Structural Variability of Pfam Domains Based on Alphafold2 Predictions Article Swipe
 YOU?
·
· 2025
· Open Access
·
· DOI: https://doi.org/10.1002/prot.70021
YOU?
·
· 2025
· Open Access
·
· DOI: https://doi.org/10.1002/prot.70021
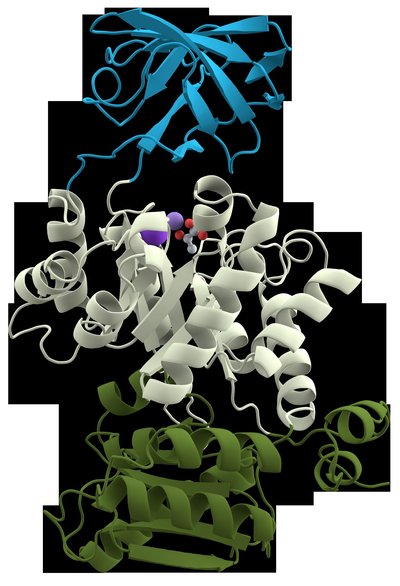
Understanding the biological functions of proteins is one of the main goals of functional genomics. Such understanding will help control and manipulate biological processes to enhance desirable traits, including improved abiotic and biotic stress resistance in humans, animals, plants, and microbes. Protein domains, regarded as the functional building blocks of proteins, have been used extensively to predict protein function. Sequence‐based approaches for protein function prediction, including the use of protein domain prediction from resources like the Pfam database, remain popular due to their reliability, low cost, and ease of use. Although the sequence variability of Pfam domains has been reported in several studies, their structural variability has been understudied. Here, we have extracted the Pfam domain structural portion from the predicted structures of the 16 model organism proteomes in the AlphaFold2 database. Our analysis revealed that many families contained between 20% and 40% members with no assigned regular secondary structures, demonstrating within‐family structural variability. To better understand this structural variability, we used FoldSeek and agglomerative clustering to identify structural variability in Pfam families. We then analyzed specific cases to provide structural details for this variability. In this study, we have used two popular prediction applications/resources, Alphafold2 and Pfam, to demonstrate inherent variability in protein domain predictions by comparing their predicted structures. Our study shows that detection of structural variability in Pfam families can facilitate curation and refinement of Pfam families, while demonstrating the need to develop more accurate protein domain prediction workflows.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.1002/prot.70021
- https://onlinelibrary.wiley.com/doi/pdfdirect/10.1002/prot.70021
- OA Status
- hybrid
- Cited By
- 1
- References
- 55
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4412585412
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4412585412Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1002/prot.70021Digital Object Identifier
- Title
-
Structural Variability of Pfam Domains Based on Alphafold2 PredictionsWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2025Year of publication
- Publication date
-
2025-07-22Full publication date if available
- Authors
-
Elly Poretsky, Carson M. Andorf, Taner Z. SenList of authors in order
- Landing page
-
https://doi.org/10.1002/prot.70021Publisher landing page
- PDF URL
-
https://onlinelibrary.wiley.com/doi/pdfdirect/10.1002/prot.70021Direct link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
hybridOpen access status per OpenAlex
- OA URL
-
https://onlinelibrary.wiley.com/doi/pdfdirect/10.1002/prot.70021Direct OA link when available
- Concepts
-
Computational biology, Protein domain, Structural genomics, Phenome, Domain (mathematical analysis), Function (biology), Proteome, Biology, Structural bioinformatics, Protein structure, Bioinformatics, Genetics, Genome, Gene, Biochemistry, Mathematical analysis, MathematicsTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
1Total citation count in OpenAlex
- Citations by year (recent)
-
2025: 1Per-year citation counts (last 5 years)
- References (count)
-
55Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4412585412 |
|---|---|
| doi | https://doi.org/10.1002/prot.70021 |
| ids.doi | https://doi.org/10.1002/prot.70021 |
| ids.pmid | https://pubmed.ncbi.nlm.nih.gov/40696837 |
| ids.openalex | https://openalex.org/W4412585412 |
| fwci | 2.68294463 |
| mesh[0].qualifier_ui | |
| mesh[0].descriptor_ui | D030562 |
| mesh[0].is_major_topic | False |
| mesh[0].qualifier_name | |
| mesh[0].descriptor_name | Databases, Protein |
| mesh[1].qualifier_ui | |
| mesh[1].descriptor_ui | D000072417 |
| mesh[1].is_major_topic | False |
| mesh[1].qualifier_name | |
| mesh[1].descriptor_name | Protein Domains |
| mesh[2].qualifier_ui | |
| mesh[2].descriptor_ui | D006801 |
| mesh[2].is_major_topic | False |
| mesh[2].qualifier_name | |
| mesh[2].descriptor_name | Humans |
| mesh[3].qualifier_ui | Q000379 |
| mesh[3].descriptor_ui | D019295 |
| mesh[3].is_major_topic | True |
| mesh[3].qualifier_name | methods |
| mesh[3].descriptor_name | Computational Biology |
| mesh[4].qualifier_ui | Q000737 |
| mesh[4].descriptor_ui | D011506 |
| mesh[4].is_major_topic | True |
| mesh[4].qualifier_name | chemistry |
| mesh[4].descriptor_name | Proteins |
| mesh[5].qualifier_ui | |
| mesh[5].descriptor_ui | D008958 |
| mesh[5].is_major_topic | False |
| mesh[5].qualifier_name | |
| mesh[5].descriptor_name | Models, Molecular |
| mesh[6].qualifier_ui | Q000737 |
| mesh[6].descriptor_ui | D020543 |
| mesh[6].is_major_topic | False |
| mesh[6].qualifier_name | chemistry |
| mesh[6].descriptor_name | Proteome |
| mesh[7].qualifier_ui | |
| mesh[7].descriptor_ui | D000595 |
| mesh[7].is_major_topic | False |
| mesh[7].qualifier_name | |
| mesh[7].descriptor_name | Amino Acid Sequence |
| mesh[8].qualifier_ui | |
| mesh[8].descriptor_ui | D012984 |
| mesh[8].is_major_topic | False |
| mesh[8].qualifier_name | |
| mesh[8].descriptor_name | Software |
| mesh[9].qualifier_ui | |
| mesh[9].descriptor_ui | D017433 |
| mesh[9].is_major_topic | False |
| mesh[9].qualifier_name | |
| mesh[9].descriptor_name | Protein Structure, Secondary |
| mesh[10].qualifier_ui | |
| mesh[10].descriptor_ui | D017510 |
| mesh[10].is_major_topic | False |
| mesh[10].qualifier_name | |
| mesh[10].descriptor_name | Protein Folding |
| mesh[11].qualifier_ui | |
| mesh[11].descriptor_ui | D000818 |
| mesh[11].is_major_topic | False |
| mesh[11].qualifier_name | |
| mesh[11].descriptor_name | Animals |
| mesh[12].qualifier_ui | |
| mesh[12].descriptor_ui | D020539 |
| mesh[12].is_major_topic | False |
| mesh[12].qualifier_name | |
| mesh[12].descriptor_name | Sequence Analysis, Protein |
| mesh[13].qualifier_ui | |
| mesh[13].descriptor_ui | D030562 |
| mesh[13].is_major_topic | False |
| mesh[13].qualifier_name | |
| mesh[13].descriptor_name | Databases, Protein |
| mesh[14].qualifier_ui | |
| mesh[14].descriptor_ui | D000072417 |
| mesh[14].is_major_topic | False |
| mesh[14].qualifier_name | |
| mesh[14].descriptor_name | Protein Domains |
| mesh[15].qualifier_ui | |
| mesh[15].descriptor_ui | D006801 |
| mesh[15].is_major_topic | False |
| mesh[15].qualifier_name | |
| mesh[15].descriptor_name | Humans |
| mesh[16].qualifier_ui | Q000379 |
| mesh[16].descriptor_ui | D019295 |
| mesh[16].is_major_topic | True |
| mesh[16].qualifier_name | methods |
| mesh[16].descriptor_name | Computational Biology |
| mesh[17].qualifier_ui | Q000737 |
| mesh[17].descriptor_ui | D011506 |
| mesh[17].is_major_topic | True |
| mesh[17].qualifier_name | chemistry |
| mesh[17].descriptor_name | Proteins |
| mesh[18].qualifier_ui | |
| mesh[18].descriptor_ui | D008958 |
| mesh[18].is_major_topic | False |
| mesh[18].qualifier_name | |
| mesh[18].descriptor_name | Models, Molecular |
| mesh[19].qualifier_ui | Q000737 |
| mesh[19].descriptor_ui | D020543 |
| mesh[19].is_major_topic | False |
| mesh[19].qualifier_name | chemistry |
| mesh[19].descriptor_name | Proteome |
| mesh[20].qualifier_ui | |
| mesh[20].descriptor_ui | D000595 |
| mesh[20].is_major_topic | False |
| mesh[20].qualifier_name | |
| mesh[20].descriptor_name | Amino Acid Sequence |
| mesh[21].qualifier_ui | |
| mesh[21].descriptor_ui | D012984 |
| mesh[21].is_major_topic | False |
| mesh[21].qualifier_name | |
| mesh[21].descriptor_name | Software |
| mesh[22].qualifier_ui | |
| mesh[22].descriptor_ui | D017433 |
| mesh[22].is_major_topic | False |
| mesh[22].qualifier_name | |
| mesh[22].descriptor_name | Protein Structure, Secondary |
| mesh[23].qualifier_ui | |
| mesh[23].descriptor_ui | D017510 |
| mesh[23].is_major_topic | False |
| mesh[23].qualifier_name | |
| mesh[23].descriptor_name | Protein Folding |
| mesh[24].qualifier_ui | |
| mesh[24].descriptor_ui | D000818 |
| mesh[24].is_major_topic | False |
| mesh[24].qualifier_name | |
| mesh[24].descriptor_name | Animals |
| mesh[25].qualifier_ui | |
| mesh[25].descriptor_ui | D020539 |
| mesh[25].is_major_topic | False |
| mesh[25].qualifier_name | |
| mesh[25].descriptor_name | Sequence Analysis, Protein |
| type | article |
| title | Structural Variability of Pfam Domains Based on Alphafold2 Predictions |
| biblio.issue | 12 |
| biblio.volume | 93 |
| biblio.last_page | 2192 |
| biblio.first_page | 2182 |
| topics[0].id | https://openalex.org/T10044 |
| topics[0].field.id | https://openalex.org/fields/13 |
| topics[0].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[0].score | 0.995199978351593 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1312 |
| topics[0].subfield.display_name | Molecular Biology |
| topics[0].display_name | Protein Structure and Dynamics |
| topics[1].id | https://openalex.org/T12254 |
| topics[1].field.id | https://openalex.org/fields/13 |
| topics[1].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[1].score | 0.9922000169754028 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1312 |
| topics[1].subfield.display_name | Molecular Biology |
| topics[1].display_name | Machine Learning in Bioinformatics |
| topics[2].id | https://openalex.org/T10932 |
| topics[2].field.id | https://openalex.org/fields/13 |
| topics[2].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[2].score | 0.9908999800682068 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1312 |
| topics[2].subfield.display_name | Molecular Biology |
| topics[2].display_name | Microbial Metabolic Engineering and Bioproduction |
| funders[0].id | https://openalex.org/F4320306114 |
| funders[0].ror | https://ror.org/01na82s61 |
| funders[0].display_name | U.S. Department of Agriculture |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C70721500 |
| concepts[0].level | 1 |
| concepts[0].score | 0.6580111384391785 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q177005 |
| concepts[0].display_name | Computational biology |
| concepts[1].id | https://openalex.org/C144292202 |
| concepts[1].level | 3 |
| concepts[1].score | 0.5989583730697632 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q898273 |
| concepts[1].display_name | Protein domain |
| concepts[2].id | https://openalex.org/C192772702 |
| concepts[2].level | 3 |
| concepts[2].score | 0.5707299709320068 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q2583975 |
| concepts[2].display_name | Structural genomics |
| concepts[3].id | https://openalex.org/C7952369 |
| concepts[3].level | 4 |
| concepts[3].score | 0.5017364025115967 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q844116 |
| concepts[3].display_name | Phenome |
| concepts[4].id | https://openalex.org/C36503486 |
| concepts[4].level | 2 |
| concepts[4].score | 0.4553152024745941 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q11235244 |
| concepts[4].display_name | Domain (mathematical analysis) |
| concepts[5].id | https://openalex.org/C14036430 |
| concepts[5].level | 2 |
| concepts[5].score | 0.44581180810928345 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q3736076 |
| concepts[5].display_name | Function (biology) |
| concepts[6].id | https://openalex.org/C104397665 |
| concepts[6].level | 2 |
| concepts[6].score | 0.44238170981407166 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q860947 |
| concepts[6].display_name | Proteome |
| concepts[7].id | https://openalex.org/C86803240 |
| concepts[7].level | 0 |
| concepts[7].score | 0.4391682744026184 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[7].display_name | Biology |
| concepts[8].id | https://openalex.org/C69131567 |
| concepts[8].level | 3 |
| concepts[8].score | 0.41063418984413147 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q2068215 |
| concepts[8].display_name | Structural bioinformatics |
| concepts[9].id | https://openalex.org/C47701112 |
| concepts[9].level | 2 |
| concepts[9].score | 0.3709801435470581 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q735188 |
| concepts[9].display_name | Protein structure |
| concepts[10].id | https://openalex.org/C60644358 |
| concepts[10].level | 1 |
| concepts[10].score | 0.3307339549064636 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q128570 |
| concepts[10].display_name | Bioinformatics |
| concepts[11].id | https://openalex.org/C54355233 |
| concepts[11].level | 1 |
| concepts[11].score | 0.241756409406662 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[11].display_name | Genetics |
| concepts[12].id | https://openalex.org/C141231307 |
| concepts[12].level | 3 |
| concepts[12].score | 0.20998939871788025 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q7020 |
| concepts[12].display_name | Genome |
| concepts[13].id | https://openalex.org/C104317684 |
| concepts[13].level | 2 |
| concepts[13].score | 0.08018743991851807 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[13].display_name | Gene |
| concepts[14].id | https://openalex.org/C55493867 |
| concepts[14].level | 1 |
| concepts[14].score | 0.0 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q7094 |
| concepts[14].display_name | Biochemistry |
| concepts[15].id | https://openalex.org/C134306372 |
| concepts[15].level | 1 |
| concepts[15].score | 0.0 |
| concepts[15].wikidata | https://www.wikidata.org/wiki/Q7754 |
| concepts[15].display_name | Mathematical analysis |
| concepts[16].id | https://openalex.org/C33923547 |
| concepts[16].level | 0 |
| concepts[16].score | 0.0 |
| concepts[16].wikidata | https://www.wikidata.org/wiki/Q395 |
| concepts[16].display_name | Mathematics |
| keywords[0].id | https://openalex.org/keywords/computational-biology |
| keywords[0].score | 0.6580111384391785 |
| keywords[0].display_name | Computational biology |
| keywords[1].id | https://openalex.org/keywords/protein-domain |
| keywords[1].score | 0.5989583730697632 |
| keywords[1].display_name | Protein domain |
| keywords[2].id | https://openalex.org/keywords/structural-genomics |
| keywords[2].score | 0.5707299709320068 |
| keywords[2].display_name | Structural genomics |
| keywords[3].id | https://openalex.org/keywords/phenome |
| keywords[3].score | 0.5017364025115967 |
| keywords[3].display_name | Phenome |
| keywords[4].id | https://openalex.org/keywords/domain |
| keywords[4].score | 0.4553152024745941 |
| keywords[4].display_name | Domain (mathematical analysis) |
| keywords[5].id | https://openalex.org/keywords/function |
| keywords[5].score | 0.44581180810928345 |
| keywords[5].display_name | Function (biology) |
| keywords[6].id | https://openalex.org/keywords/proteome |
| keywords[6].score | 0.44238170981407166 |
| keywords[6].display_name | Proteome |
| keywords[7].id | https://openalex.org/keywords/biology |
| keywords[7].score | 0.4391682744026184 |
| keywords[7].display_name | Biology |
| keywords[8].id | https://openalex.org/keywords/structural-bioinformatics |
| keywords[8].score | 0.41063418984413147 |
| keywords[8].display_name | Structural bioinformatics |
| keywords[9].id | https://openalex.org/keywords/protein-structure |
| keywords[9].score | 0.3709801435470581 |
| keywords[9].display_name | Protein structure |
| keywords[10].id | https://openalex.org/keywords/bioinformatics |
| keywords[10].score | 0.3307339549064636 |
| keywords[10].display_name | Bioinformatics |
| keywords[11].id | https://openalex.org/keywords/genetics |
| keywords[11].score | 0.241756409406662 |
| keywords[11].display_name | Genetics |
| keywords[12].id | https://openalex.org/keywords/genome |
| keywords[12].score | 0.20998939871788025 |
| keywords[12].display_name | Genome |
| keywords[13].id | https://openalex.org/keywords/gene |
| keywords[13].score | 0.08018743991851807 |
| keywords[13].display_name | Gene |
| language | en |
| locations[0].id | doi:10.1002/prot.70021 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S121161810 |
| locations[0].source.issn | 0887-3585, 1097-0134 |
| locations[0].source.type | journal |
| locations[0].source.is_oa | False |
| locations[0].source.issn_l | 0887-3585 |
| locations[0].source.is_core | True |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | Proteins Structure Function and Bioinformatics |
| locations[0].source.host_organization | https://openalex.org/P4310320595 |
| locations[0].source.host_organization_name | Wiley |
| locations[0].source.host_organization_lineage | https://openalex.org/P4310320595 |
| locations[0].source.host_organization_lineage_names | Wiley |
| locations[0].license | cc-by |
| locations[0].pdf_url | https://onlinelibrary.wiley.com/doi/pdfdirect/10.1002/prot.70021 |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | Proteins: Structure, Function, and Bioinformatics |
| locations[0].landing_page_url | https://doi.org/10.1002/prot.70021 |
| locations[1].id | pmid:40696837 |
| locations[1].is_oa | False |
| locations[1].source.id | https://openalex.org/S4306525036 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | PubMed |
| locations[1].source.host_organization | https://openalex.org/I1299303238 |
| locations[1].source.host_organization_name | National Institutes of Health |
| locations[1].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[1].license | |
| locations[1].pdf_url | |
| locations[1].version | publishedVersion |
| locations[1].raw_type | |
| locations[1].license_id | |
| locations[1].is_accepted | True |
| locations[1].is_published | True |
| locations[1].raw_source_name | Proteins |
| locations[1].landing_page_url | https://pubmed.ncbi.nlm.nih.gov/40696837 |
| locations[2].id | pmh:oai:escholarship.org:ark:/13030/qt4ds3g6xj |
| locations[2].is_oa | True |
| locations[2].source | |
| locations[2].license | cc-by |
| locations[2].pdf_url | |
| locations[2].version | submittedVersion |
| locations[2].raw_type | article |
| locations[2].license_id | https://openalex.org/licenses/cc-by |
| locations[2].is_accepted | False |
| locations[2].is_published | False |
| locations[2].raw_source_name | |
| locations[2].landing_page_url | https://escholarship.org/uc/item/4ds3g6xj |
| locations[3].id | pmh:oai:europepmc.org:11403245 |
| locations[3].is_oa | True |
| locations[3].source.id | https://openalex.org/S4306400806 |
| locations[3].source.issn | |
| locations[3].source.type | repository |
| locations[3].source.is_oa | False |
| locations[3].source.issn_l | |
| locations[3].source.is_core | False |
| locations[3].source.is_in_doaj | False |
| locations[3].source.display_name | Europe PMC (PubMed Central) |
| locations[3].source.host_organization | https://openalex.org/I1303153112 |
| locations[3].source.host_organization_name | European Bioinformatics Institute |
| locations[3].source.host_organization_lineage | https://openalex.org/I1303153112 |
| locations[3].license | other-oa |
| locations[3].pdf_url | |
| locations[3].version | submittedVersion |
| locations[3].raw_type | Text |
| locations[3].license_id | https://openalex.org/licenses/other-oa |
| locations[3].is_accepted | False |
| locations[3].is_published | False |
| locations[3].raw_source_name | |
| locations[3].landing_page_url | https://www.ncbi.nlm.nih.gov/pmc/articles/12594177 |
| indexed_in | crossref, pubmed |
| authorships[0].author.id | https://openalex.org/A5085736340 |
| authorships[0].author.orcid | https://orcid.org/0000-0002-6116-6827 |
| authorships[0].author.display_name | Elly Poretsky |
| authorships[0].countries | US |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I1312222531, https://openalex.org/I1336096307, https://openalex.org/I4210140558 |
| authorships[0].affiliations[0].raw_affiliation_string | Crop Improvement and Genetics Research Unit, United States Department of Agriculture, Agricultural Research Service, Western Regional Research Center, Albany, California, USA |
| authorships[0].institutions[0].id | https://openalex.org/I1312222531 |
| authorships[0].institutions[0].ror | https://ror.org/02d2m2044 |
| authorships[0].institutions[0].type | government |
| authorships[0].institutions[0].lineage | https://openalex.org/I1312222531, https://openalex.org/I1336096307 |
| authorships[0].institutions[0].country_code | US |
| authorships[0].institutions[0].display_name | Agricultural Research Service |
| authorships[0].institutions[1].id | https://openalex.org/I1336096307 |
| authorships[0].institutions[1].ror | https://ror.org/01na82s61 |
| authorships[0].institutions[1].type | government |
| authorships[0].institutions[1].lineage | https://openalex.org/I1336096307 |
| authorships[0].institutions[1].country_code | US |
| authorships[0].institutions[1].display_name | United States Department of Agriculture |
| authorships[0].institutions[2].id | https://openalex.org/I4210140558 |
| authorships[0].institutions[2].ror | https://ror.org/03x7fn667 |
| authorships[0].institutions[2].type | government |
| authorships[0].institutions[2].lineage | https://openalex.org/I1312222531, https://openalex.org/I1336096307, https://openalex.org/I4210094889, https://openalex.org/I4210140558 |
| authorships[0].institutions[2].country_code | US |
| authorships[0].institutions[2].display_name | Western Regional Research Center |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Elly Poretsky |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Crop Improvement and Genetics Research Unit, United States Department of Agriculture, Agricultural Research Service, Western Regional Research Center, Albany, California, USA |
| authorships[1].author.id | https://openalex.org/A5045214155 |
| authorships[1].author.orcid | https://orcid.org/0000-0003-2380-6704 |
| authorships[1].author.display_name | Carson M. Andorf |
| authorships[1].countries | US |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I173911158 |
| authorships[1].affiliations[0].raw_affiliation_string | Department of Computer Science, Iowa State University, Ames, Iowa, USA |
| authorships[1].affiliations[1].institution_ids | https://openalex.org/I1312222531, https://openalex.org/I1336096307 |
| authorships[1].affiliations[1].raw_affiliation_string | Corn Insects and Crop Genetics Research Unit, United States Department of Agriculture, Agricultural Research Service, Ames, Iowa, USA |
| authorships[1].institutions[0].id | https://openalex.org/I1312222531 |
| authorships[1].institutions[0].ror | https://ror.org/02d2m2044 |
| authorships[1].institutions[0].type | government |
| authorships[1].institutions[0].lineage | https://openalex.org/I1312222531, https://openalex.org/I1336096307 |
| authorships[1].institutions[0].country_code | US |
| authorships[1].institutions[0].display_name | Agricultural Research Service |
| authorships[1].institutions[1].id | https://openalex.org/I173911158 |
| authorships[1].institutions[1].ror | https://ror.org/04rswrd78 |
| authorships[1].institutions[1].type | education |
| authorships[1].institutions[1].lineage | https://openalex.org/I173911158 |
| authorships[1].institutions[1].country_code | US |
| authorships[1].institutions[1].display_name | Iowa State University |
| authorships[1].institutions[2].id | https://openalex.org/I1336096307 |
| authorships[1].institutions[2].ror | https://ror.org/01na82s61 |
| authorships[1].institutions[2].type | government |
| authorships[1].institutions[2].lineage | https://openalex.org/I1336096307 |
| authorships[1].institutions[2].country_code | US |
| authorships[1].institutions[2].display_name | United States Department of Agriculture |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Carson M. Andorf |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | Corn Insects and Crop Genetics Research Unit, United States Department of Agriculture, Agricultural Research Service, Ames, Iowa, USA, Department of Computer Science, Iowa State University, Ames, Iowa, USA |
| authorships[2].author.id | https://openalex.org/A5022049098 |
| authorships[2].author.orcid | https://orcid.org/0000-0002-5553-6190 |
| authorships[2].author.display_name | Taner Z. Sen |
| authorships[2].countries | US |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I1312222531, https://openalex.org/I1336096307, https://openalex.org/I4210140558 |
| authorships[2].affiliations[0].raw_affiliation_string | Crop Improvement and Genetics Research Unit, United States Department of Agriculture, Agricultural Research Service, Western Regional Research Center, Albany, California, USA |
| authorships[2].affiliations[1].institution_ids | https://openalex.org/I95457486 |
| authorships[2].affiliations[1].raw_affiliation_string | Department of Bioengineering, University of California, Berkeley, California, USA |
| authorships[2].institutions[0].id | https://openalex.org/I1312222531 |
| authorships[2].institutions[0].ror | https://ror.org/02d2m2044 |
| authorships[2].institutions[0].type | government |
| authorships[2].institutions[0].lineage | https://openalex.org/I1312222531, https://openalex.org/I1336096307 |
| authorships[2].institutions[0].country_code | US |
| authorships[2].institutions[0].display_name | Agricultural Research Service |
| authorships[2].institutions[1].id | https://openalex.org/I1336096307 |
| authorships[2].institutions[1].ror | https://ror.org/01na82s61 |
| authorships[2].institutions[1].type | government |
| authorships[2].institutions[1].lineage | https://openalex.org/I1336096307 |
| authorships[2].institutions[1].country_code | US |
| authorships[2].institutions[1].display_name | United States Department of Agriculture |
| authorships[2].institutions[2].id | https://openalex.org/I95457486 |
| authorships[2].institutions[2].ror | https://ror.org/01an7q238 |
| authorships[2].institutions[2].type | education |
| authorships[2].institutions[2].lineage | https://openalex.org/I95457486 |
| authorships[2].institutions[2].country_code | US |
| authorships[2].institutions[2].display_name | University of California, Berkeley |
| authorships[2].institutions[3].id | https://openalex.org/I4210140558 |
| authorships[2].institutions[3].ror | https://ror.org/03x7fn667 |
| authorships[2].institutions[3].type | government |
| authorships[2].institutions[3].lineage | https://openalex.org/I1312222531, https://openalex.org/I1336096307, https://openalex.org/I4210094889, https://openalex.org/I4210140558 |
| authorships[2].institutions[3].country_code | US |
| authorships[2].institutions[3].display_name | Western Regional Research Center |
| authorships[2].author_position | last |
| authorships[2].raw_author_name | Taner Z. Sen |
| authorships[2].is_corresponding | True |
| authorships[2].raw_affiliation_strings | Crop Improvement and Genetics Research Unit, United States Department of Agriculture, Agricultural Research Service, Western Regional Research Center, Albany, California, USA, Department of Bioengineering, University of California, Berkeley, California, USA |
| has_content.pdf | True |
| has_content.grobid_xml | True |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://onlinelibrary.wiley.com/doi/pdfdirect/10.1002/prot.70021 |
| open_access.oa_status | hybrid |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Structural Variability of Pfam Domains Based on Alphafold2 Predictions |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-08T23:21:52.890332 |
| primary_topic.id | https://openalex.org/T10044 |
| primary_topic.field.id | https://openalex.org/fields/13 |
| primary_topic.field.display_name | Biochemistry, Genetics and Molecular Biology |
| primary_topic.score | 0.995199978351593 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1312 |
| primary_topic.subfield.display_name | Molecular Biology |
| primary_topic.display_name | Protein Structure and Dynamics |
| related_works | https://openalex.org/W2611147784, https://openalex.org/W2014011595, https://openalex.org/W2346580382, https://openalex.org/W2015390241, https://openalex.org/W2166261803, https://openalex.org/W2089999388, https://openalex.org/W2610336742, https://openalex.org/W2118652015, https://openalex.org/W1972763041, https://openalex.org/W853003716 |
| cited_by_count | 1 |
| counts_by_year[0].year | 2025 |
| counts_by_year[0].cited_by_count | 1 |
| locations_count | 4 |
| best_oa_location.id | doi:10.1002/prot.70021 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S121161810 |
| best_oa_location.source.issn | 0887-3585, 1097-0134 |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | False |
| best_oa_location.source.issn_l | 0887-3585 |
| best_oa_location.source.is_core | True |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | Proteins Structure Function and Bioinformatics |
| best_oa_location.source.host_organization | https://openalex.org/P4310320595 |
| best_oa_location.source.host_organization_name | Wiley |
| best_oa_location.source.host_organization_lineage | https://openalex.org/P4310320595 |
| best_oa_location.source.host_organization_lineage_names | Wiley |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | https://onlinelibrary.wiley.com/doi/pdfdirect/10.1002/prot.70021 |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | journal-article |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | Proteins: Structure, Function, and Bioinformatics |
| best_oa_location.landing_page_url | https://doi.org/10.1002/prot.70021 |
| primary_location.id | doi:10.1002/prot.70021 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S121161810 |
| primary_location.source.issn | 0887-3585, 1097-0134 |
| primary_location.source.type | journal |
| primary_location.source.is_oa | False |
| primary_location.source.issn_l | 0887-3585 |
| primary_location.source.is_core | True |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | Proteins Structure Function and Bioinformatics |
| primary_location.source.host_organization | https://openalex.org/P4310320595 |
| primary_location.source.host_organization_name | Wiley |
| primary_location.source.host_organization_lineage | https://openalex.org/P4310320595 |
| primary_location.source.host_organization_lineage_names | Wiley |
| primary_location.license | cc-by |
| primary_location.pdf_url | https://onlinelibrary.wiley.com/doi/pdfdirect/10.1002/prot.70021 |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | Proteins: Structure, Function, and Bioinformatics |
| primary_location.landing_page_url | https://doi.org/10.1002/prot.70021 |
| publication_date | 2025-07-22 |
| publication_year | 2025 |
| referenced_works | https://openalex.org/W4310951880, https://openalex.org/W4404531735, https://openalex.org/W2101291993, https://openalex.org/W2132122858, https://openalex.org/W4388464011, https://openalex.org/W4403925091, https://openalex.org/W4387002923, https://openalex.org/W3186179742, https://openalex.org/W4406063185, https://openalex.org/W4384500503, https://openalex.org/W4323309784, https://openalex.org/W4280512557, https://openalex.org/W4327566741, https://openalex.org/W1969327849, https://openalex.org/W4292112379, https://openalex.org/W4410429680, https://openalex.org/W1974970687, https://openalex.org/W4406714966, https://openalex.org/W4382601731, https://openalex.org/W4392844094, https://openalex.org/W4404343451, https://openalex.org/W2114850508, https://openalex.org/W2141920771, https://openalex.org/W4205122286, https://openalex.org/W6675354045, https://openalex.org/W3101667854, https://openalex.org/W3107541694, https://openalex.org/W1600438564, https://openalex.org/W2791753912, https://openalex.org/W3095583226, https://openalex.org/W2898402099, https://openalex.org/W4394694796, https://openalex.org/W2102245393, https://openalex.org/W4404693690, https://openalex.org/W2151651469, https://openalex.org/W1554728849, https://openalex.org/W2885419062, https://openalex.org/W2138122982, https://openalex.org/W1981930726, https://openalex.org/W2956397917, https://openalex.org/W4389174567, https://openalex.org/W2068072987, https://openalex.org/W2951320290, https://openalex.org/W4396721167, https://openalex.org/W3177828909, https://openalex.org/W4386709418, https://openalex.org/W2108067237, https://openalex.org/W3108943181, https://openalex.org/W4406669122, https://openalex.org/W2127921600, https://openalex.org/W3208408872, https://openalex.org/W2997591727, https://openalex.org/W4375858802, https://openalex.org/W2319660501, https://openalex.org/W4386708656 |
| referenced_works_count | 55 |
| abstract_inverted_index.16 | 125 |
| abstract_inverted_index.In | 186 |
| abstract_inverted_index.To | 155 |
| abstract_inverted_index.We | 174 |
| abstract_inverted_index.as | 45 |
| abstract_inverted_index.by | 207 |
| abstract_inverted_index.in | 36, 101, 129, 171, 203, 220 |
| abstract_inverted_index.is | 7 |
| abstract_inverted_index.no | 146 |
| abstract_inverted_index.of | 5, 9, 13, 50, 69, 89, 95, 123, 217, 228 |
| abstract_inverted_index.to | 25, 56, 82, 167, 179, 199, 235 |
| abstract_inverted_index.we | 111, 161, 189 |
| abstract_inverted_index.20% | 141 |
| abstract_inverted_index.40% | 143 |
| abstract_inverted_index.Our | 133, 212 |
| abstract_inverted_index.and | 21, 32, 40, 87, 142, 164, 197, 226 |
| abstract_inverted_index.can | 223 |
| abstract_inverted_index.due | 81 |
| abstract_inverted_index.for | 62, 183 |
| abstract_inverted_index.has | 98, 107 |
| abstract_inverted_index.low | 85 |
| abstract_inverted_index.one | 8 |
| abstract_inverted_index.the | 2, 10, 46, 67, 76, 92, 114, 120, 124, 130, 233 |
| abstract_inverted_index.two | 192 |
| abstract_inverted_index.use | 68 |
| abstract_inverted_index.Pfam | 77, 96, 115, 172, 221, 229 |
| abstract_inverted_index.Such | 16 |
| abstract_inverted_index.been | 53, 99, 108 |
| abstract_inverted_index.ease | 88 |
| abstract_inverted_index.from | 73, 119 |
| abstract_inverted_index.have | 52, 112, 190 |
| abstract_inverted_index.help | 19 |
| abstract_inverted_index.like | 75 |
| abstract_inverted_index.main | 11 |
| abstract_inverted_index.many | 137 |
| abstract_inverted_index.more | 237 |
| abstract_inverted_index.need | 234 |
| abstract_inverted_index.that | 136, 215 |
| abstract_inverted_index.then | 175 |
| abstract_inverted_index.this | 158, 184, 187 |
| abstract_inverted_index.use. | 90 |
| abstract_inverted_index.used | 54, 162, 191 |
| abstract_inverted_index.will | 18 |
| abstract_inverted_index.with | 145 |
| abstract_inverted_index.Here, | 110 |
| abstract_inverted_index.Pfam, | 198 |
| abstract_inverted_index.cases | 178 |
| abstract_inverted_index.cost, | 86 |
| abstract_inverted_index.goals | 12 |
| abstract_inverted_index.model | 126 |
| abstract_inverted_index.shows | 214 |
| abstract_inverted_index.study | 213 |
| abstract_inverted_index.their | 83, 104, 209 |
| abstract_inverted_index.while | 231 |
| abstract_inverted_index.better | 156 |
| abstract_inverted_index.biotic | 33 |
| abstract_inverted_index.blocks | 49 |
| abstract_inverted_index.domain | 71, 116, 205, 240 |
| abstract_inverted_index.remain | 79 |
| abstract_inverted_index.stress | 34 |
| abstract_inverted_index.study, | 188 |
| abstract_inverted_index.Protein | 42 |
| abstract_inverted_index.abiotic | 31 |
| abstract_inverted_index.between | 140 |
| abstract_inverted_index.control | 20 |
| abstract_inverted_index.details | 182 |
| abstract_inverted_index.develop | 236 |
| abstract_inverted_index.domains | 97 |
| abstract_inverted_index.enhance | 26 |
| abstract_inverted_index.humans, | 37 |
| abstract_inverted_index.members | 144 |
| abstract_inverted_index.plants, | 39 |
| abstract_inverted_index.popular | 80, 193 |
| abstract_inverted_index.portion | 118 |
| abstract_inverted_index.predict | 57 |
| abstract_inverted_index.protein | 58, 63, 70, 204, 239 |
| abstract_inverted_index.provide | 180 |
| abstract_inverted_index.regular | 148 |
| abstract_inverted_index.several | 102 |
| abstract_inverted_index.traits, | 28 |
| abstract_inverted_index.ABSTRACT | 0 |
| abstract_inverted_index.Although | 91 |
| abstract_inverted_index.FoldSeek | 163 |
| abstract_inverted_index.accurate | 238 |
| abstract_inverted_index.analysis | 134 |
| abstract_inverted_index.analyzed | 176 |
| abstract_inverted_index.animals, | 38 |
| abstract_inverted_index.assigned | 147 |
| abstract_inverted_index.building | 48 |
| abstract_inverted_index.curation | 225 |
| abstract_inverted_index.domains, | 43 |
| abstract_inverted_index.families | 138, 222 |
| abstract_inverted_index.function | 64 |
| abstract_inverted_index.identify | 168 |
| abstract_inverted_index.improved | 30 |
| abstract_inverted_index.inherent | 201 |
| abstract_inverted_index.organism | 127 |
| abstract_inverted_index.proteins | 6 |
| abstract_inverted_index.regarded | 44 |
| abstract_inverted_index.reported | 100 |
| abstract_inverted_index.revealed | 135 |
| abstract_inverted_index.sequence | 93 |
| abstract_inverted_index.specific | 177 |
| abstract_inverted_index.studies, | 103 |
| abstract_inverted_index.comparing | 208 |
| abstract_inverted_index.contained | 139 |
| abstract_inverted_index.database, | 78 |
| abstract_inverted_index.database. | 132 |
| abstract_inverted_index.desirable | 27 |
| abstract_inverted_index.detection | 216 |
| abstract_inverted_index.extracted | 113 |
| abstract_inverted_index.families, | 230 |
| abstract_inverted_index.families. | 173 |
| abstract_inverted_index.function. | 59 |
| abstract_inverted_index.functions | 4 |
| abstract_inverted_index.genomics. | 15 |
| abstract_inverted_index.including | 29, 66 |
| abstract_inverted_index.microbes. | 41 |
| abstract_inverted_index.predicted | 121, 210 |
| abstract_inverted_index.processes | 24 |
| abstract_inverted_index.proteins, | 51 |
| abstract_inverted_index.proteomes | 128 |
| abstract_inverted_index.resources | 74 |
| abstract_inverted_index.secondary | 149 |
| abstract_inverted_index.AlphaFold2 | 131 |
| abstract_inverted_index.Alphafold2 | 196 |
| abstract_inverted_index.approaches | 61 |
| abstract_inverted_index.biological | 3, 23 |
| abstract_inverted_index.clustering | 166 |
| abstract_inverted_index.facilitate | 224 |
| abstract_inverted_index.functional | 14, 47 |
| abstract_inverted_index.manipulate | 22 |
| abstract_inverted_index.prediction | 72, 194, 241 |
| abstract_inverted_index.refinement | 227 |
| abstract_inverted_index.resistance | 35 |
| abstract_inverted_index.structural | 105, 117, 153, 159, 169, 181, 218 |
| abstract_inverted_index.structures | 122 |
| abstract_inverted_index.understand | 157 |
| abstract_inverted_index.workflows. | 242 |
| abstract_inverted_index.demonstrate | 200 |
| abstract_inverted_index.extensively | 55 |
| abstract_inverted_index.prediction, | 65 |
| abstract_inverted_index.predictions | 206 |
| abstract_inverted_index.structures, | 150 |
| abstract_inverted_index.structures. | 211 |
| abstract_inverted_index.variability | 94, 106, 170, 202, 219 |
| abstract_inverted_index.reliability, | 84 |
| abstract_inverted_index.variability, | 160 |
| abstract_inverted_index.variability. | 154, 185 |
| abstract_inverted_index.Understanding | 1 |
| abstract_inverted_index.agglomerative | 165 |
| abstract_inverted_index.demonstrating | 151, 232 |
| abstract_inverted_index.understanding | 17 |
| abstract_inverted_index.understudied. | 109 |
| abstract_inverted_index.within‐family | 152 |
| abstract_inverted_index.Sequence‐based | 60 |
| abstract_inverted_index.applications/resources, | 195 |
| cited_by_percentile_year.max | 95 |
| cited_by_percentile_year.min | 91 |
| corresponding_author_ids | https://openalex.org/A5022049098 |
| countries_distinct_count | 1 |
| institutions_distinct_count | 3 |
| corresponding_institution_ids | https://openalex.org/I1312222531, https://openalex.org/I1336096307, https://openalex.org/I4210140558, https://openalex.org/I95457486 |
| citation_normalized_percentile.value | 0.83661246 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | True |