are | 104 |
| abstract_inverted_index.its | 28 |
| abstract_inverted_index.may | 111 |
| abstract_inverted_index.not | 2 |
| abstract_inverted_index.one | 45 |
| abstract_inverted_index.the | 5, 47, 53, 71, 83, 89, 117, 125, 177, 181, 192, 196, 198, 201 |
| abstract_inverted_index.3’ | 29 |
| abstract_inverted_index.Lung | 12 |
| abstract_inverted_index.both | 52 |
| abstract_inverted_index.cell | 109, 119, 121, 140, 157, 162, 214 |
| abstract_inverted_index.data | 147 |
| abstract_inverted_index.end. | 30 |
| abstract_inverted_index.even | 165 |
| abstract_inverted_index.free | 122 |
| abstract_inverted_index.here | 116 |
| abstract_inverted_index.long | 6, 39 |
| abstract_inverted_index.most | 48 |
| abstract_inverted_index.only | 190 |
| abstract_inverted_index.over | 36 |
| abstract_inverted_index.rest | 199 |
| abstract_inverted_index.that | 103 |
| abstract_inverted_index.8,000 | 37 |
| abstract_inverted_index.SHAPE | 143 |
| abstract_inverted_index.alone | 80 |
| abstract_inverted_index.alter | 112 |
| abstract_inverted_index.cells | 160 |
| abstract_inverted_index.free, | 163 |
| abstract_inverted_index.green | 129 |
| abstract_inverted_index.helix | 25, 73 |
| abstract_inverted_index.human | 127 |
| abstract_inverted_index.least | 208 |
| abstract_inverted_index.lines | 141 |
| abstract_inverted_index.often | 97 |
| abstract_inverted_index.that, | 185 |
| abstract_inverted_index.types | 110, 215 |
| abstract_inverted_index.using | 142 |
| abstract_inverted_index.MALAT1 | 9, 33, 135 |
| abstract_inverted_index.across | 180, 212 |
| abstract_inverted_index.cannot | 81 |
| abstract_inverted_index.degree | 84 |
| abstract_inverted_index.entire | 32, 90, 182 |
| abstract_inverted_index.highly | 20 |
| abstract_inverted_index.humans | 41 |
| abstract_inverted_index.levels | 150 |
| abstract_inverted_index.lines, | 158 |
| abstract_inverted_index.monkey | 130 |
| abstract_inverted_index.seeing | 187 |
| abstract_inverted_index.signal | 62 |
| abstract_inverted_index.triple | 24, 72 |
| abstract_inverted_index.Primary | 75 |
| abstract_inverted_index.between | 166 |
| abstract_inverted_index.binding | 101 |
| abstract_inverted_index.confirm | 82 |
| abstract_inverted_index.context | 98 |
| abstract_inverted_index.despite | 168, 186 |
| abstract_inverted_index.levels. | 57 |
| abstract_inverted_index.lncRNA, | 197 |
| abstract_inverted_index.primary | 172 |
| abstract_inverted_index.reveals | 148 |
| abstract_inverted_index.sabaeus | 133 |
| abstract_inverted_index.signals | 189 |
| abstract_inverted_index.uniform | 152 |
| abstract_inverted_index.Abstract | 0 |
| abstract_inverted_index.Although | 1 |
| abstract_inverted_index.analysis | 77 |
| abstract_inverted_index.chemical | 144 |
| abstract_inverted_index.junction | 194 |
| abstract_inverted_index.lncRNAs, | 50 |
| abstract_inverted_index.measured | 64 |
| abstract_inverted_index.multiple | 138, 213 |
| abstract_inverted_index.primates | 210 |
| abstract_inverted_index.probing. | 145 |
| abstract_inverted_index.proteins | 102 |
| abstract_inverted_index.sequence | 54, 76 |
| abstract_inverted_index.species, | 167 |
| abstract_inverted_index.suggests | 184 |
| abstract_inverted_index.conserved | 21, 49, 206 |
| abstract_inverted_index.different | 108, 156 |
| abstract_inverted_index.expressed | 106 |
| abstract_inverted_index.sequence. | 173 |
| abstract_inverted_index.strongest | 59 |
| abstract_inverted_index.structure | 26, 56, 95, 203 |
| abstract_inverted_index.Associated | 11 |
| abstract_inverted_index.Transcript | 14 |
| abstract_inverted_index.considered | 44 |
| abstract_inverted_index.dependent; | 99 |
| abstract_inverted_index.non-coding | 7 |
| abstract_inverted_index.nucleotide | 23 |
| abstract_inverted_index.remarkably | 205 |
| abstract_inverted_index.stabilized | 17 |
| abstract_inverted_index.structural | 60, 86, 153, 178 |
| abstract_inverted_index.structure. | 74, 113 |
| abstract_inverted_index.structures | 123 |
| abstract_inverted_index.surprising | 149 |
| abstract_inverted_index.transcript | 34, 183 |
| abstract_inverted_index.uniformity | 175 |
| abstract_inverted_index.(Metastasis | 10 |
| abstract_inverted_index.Chlorocebus | 132 |
| abstract_inverted_index.base-pairs) | 68 |
| abstract_inverted_index.canonically | 3 |
| abstract_inverted_index.conditions. | 217 |
| abstract_inverted_index.differences | 170 |
| abstract_inverted_index.full-length | 91, 126 |
| abstract_inverted_index.investigate | 115 |
| abstract_inverted_index.nucleotides | 38 |
| abstract_inverted_index.significant | 169 |
| abstract_inverted_index.three-helix | 193 |
| abstract_inverted_index.transcript. | 92 |
| abstract_inverted_index.transcripts | 136 |
| abstract_inverted_index.Furthermore, | 93 |
| abstract_inverted_index.co-variation | 66, 79, 188 |
| abstract_inverted_index.conservation | 61, 87, 154, 179 |
| abstract_inverted_index.Adenocarcinoma | 13 |
| abstract_inverted_index.differentially | 105 |
| abstract_inverted_index.tissue-derived | 139 |
| abstract_inverted_index.transcript’s | 202 |
| abstract_inverted_index.polyadenylated, | 4 |
| cited_by_percentile_year.max | 94 |
| cited_by_percentile_year.min | 89 |
| corresponding_author_ids | https://openalex.org/A5026069604 |
| countries_distinct_count | 1 |
| institutions_distinct_count | 5 |
| sustainable_development_goals[0].id | https://metadata.un.org/sdg/15 |
| sustainable_development_goals[0].score | 0.7300000190734863 |
| sustainable_development_goals[0].display_name | Life in Land |
| citation_normalized_percentile.value | 0.41970612 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |