Structure-Function Analysis of the Transmembrane Protein AmpG from Pseudomonas aeruginosa Article Swipe
 YOU?
·
· 2016
· Open Access
·
· DOI: https://doi.org/10.1371/journal.pone.0168060
YOU?
·
· 2016
· Open Access
·
· DOI: https://doi.org/10.1371/journal.pone.0168060
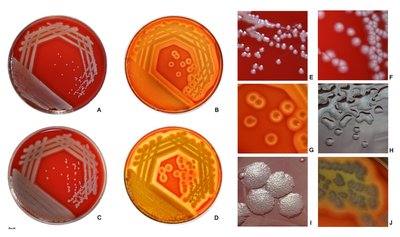
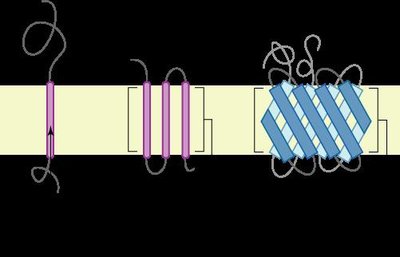
AmpG is a transmembrane protein with permease activity that transports meuropeptide from the periplasm to the cytoplasm, which is essential for the induction of the ampC encoding β-lactamase. To obtain new insights into the relationship between AmpG structure and function, comparative genomics analysis, secondary and tertiary structure modeling, site-directed mutational analyses and genetic complementation experiments were performed in this study. AmpGs from different genera of bacteria (Escherichia coli, Vibrio cholerae and Acinetobacter baumannii) could complement AmpG function in Pseudomonas aeruginosa. The minimal inhibitory concentration (MIC) to ampicillin is 512 μg/ml for wild type strain PAO1, while it is 32 μg/ml for an ampG deletion mutant strain (PAO1ΔampG) with a corresponding decrease in the activity of the ampC-encoded β-lactamase. Site-directed mutagenesis of conserved AmpG residues (G29, A129, Q131 and A197) resulted in a loss of function, resulting in a loss of resistance to ampicillin in PAO1ΔampG. The G29A, G29V, A129T, A129V, A129D, A197S and A197D mutants had lower resistance to ampicillin and significantly decreased activity of the AmpC β-lactamase. The G29A, G29V, A129V, A197S and A197D mutants had decreased ampG mRNA transcript levels. The A129T and A129D mutants had normal ampG mRNA transcript levels, but the function of the protein was drastically reduced. Our experimental results demonstrate that the conserved amino acids played essential roles in maintaining the function of AmpG. Combined with the AmpG structural information, these critical amino acids can be targeted for the development of new anti-bacterial agents.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.1371/journal.pone.0168060
- https://journals.plos.org/plosone/article/file?id=10.1371/journal.pone.0168060&type=printable
- OA Status
- gold
- Cited By
- 14
- References
- 35
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W2563221447
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W2563221447Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1371/journal.pone.0168060Digital Object Identifier
- Title
-
Structure-Function Analysis of the Transmembrane Protein AmpG from Pseudomonas aeruginosaWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2016Year of publication
- Publication date
-
2016-12-13Full publication date if available
- Authors
-
Peizhen Li, Jun Ying, Guangjian Yang, Aifang Li, Jian Wang, Junwan Lu, Junrong Wang, Teng Xu, Huiguang Yi, Kewei Li, Shouguang Jin, Qiyu Bao, Kaibo ZhangList of authors in order
- Landing page
-
https://doi.org/10.1371/journal.pone.0168060Publisher landing page
- PDF URL
-
https://journals.plos.org/plosone/article/file?id=10.1371/journal.pone.0168060&type=printableDirect link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
goldOpen access status per OpenAlex
- OA URL
-
https://journals.plos.org/plosone/article/file?id=10.1371/journal.pone.0168060&type=printableDirect OA link when available
- Concepts
-
Periplasmic space, Complementation, Biology, Mutant, Pseudomonas aeruginosa, Vibrio cholerae, Escherichia coli, Mutagenesis, Microbiology, Bacterial outer membrane, Amp resistance, Transmembrane protein, Biochemistry, Genetics, Gene, Bacteria, ReceptorTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
14Total citation count in OpenAlex
- Citations by year (recent)
-
2025: 3, 2023: 1, 2022: 3, 2021: 1, 2019: 1Per-year citation counts (last 5 years)
- References (count)
-
35Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W2563221447 |
|---|---|
| doi | https://doi.org/10.1371/journal.pone.0168060 |
| ids.doi | https://doi.org/10.1371/journal.pone.0168060 |
| ids.mag | 2563221447 |
| ids.pmid | https://pubmed.ncbi.nlm.nih.gov/27959942 |
| ids.openalex | https://openalex.org/W2563221447 |
| fwci | 0.86532241 |
| mesh[0].qualifier_ui | |
| mesh[0].descriptor_ui | D020816 |
| mesh[0].is_major_topic | False |
| mesh[0].qualifier_name | |
| mesh[0].descriptor_name | Amino Acid Motifs |
| mesh[1].qualifier_ui | Q000737 |
| mesh[1].descriptor_ui | D001426 |
| mesh[1].is_major_topic | False |
| mesh[1].qualifier_name | chemistry |
| mesh[1].descriptor_name | Bacterial Proteins |
| mesh[2].qualifier_ui | Q000235 |
| mesh[2].descriptor_ui | D001426 |
| mesh[2].is_major_topic | False |
| mesh[2].qualifier_name | genetics |
| mesh[2].descriptor_name | Bacterial Proteins |
| mesh[3].qualifier_ui | Q000378 |
| mesh[3].descriptor_ui | D001426 |
| mesh[3].is_major_topic | False |
| mesh[3].qualifier_name | metabolism |
| mesh[3].descriptor_name | Bacterial Proteins |
| mesh[4].qualifier_ui | Q000737 |
| mesh[4].descriptor_ui | D026901 |
| mesh[4].is_major_topic | False |
| mesh[4].qualifier_name | chemistry |
| mesh[4].descriptor_name | Membrane Transport Proteins |
| mesh[5].qualifier_ui | Q000235 |
| mesh[5].descriptor_ui | D026901 |
| mesh[5].is_major_topic | False |
| mesh[5].qualifier_name | genetics |
| mesh[5].descriptor_name | Membrane Transport Proteins |
| mesh[6].qualifier_ui | Q000378 |
| mesh[6].descriptor_ui | D026901 |
| mesh[6].is_major_topic | False |
| mesh[6].qualifier_name | metabolism |
| mesh[6].descriptor_name | Membrane Transport Proteins |
| mesh[7].qualifier_ui | |
| mesh[7].descriptor_ui | D020125 |
| mesh[7].is_major_topic | False |
| mesh[7].qualifier_name | |
| mesh[7].descriptor_name | Mutation, Missense |
| mesh[8].qualifier_ui | Q000201 |
| mesh[8].descriptor_ui | D011550 |
| mesh[8].is_major_topic | False |
| mesh[8].qualifier_name | enzymology |
| mesh[8].descriptor_name | Pseudomonas aeruginosa |
| mesh[9].qualifier_ui | |
| mesh[9].descriptor_ui | D021281 |
| mesh[9].is_major_topic | True |
| mesh[9].qualifier_name | |
| mesh[9].descriptor_name | Quantitative Structure-Activity Relationship |
| mesh[10].qualifier_ui | |
| mesh[10].descriptor_ui | D020816 |
| mesh[10].is_major_topic | False |
| mesh[10].qualifier_name | |
| mesh[10].descriptor_name | Amino Acid Motifs |
| mesh[11].qualifier_ui | Q000737 |
| mesh[11].descriptor_ui | D001426 |
| mesh[11].is_major_topic | False |
| mesh[11].qualifier_name | chemistry |
| mesh[11].descriptor_name | Bacterial Proteins |
| mesh[12].qualifier_ui | Q000235 |
| mesh[12].descriptor_ui | D001426 |
| mesh[12].is_major_topic | False |
| mesh[12].qualifier_name | genetics |
| mesh[12].descriptor_name | Bacterial Proteins |
| mesh[13].qualifier_ui | Q000378 |
| mesh[13].descriptor_ui | D001426 |
| mesh[13].is_major_topic | False |
| mesh[13].qualifier_name | metabolism |
| mesh[13].descriptor_name | Bacterial Proteins |
| mesh[14].qualifier_ui | Q000737 |
| mesh[14].descriptor_ui | D026901 |
| mesh[14].is_major_topic | False |
| mesh[14].qualifier_name | chemistry |
| mesh[14].descriptor_name | Membrane Transport Proteins |
| mesh[15].qualifier_ui | Q000235 |
| mesh[15].descriptor_ui | D026901 |
| mesh[15].is_major_topic | False |
| mesh[15].qualifier_name | genetics |
| mesh[15].descriptor_name | Membrane Transport Proteins |
| mesh[16].qualifier_ui | Q000378 |
| mesh[16].descriptor_ui | D026901 |
| mesh[16].is_major_topic | False |
| mesh[16].qualifier_name | metabolism |
| mesh[16].descriptor_name | Membrane Transport Proteins |
| mesh[17].qualifier_ui | |
| mesh[17].descriptor_ui | D020125 |
| mesh[17].is_major_topic | False |
| mesh[17].qualifier_name | |
| mesh[17].descriptor_name | Mutation, Missense |
| mesh[18].qualifier_ui | Q000201 |
| mesh[18].descriptor_ui | D011550 |
| mesh[18].is_major_topic | False |
| mesh[18].qualifier_name | enzymology |
| mesh[18].descriptor_name | Pseudomonas aeruginosa |
| mesh[19].qualifier_ui | |
| mesh[19].descriptor_ui | D021281 |
| mesh[19].is_major_topic | True |
| mesh[19].qualifier_name | |
| mesh[19].descriptor_name | Quantitative Structure-Activity Relationship |
| type | article |
| title | Structure-Function Analysis of the Transmembrane Protein AmpG from Pseudomonas aeruginosa |
| biblio.issue | 12 |
| biblio.volume | 11 |
| biblio.last_page | e0168060 |
| biblio.first_page | e0168060 |
| topics[0].id | https://openalex.org/T10147 |
| topics[0].field.id | https://openalex.org/fields/13 |
| topics[0].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[0].score | 1.0 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1313 |
| topics[0].subfield.display_name | Molecular Medicine |
| topics[0].display_name | Antibiotic Resistance in Bacteria |
| topics[1].id | https://openalex.org/T10120 |
| topics[1].field.id | https://openalex.org/fields/13 |
| topics[1].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[1].score | 0.9987999796867371 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1311 |
| topics[1].subfield.display_name | Genetics |
| topics[1].display_name | Bacterial Genetics and Biotechnology |
| topics[2].id | https://openalex.org/T10593 |
| topics[2].field.id | https://openalex.org/fields/13 |
| topics[2].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[2].score | 0.9980999827384949 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1312 |
| topics[2].subfield.display_name | Molecular Biology |
| topics[2].display_name | Bacterial biofilms and quorum sensing |
| is_xpac | False |
| apc_list.value | 1805 |
| apc_list.currency | USD |
| apc_list.value_usd | 1805 |
| apc_paid.value | 1805 |
| apc_paid.currency | USD |
| apc_paid.value_usd | 1805 |
| concepts[0].id | https://openalex.org/C201663137 |
| concepts[0].level | 4 |
| concepts[0].score | 0.7595789432525635 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q21198990 |
| concepts[0].display_name | Periplasmic space |
| concepts[1].id | https://openalex.org/C188082640 |
| concepts[1].level | 4 |
| concepts[1].score | 0.6797581911087036 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q1780899 |
| concepts[1].display_name | Complementation |
| concepts[2].id | https://openalex.org/C86803240 |
| concepts[2].level | 0 |
| concepts[2].score | 0.6578990817070007 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[2].display_name | Biology |
| concepts[3].id | https://openalex.org/C143065580 |
| concepts[3].level | 3 |
| concepts[3].score | 0.645902693271637 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q3285695 |
| concepts[3].display_name | Mutant |
| concepts[4].id | https://openalex.org/C2777637488 |
| concepts[4].level | 3 |
| concepts[4].score | 0.5899717807769775 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q31856 |
| concepts[4].display_name | Pseudomonas aeruginosa |
| concepts[5].id | https://openalex.org/C2778643871 |
| concepts[5].level | 3 |
| concepts[5].score | 0.5126672983169556 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q160821 |
| concepts[5].display_name | Vibrio cholerae |
| concepts[6].id | https://openalex.org/C547475151 |
| concepts[6].level | 3 |
| concepts[6].score | 0.4887830913066864 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q25419 |
| concepts[6].display_name | Escherichia coli |
| concepts[7].id | https://openalex.org/C16318435 |
| concepts[7].level | 4 |
| concepts[7].score | 0.4486994445323944 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q149299 |
| concepts[7].display_name | Mutagenesis |
| concepts[8].id | https://openalex.org/C89423630 |
| concepts[8].level | 1 |
| concepts[8].score | 0.44353073835372925 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q7193 |
| concepts[8].display_name | Microbiology |
| concepts[9].id | https://openalex.org/C146587185 |
| concepts[9].level | 4 |
| concepts[9].score | 0.4337732195854187 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q258248 |
| concepts[9].display_name | Bacterial outer membrane |
| concepts[10].id | https://openalex.org/C183641309 |
| concepts[10].level | 4 |
| concepts[10].score | 0.4300363063812256 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q306264 |
| concepts[10].display_name | Amp resistance |
| concepts[11].id | https://openalex.org/C24530287 |
| concepts[11].level | 3 |
| concepts[11].score | 0.4134621322154999 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q424204 |
| concepts[11].display_name | Transmembrane protein |
| concepts[12].id | https://openalex.org/C55493867 |
| concepts[12].level | 1 |
| concepts[12].score | 0.37188416719436646 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q7094 |
| concepts[12].display_name | Biochemistry |
| concepts[13].id | https://openalex.org/C54355233 |
| concepts[13].level | 1 |
| concepts[13].score | 0.23612743616104126 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[13].display_name | Genetics |
| concepts[14].id | https://openalex.org/C104317684 |
| concepts[14].level | 2 |
| concepts[14].score | 0.23023942112922668 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[14].display_name | Gene |
| concepts[15].id | https://openalex.org/C523546767 |
| concepts[15].level | 2 |
| concepts[15].score | 0.22322821617126465 |
| concepts[15].wikidata | https://www.wikidata.org/wiki/Q10876 |
| concepts[15].display_name | Bacteria |
| concepts[16].id | https://openalex.org/C170493617 |
| concepts[16].level | 2 |
| concepts[16].score | 0.0 |
| concepts[16].wikidata | https://www.wikidata.org/wiki/Q208467 |
| concepts[16].display_name | Receptor |
| keywords[0].id | https://openalex.org/keywords/periplasmic-space |
| keywords[0].score | 0.7595789432525635 |
| keywords[0].display_name | Periplasmic space |
| keywords[1].id | https://openalex.org/keywords/complementation |
| keywords[1].score | 0.6797581911087036 |
| keywords[1].display_name | Complementation |
| keywords[2].id | https://openalex.org/keywords/biology |
| keywords[2].score | 0.6578990817070007 |
| keywords[2].display_name | Biology |
| keywords[3].id | https://openalex.org/keywords/mutant |
| keywords[3].score | 0.645902693271637 |
| keywords[3].display_name | Mutant |
| keywords[4].id | https://openalex.org/keywords/pseudomonas-aeruginosa |
| keywords[4].score | 0.5899717807769775 |
| keywords[4].display_name | Pseudomonas aeruginosa |
| keywords[5].id | https://openalex.org/keywords/vibrio-cholerae |
| keywords[5].score | 0.5126672983169556 |
| keywords[5].display_name | Vibrio cholerae |
| keywords[6].id | https://openalex.org/keywords/escherichia-coli |
| keywords[6].score | 0.4887830913066864 |
| keywords[6].display_name | Escherichia coli |
| keywords[7].id | https://openalex.org/keywords/mutagenesis |
| keywords[7].score | 0.4486994445323944 |
| keywords[7].display_name | Mutagenesis |
| keywords[8].id | https://openalex.org/keywords/microbiology |
| keywords[8].score | 0.44353073835372925 |
| keywords[8].display_name | Microbiology |
| keywords[9].id | https://openalex.org/keywords/bacterial-outer-membrane |
| keywords[9].score | 0.4337732195854187 |
| keywords[9].display_name | Bacterial outer membrane |
| keywords[10].id | https://openalex.org/keywords/amp-resistance |
| keywords[10].score | 0.4300363063812256 |
| keywords[10].display_name | Amp resistance |
| keywords[11].id | https://openalex.org/keywords/transmembrane-protein |
| keywords[11].score | 0.4134621322154999 |
| keywords[11].display_name | Transmembrane protein |
| keywords[12].id | https://openalex.org/keywords/biochemistry |
| keywords[12].score | 0.37188416719436646 |
| keywords[12].display_name | Biochemistry |
| keywords[13].id | https://openalex.org/keywords/genetics |
| keywords[13].score | 0.23612743616104126 |
| keywords[13].display_name | Genetics |
| keywords[14].id | https://openalex.org/keywords/gene |
| keywords[14].score | 0.23023942112922668 |
| keywords[14].display_name | Gene |
| keywords[15].id | https://openalex.org/keywords/bacteria |
| keywords[15].score | 0.22322821617126465 |
| keywords[15].display_name | Bacteria |
| language | en |
| locations[0].id | doi:10.1371/journal.pone.0168060 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S202381698 |
| locations[0].source.issn | 1932-6203 |
| locations[0].source.type | journal |
| locations[0].source.is_oa | True |
| locations[0].source.issn_l | 1932-6203 |
| locations[0].source.is_core | True |
| locations[0].source.is_in_doaj | True |
| locations[0].source.display_name | PLoS ONE |
| locations[0].source.host_organization | https://openalex.org/P4310315706 |
| locations[0].source.host_organization_name | Public Library of Science |
| locations[0].source.host_organization_lineage | https://openalex.org/P4310315706 |
| locations[0].license | cc-by |
| locations[0].pdf_url | https://journals.plos.org/plosone/article/file?id=10.1371/journal.pone.0168060&type=printable |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | PLOS ONE |
| locations[0].landing_page_url | https://doi.org/10.1371/journal.pone.0168060 |
| locations[1].id | pmid:27959942 |
| locations[1].is_oa | False |
| locations[1].source.id | https://openalex.org/S4306525036 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | PubMed |
| locations[1].source.host_organization | https://openalex.org/I1299303238 |
| locations[1].source.host_organization_name | National Institutes of Health |
| locations[1].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[1].license | |
| locations[1].pdf_url | |
| locations[1].version | publishedVersion |
| locations[1].raw_type | |
| locations[1].license_id | |
| locations[1].is_accepted | True |
| locations[1].is_published | True |
| locations[1].raw_source_name | PloS one |
| locations[1].landing_page_url | https://pubmed.ncbi.nlm.nih.gov/27959942 |
| locations[2].id | pmh:oai:doaj.org/article:c78894465d1c4cefa8913a0e02d7f9fe |
| locations[2].is_oa | True |
| locations[2].source.id | https://openalex.org/S4306401280 |
| locations[2].source.issn | |
| locations[2].source.type | repository |
| locations[2].source.is_oa | False |
| locations[2].source.issn_l | |
| locations[2].source.is_core | False |
| locations[2].source.is_in_doaj | False |
| locations[2].source.display_name | DOAJ (DOAJ: Directory of Open Access Journals) |
| locations[2].source.host_organization | |
| locations[2].source.host_organization_name | |
| locations[2].source.host_organization_lineage | |
| locations[2].license | cc-by-sa |
| locations[2].pdf_url | |
| locations[2].version | submittedVersion |
| locations[2].raw_type | article |
| locations[2].license_id | https://openalex.org/licenses/cc-by-sa |
| locations[2].is_accepted | False |
| locations[2].is_published | False |
| locations[2].raw_source_name | PLoS ONE, Vol 11, Iss 12, p e0168060 (2016) |
| locations[2].landing_page_url | https://doaj.org/article/c78894465d1c4cefa8913a0e02d7f9fe |
| locations[3].id | pmh:oai:pubmedcentral.nih.gov:5154545 |
| locations[3].is_oa | True |
| locations[3].source.id | https://openalex.org/S2764455111 |
| locations[3].source.issn | |
| locations[3].source.type | repository |
| locations[3].source.is_oa | False |
| locations[3].source.issn_l | |
| locations[3].source.is_core | False |
| locations[3].source.is_in_doaj | False |
| locations[3].source.display_name | PubMed Central |
| locations[3].source.host_organization | https://openalex.org/I1299303238 |
| locations[3].source.host_organization_name | National Institutes of Health |
| locations[3].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[3].license | cc-by |
| locations[3].pdf_url | |
| locations[3].version | submittedVersion |
| locations[3].raw_type | Text |
| locations[3].license_id | https://openalex.org/licenses/cc-by |
| locations[3].is_accepted | False |
| locations[3].is_published | False |
| locations[3].raw_source_name | |
| locations[3].landing_page_url | http://doi.org/10.1371/journal.pone.0168060 |
| locations[4].id | pmh:oai:figshare.com:article/4314026 |
| locations[4].is_oa | True |
| locations[4].source.id | https://openalex.org/S4306400572 |
| locations[4].source.issn | |
| locations[4].source.type | repository |
| locations[4].source.is_oa | False |
| locations[4].source.issn_l | |
| locations[4].source.is_core | False |
| locations[4].source.is_in_doaj | False |
| locations[4].source.display_name | OPAL (Open@LaTrobe) (La Trobe University) |
| locations[4].source.host_organization | https://openalex.org/I196829312 |
| locations[4].source.host_organization_name | La Trobe University |
| locations[4].source.host_organization_lineage | https://openalex.org/I196829312 |
| locations[4].license | cc-by |
| locations[4].pdf_url | |
| locations[4].version | submittedVersion |
| locations[4].raw_type | Dataset |
| locations[4].license_id | https://openalex.org/licenses/cc-by |
| locations[4].is_accepted | False |
| locations[4].is_published | False |
| locations[4].raw_source_name | |
| locations[4].landing_page_url | https://figshare.com/articles/dataset/Structure-Function_Analysis_of_the_Transmembrane_Protein_AmpG_from_i_Pseudomonas_aeruginosa_i_/4314026 |
| indexed_in | crossref, doaj, pubmed |
| authorships[0].author.id | https://openalex.org/A5087478830 |
| authorships[0].author.orcid | https://orcid.org/0000-0003-1673-6031 |
| authorships[0].author.display_name | Peizhen Li |
| authorships[0].countries | CN |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I27781120 |
| authorships[0].affiliations[0].raw_affiliation_string | School of Laboratory Medicine and Life Science/Institute of Biomedical Informatics, Wenzhou Medical University, Wenzhou, China |
| authorships[0].institutions[0].id | https://openalex.org/I27781120 |
| authorships[0].institutions[0].ror | https://ror.org/00rd5t069 |
| authorships[0].institutions[0].type | education |
| authorships[0].institutions[0].lineage | https://openalex.org/I27781120 |
| authorships[0].institutions[0].country_code | CN |
| authorships[0].institutions[0].display_name | Wenzhou Medical University |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Peizhen Li |
| authorships[0].is_corresponding | True |
| authorships[0].raw_affiliation_strings | School of Laboratory Medicine and Life Science/Institute of Biomedical Informatics, Wenzhou Medical University, Wenzhou, China |
| authorships[1].author.id | https://openalex.org/A5006942289 |
| authorships[1].author.orcid | https://orcid.org/0000-0003-1439-8067 |
| authorships[1].author.display_name | Jun Ying |
| authorships[1].countries | CN |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I27781120 |
| authorships[1].affiliations[0].raw_affiliation_string | School of Laboratory Medicine and Life Science/Institute of Biomedical Informatics, Wenzhou Medical University, Wenzhou, China |
| authorships[1].institutions[0].id | https://openalex.org/I27781120 |
| authorships[1].institutions[0].ror | https://ror.org/00rd5t069 |
| authorships[1].institutions[0].type | education |
| authorships[1].institutions[0].lineage | https://openalex.org/I27781120 |
| authorships[1].institutions[0].country_code | CN |
| authorships[1].institutions[0].display_name | Wenzhou Medical University |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Jun Ying |
| authorships[1].is_corresponding | True |
| authorships[1].raw_affiliation_strings | School of Laboratory Medicine and Life Science/Institute of Biomedical Informatics, Wenzhou Medical University, Wenzhou, China |
| authorships[2].author.id | https://openalex.org/A5101953347 |
| authorships[2].author.orcid | https://orcid.org/0000-0002-7611-2370 |
| authorships[2].author.display_name | Guangjian Yang |
| authorships[2].countries | CN |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I27781120 |
| authorships[2].affiliations[0].raw_affiliation_string | School of Laboratory Medicine and Life Science/Institute of Biomedical Informatics, Wenzhou Medical University, Wenzhou, China |
| authorships[2].institutions[0].id | https://openalex.org/I27781120 |
| authorships[2].institutions[0].ror | https://ror.org/00rd5t069 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I27781120 |
| authorships[2].institutions[0].country_code | CN |
| authorships[2].institutions[0].display_name | Wenzhou Medical University |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Guangjian Yang |
| authorships[2].is_corresponding | True |
| authorships[2].raw_affiliation_strings | School of Laboratory Medicine and Life Science/Institute of Biomedical Informatics, Wenzhou Medical University, Wenzhou, China |
| authorships[3].author.id | https://openalex.org/A5087461678 |
| authorships[3].author.orcid | https://orcid.org/0000-0003-4036-9841 |
| authorships[3].author.display_name | Aifang Li |
| authorships[3].countries | CN |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I27781120 |
| authorships[3].affiliations[0].raw_affiliation_string | School of Laboratory Medicine and Life Science/Institute of Biomedical Informatics, Wenzhou Medical University, Wenzhou, China |
| authorships[3].institutions[0].id | https://openalex.org/I27781120 |
| authorships[3].institutions[0].ror | https://ror.org/00rd5t069 |
| authorships[3].institutions[0].type | education |
| authorships[3].institutions[0].lineage | https://openalex.org/I27781120 |
| authorships[3].institutions[0].country_code | CN |
| authorships[3].institutions[0].display_name | Wenzhou Medical University |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Aifang Li |
| authorships[3].is_corresponding | True |
| authorships[3].raw_affiliation_strings | School of Laboratory Medicine and Life Science/Institute of Biomedical Informatics, Wenzhou Medical University, Wenzhou, China |
| authorships[4].author.id | https://openalex.org/A5100370474 |
| authorships[4].author.orcid | https://orcid.org/0000-0002-6545-3663 |
| authorships[4].author.display_name | Jian Wang |
| authorships[4].countries | CN |
| authorships[4].affiliations[0].institution_ids | https://openalex.org/I27781120 |
| authorships[4].affiliations[0].raw_affiliation_string | School of Laboratory Medicine and Life Science/Institute of Biomedical Informatics, Wenzhou Medical University, Wenzhou, China |
| authorships[4].institutions[0].id | https://openalex.org/I27781120 |
| authorships[4].institutions[0].ror | https://ror.org/00rd5t069 |
| authorships[4].institutions[0].type | education |
| authorships[4].institutions[0].lineage | https://openalex.org/I27781120 |
| authorships[4].institutions[0].country_code | CN |
| authorships[4].institutions[0].display_name | Wenzhou Medical University |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Jian Wang |
| authorships[4].is_corresponding | True |
| authorships[4].raw_affiliation_strings | School of Laboratory Medicine and Life Science/Institute of Biomedical Informatics, Wenzhou Medical University, Wenzhou, China |
| authorships[5].author.id | https://openalex.org/A5102016185 |
| authorships[5].author.orcid | https://orcid.org/0000-0002-7378-6783 |
| authorships[5].author.display_name | Junwan Lu |
| authorships[5].countries | CN |
| authorships[5].affiliations[0].institution_ids | https://openalex.org/I3129496460 |
| authorships[5].affiliations[0].raw_affiliation_string | School of Medicine, Lishui College, Lishui, China |
| authorships[5].institutions[0].id | https://openalex.org/I3129496460 |
| authorships[5].institutions[0].ror | https://ror.org/0418kp584 |
| authorships[5].institutions[0].type | education |
| authorships[5].institutions[0].lineage | https://openalex.org/I3129496460 |
| authorships[5].institutions[0].country_code | CN |
| authorships[5].institutions[0].display_name | Lishui University |
| authorships[5].author_position | middle |
| authorships[5].raw_author_name | Junwan Lu |
| authorships[5].is_corresponding | True |
| authorships[5].raw_affiliation_strings | School of Medicine, Lishui College, Lishui, China |
| authorships[6].author.id | https://openalex.org/A5101760487 |
| authorships[6].author.orcid | https://orcid.org/0000-0001-7430-9308 |
| authorships[6].author.display_name | Junrong Wang |
| authorships[6].countries | CN |
| authorships[6].affiliations[0].institution_ids | https://openalex.org/I4210144189 |
| authorships[6].affiliations[0].raw_affiliation_string | Wenling Women's & Children's Hospital, Wenling, China |
| authorships[6].institutions[0].id | https://openalex.org/I4210144189 |
| authorships[6].institutions[0].ror | https://ror.org/04e3jvd14 |
| authorships[6].institutions[0].type | healthcare |
| authorships[6].institutions[0].lineage | https://openalex.org/I4210144189 |
| authorships[6].institutions[0].country_code | CN |
| authorships[6].institutions[0].display_name | The First People's Hospital of Wenling |
| authorships[6].author_position | middle |
| authorships[6].raw_author_name | Junrong Wang |
| authorships[6].is_corresponding | True |
| authorships[6].raw_affiliation_strings | Wenling Women's & Children's Hospital, Wenling, China |
| authorships[7].author.id | https://openalex.org/A5003791054 |
| authorships[7].author.orcid | https://orcid.org/0000-0001-6859-4202 |
| authorships[7].author.display_name | Teng Xu |
| authorships[7].countries | CN |
| authorships[7].affiliations[0].institution_ids | https://openalex.org/I27781120 |
| authorships[7].affiliations[0].raw_affiliation_string | School of Laboratory Medicine and Life Science/Institute of Biomedical Informatics, Wenzhou Medical University, Wenzhou, China |
| authorships[7].institutions[0].id | https://openalex.org/I27781120 |
| authorships[7].institutions[0].ror | https://ror.org/00rd5t069 |
| authorships[7].institutions[0].type | education |
| authorships[7].institutions[0].lineage | https://openalex.org/I27781120 |
| authorships[7].institutions[0].country_code | CN |
| authorships[7].institutions[0].display_name | Wenzhou Medical University |
| authorships[7].author_position | middle |
| authorships[7].raw_author_name | Teng Xu |
| authorships[7].is_corresponding | True |
| authorships[7].raw_affiliation_strings | School of Laboratory Medicine and Life Science/Institute of Biomedical Informatics, Wenzhou Medical University, Wenzhou, China |
| authorships[8].author.id | https://openalex.org/A5040873566 |
| authorships[8].author.orcid | https://orcid.org/0000-0002-9676-7438 |
| authorships[8].author.display_name | Huiguang Yi |
| authorships[8].countries | CN |
| authorships[8].affiliations[0].institution_ids | https://openalex.org/I27781120 |
| authorships[8].affiliations[0].raw_affiliation_string | School of Laboratory Medicine and Life Science/Institute of Biomedical Informatics, Wenzhou Medical University, Wenzhou, China |
| authorships[8].institutions[0].id | https://openalex.org/I27781120 |
| authorships[8].institutions[0].ror | https://ror.org/00rd5t069 |
| authorships[8].institutions[0].type | education |
| authorships[8].institutions[0].lineage | https://openalex.org/I27781120 |
| authorships[8].institutions[0].country_code | CN |
| authorships[8].institutions[0].display_name | Wenzhou Medical University |
| authorships[8].author_position | middle |
| authorships[8].raw_author_name | Huiguang Yi |
| authorships[8].is_corresponding | True |
| authorships[8].raw_affiliation_strings | School of Laboratory Medicine and Life Science/Institute of Biomedical Informatics, Wenzhou Medical University, Wenzhou, China |
| authorships[9].author.id | https://openalex.org/A5102006839 |
| authorships[9].author.orcid | https://orcid.org/0000-0002-2797-576X |
| authorships[9].author.display_name | Kewei Li |
| authorships[9].countries | CN |
| authorships[9].affiliations[0].institution_ids | https://openalex.org/I27781120 |
| authorships[9].affiliations[0].raw_affiliation_string | School of Laboratory Medicine and Life Science/Institute of Biomedical Informatics, Wenzhou Medical University, Wenzhou, China |
| authorships[9].institutions[0].id | https://openalex.org/I27781120 |
| authorships[9].institutions[0].ror | https://ror.org/00rd5t069 |
| authorships[9].institutions[0].type | education |
| authorships[9].institutions[0].lineage | https://openalex.org/I27781120 |
| authorships[9].institutions[0].country_code | CN |
| authorships[9].institutions[0].display_name | Wenzhou Medical University |
| authorships[9].author_position | middle |
| authorships[9].raw_author_name | Kewei Li |
| authorships[9].is_corresponding | True |
| authorships[9].raw_affiliation_strings | School of Laboratory Medicine and Life Science/Institute of Biomedical Informatics, Wenzhou Medical University, Wenzhou, China |
| authorships[10].author.id | https://openalex.org/A5036438615 |
| authorships[10].author.orcid | https://orcid.org/0000-0002-6447-6978 |
| authorships[10].author.display_name | Shouguang Jin |
| authorships[10].countries | US |
| authorships[10].affiliations[0].institution_ids | https://openalex.org/I33213144 |
| authorships[10].affiliations[0].raw_affiliation_string | Department of Molecular Genetics and Microbiology, University of Florida, Gainesville, Florida, United States of America |
| authorships[10].institutions[0].id | https://openalex.org/I33213144 |
| authorships[10].institutions[0].ror | https://ror.org/02y3ad647 |
| authorships[10].institutions[0].type | education |
| authorships[10].institutions[0].lineage | https://openalex.org/I33213144 |
| authorships[10].institutions[0].country_code | US |
| authorships[10].institutions[0].display_name | University of Florida |
| authorships[10].author_position | middle |
| authorships[10].raw_author_name | Shouguang Jin |
| authorships[10].is_corresponding | True |
| authorships[10].raw_affiliation_strings | Department of Molecular Genetics and Microbiology, University of Florida, Gainesville, Florida, United States of America |
| authorships[11].author.id | https://openalex.org/A5101943568 |
| authorships[11].author.orcid | https://orcid.org/0000-0001-6523-8763 |
| authorships[11].author.display_name | Qiyu Bao |
| authorships[11].countries | CN |
| authorships[11].affiliations[0].institution_ids | https://openalex.org/I27781120 |
| authorships[11].affiliations[0].raw_affiliation_string | School of Laboratory Medicine and Life Science/Institute of Biomedical Informatics, Wenzhou Medical University, Wenzhou, China |
| authorships[11].institutions[0].id | https://openalex.org/I27781120 |
| authorships[11].institutions[0].ror | https://ror.org/00rd5t069 |
| authorships[11].institutions[0].type | education |
| authorships[11].institutions[0].lineage | https://openalex.org/I27781120 |
| authorships[11].institutions[0].country_code | CN |
| authorships[11].institutions[0].display_name | Wenzhou Medical University |
| authorships[11].author_position | middle |
| authorships[11].raw_author_name | Qiyu Bao |
| authorships[11].is_corresponding | True |
| authorships[11].raw_affiliation_strings | School of Laboratory Medicine and Life Science/Institute of Biomedical Informatics, Wenzhou Medical University, Wenzhou, China |
| authorships[12].author.id | https://openalex.org/A5009099198 |
| authorships[12].author.orcid | https://orcid.org/0000-0002-0732-9218 |
| authorships[12].author.display_name | Kaibo Zhang |
| authorships[12].countries | CN |
| authorships[12].affiliations[0].institution_ids | https://openalex.org/I3129496460 |
| authorships[12].affiliations[0].raw_affiliation_string | School of Medicine, Lishui College, Lishui, China |
| authorships[12].affiliations[1].institution_ids | https://openalex.org/I27781120 |
| authorships[12].affiliations[1].raw_affiliation_string | School of Laboratory Medicine and Life Science/Institute of Biomedical Informatics, Wenzhou Medical University, Wenzhou, China |
| authorships[12].institutions[0].id | https://openalex.org/I3129496460 |
| authorships[12].institutions[0].ror | https://ror.org/0418kp584 |
| authorships[12].institutions[0].type | education |
| authorships[12].institutions[0].lineage | https://openalex.org/I3129496460 |
| authorships[12].institutions[0].country_code | CN |
| authorships[12].institutions[0].display_name | Lishui University |
| authorships[12].institutions[1].id | https://openalex.org/I27781120 |
| authorships[12].institutions[1].ror | https://ror.org/00rd5t069 |
| authorships[12].institutions[1].type | education |
| authorships[12].institutions[1].lineage | https://openalex.org/I27781120 |
| authorships[12].institutions[1].country_code | CN |
| authorships[12].institutions[1].display_name | Wenzhou Medical University |
| authorships[12].author_position | last |
| authorships[12].raw_author_name | Kaibo Zhang |
| authorships[12].is_corresponding | True |
| authorships[12].raw_affiliation_strings | School of Laboratory Medicine and Life Science/Institute of Biomedical Informatics, Wenzhou Medical University, Wenzhou, China, School of Medicine, Lishui College, Lishui, China |
| has_content.pdf | True |
| has_content.grobid_xml | True |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://journals.plos.org/plosone/article/file?id=10.1371/journal.pone.0168060&type=printable |
| open_access.oa_status | gold |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Structure-Function Analysis of the Transmembrane Protein AmpG from Pseudomonas aeruginosa |
| has_fulltext | True |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T10147 |
| primary_topic.field.id | https://openalex.org/fields/13 |
| primary_topic.field.display_name | Biochemistry, Genetics and Molecular Biology |
| primary_topic.score | 1.0 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1313 |
| primary_topic.subfield.display_name | Molecular Medicine |
| primary_topic.display_name | Antibiotic Resistance in Bacteria |
| related_works | https://openalex.org/W2035118599, https://openalex.org/W2177070127, https://openalex.org/W1566092905, https://openalex.org/W2137745451, https://openalex.org/W2099486582, https://openalex.org/W2745981626, https://openalex.org/W2214458739, https://openalex.org/W2355766117, https://openalex.org/W2140228345, https://openalex.org/W4366290807 |
| cited_by_count | 14 |
| counts_by_year[0].year | 2025 |
| counts_by_year[0].cited_by_count | 3 |
| counts_by_year[1].year | 2023 |
| counts_by_year[1].cited_by_count | 1 |
| counts_by_year[2].year | 2022 |
| counts_by_year[2].cited_by_count | 3 |
| counts_by_year[3].year | 2021 |
| counts_by_year[3].cited_by_count | 1 |
| counts_by_year[4].year | 2019 |
| counts_by_year[4].cited_by_count | 1 |
| counts_by_year[5].year | 2018 |
| counts_by_year[5].cited_by_count | 3 |
| counts_by_year[6].year | 2017 |
| counts_by_year[6].cited_by_count | 1 |
| counts_by_year[7].year | 2016 |
| counts_by_year[7].cited_by_count | 1 |
| locations_count | 5 |
| best_oa_location.id | doi:10.1371/journal.pone.0168060 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S202381698 |
| best_oa_location.source.issn | 1932-6203 |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | True |
| best_oa_location.source.issn_l | 1932-6203 |
| best_oa_location.source.is_core | True |
| best_oa_location.source.is_in_doaj | True |
| best_oa_location.source.display_name | PLoS ONE |
| best_oa_location.source.host_organization | https://openalex.org/P4310315706 |
| best_oa_location.source.host_organization_name | Public Library of Science |
| best_oa_location.source.host_organization_lineage | https://openalex.org/P4310315706 |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | https://journals.plos.org/plosone/article/file?id=10.1371/journal.pone.0168060&type=printable |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | journal-article |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | PLOS ONE |
| best_oa_location.landing_page_url | https://doi.org/10.1371/journal.pone.0168060 |
| primary_location.id | doi:10.1371/journal.pone.0168060 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S202381698 |
| primary_location.source.issn | 1932-6203 |
| primary_location.source.type | journal |
| primary_location.source.is_oa | True |
| primary_location.source.issn_l | 1932-6203 |
| primary_location.source.is_core | True |
| primary_location.source.is_in_doaj | True |
| primary_location.source.display_name | PLoS ONE |
| primary_location.source.host_organization | https://openalex.org/P4310315706 |
| primary_location.source.host_organization_name | Public Library of Science |
| primary_location.source.host_organization_lineage | https://openalex.org/P4310315706 |
| primary_location.license | cc-by |
| primary_location.pdf_url | https://journals.plos.org/plosone/article/file?id=10.1371/journal.pone.0168060&type=printable |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | PLOS ONE |
| primary_location.landing_page_url | https://doi.org/10.1371/journal.pone.0168060 |
| publication_date | 2016-12-13 |
| publication_year | 2016 |
| referenced_works | https://openalex.org/W2139806506, https://openalex.org/W2134352676, https://openalex.org/W2144216136, https://openalex.org/W2097432034, https://openalex.org/W1975146889, https://openalex.org/W2119704151, https://openalex.org/W2061826699, https://openalex.org/W2070002440, https://openalex.org/W2153280373, https://openalex.org/W2161706773, https://openalex.org/W2157436998, https://openalex.org/W2131615228, https://openalex.org/W2107277218, https://openalex.org/W2156396808, https://openalex.org/W2044816911, https://openalex.org/W2003852785, https://openalex.org/W2037312364, https://openalex.org/W2110120447, https://openalex.org/W2007278459, https://openalex.org/W2152419408, https://openalex.org/W2096970090, https://openalex.org/W2106884274, https://openalex.org/W2156295601, https://openalex.org/W2123750465, https://openalex.org/W2105390055, https://openalex.org/W2157824544, https://openalex.org/W2132569635, https://openalex.org/W2006964441, https://openalex.org/W2063469071, https://openalex.org/W6681170628, https://openalex.org/W2149353640, https://openalex.org/W2075893586, https://openalex.org/W2144634347, https://openalex.org/W2314225431, https://openalex.org/W2036234525 |
| referenced_works_count | 35 |
| abstract_inverted_index.a | 2, 108, 131, 137 |
| abstract_inverted_index.32 | 98 |
| abstract_inverted_index.To | 28 |
| abstract_inverted_index.an | 101 |
| abstract_inverted_index.be | 231 |
| abstract_inverted_index.in | 57, 77, 111, 130, 136, 143, 214 |
| abstract_inverted_index.is | 1, 18, 87, 97 |
| abstract_inverted_index.it | 96 |
| abstract_inverted_index.of | 23, 64, 114, 120, 133, 139, 164, 196, 218, 236 |
| abstract_inverted_index.to | 14, 85, 141, 158 |
| abstract_inverted_index.512 | 88 |
| abstract_inverted_index.Our | 202 |
| abstract_inverted_index.The | 80, 145, 168, 182 |
| abstract_inverted_index.and | 38, 44, 51, 70, 127, 152, 160, 173, 184 |
| abstract_inverted_index.but | 193 |
| abstract_inverted_index.can | 230 |
| abstract_inverted_index.for | 20, 90, 100, 233 |
| abstract_inverted_index.had | 155, 176, 187 |
| abstract_inverted_index.new | 30, 237 |
| abstract_inverted_index.the | 12, 15, 21, 24, 33, 112, 115, 165, 194, 197, 207, 216, 222, 234 |
| abstract_inverted_index.was | 199 |
| abstract_inverted_index.AmpC | 166 |
| abstract_inverted_index.AmpG | 0, 36, 75, 122, 223 |
| abstract_inverted_index.Q131 | 126 |
| abstract_inverted_index.ampC | 25 |
| abstract_inverted_index.ampG | 102, 178, 189 |
| abstract_inverted_index.from | 11, 61 |
| abstract_inverted_index.into | 32 |
| abstract_inverted_index.loss | 132, 138 |
| abstract_inverted_index.mRNA | 179, 190 |
| abstract_inverted_index.that | 8, 206 |
| abstract_inverted_index.this | 58 |
| abstract_inverted_index.type | 92 |
| abstract_inverted_index.were | 55 |
| abstract_inverted_index.wild | 91 |
| abstract_inverted_index.with | 5, 107, 221 |
| abstract_inverted_index.(G29, | 124 |
| abstract_inverted_index.(MIC) | 84 |
| abstract_inverted_index.A129, | 125 |
| abstract_inverted_index.A129D | 185 |
| abstract_inverted_index.A129T | 183 |
| abstract_inverted_index.A197) | 128 |
| abstract_inverted_index.A197D | 153, 174 |
| abstract_inverted_index.A197S | 151, 172 |
| abstract_inverted_index.AmpG. | 219 |
| abstract_inverted_index.AmpGs | 60 |
| abstract_inverted_index.G29A, | 146, 169 |
| abstract_inverted_index.G29V, | 147, 170 |
| abstract_inverted_index.PAO1, | 94 |
| abstract_inverted_index.acids | 210, 229 |
| abstract_inverted_index.amino | 209, 228 |
| abstract_inverted_index.coli, | 67 |
| abstract_inverted_index.could | 73 |
| abstract_inverted_index.lower | 156 |
| abstract_inverted_index.roles | 213 |
| abstract_inverted_index.these | 226 |
| abstract_inverted_index.which | 17 |
| abstract_inverted_index.while | 95 |
| abstract_inverted_index.A129D, | 150 |
| abstract_inverted_index.A129T, | 148 |
| abstract_inverted_index.A129V, | 149, 171 |
| abstract_inverted_index.Vibrio | 68 |
| abstract_inverted_index.genera | 63 |
| abstract_inverted_index.mutant | 104 |
| abstract_inverted_index.normal | 188 |
| abstract_inverted_index.obtain | 29 |
| abstract_inverted_index.played | 211 |
| abstract_inverted_index.strain | 93, 105 |
| abstract_inverted_index.study. | 59 |
| abstract_inverted_index.μg/ml | 89, 99 |
| abstract_inverted_index.agents. | 239 |
| abstract_inverted_index.between | 35 |
| abstract_inverted_index.genetic | 52 |
| abstract_inverted_index.levels, | 192 |
| abstract_inverted_index.levels. | 181 |
| abstract_inverted_index.minimal | 81 |
| abstract_inverted_index.mutants | 154, 175, 186 |
| abstract_inverted_index.protein | 4, 198 |
| abstract_inverted_index.results | 204 |
| abstract_inverted_index.Combined | 220 |
| abstract_inverted_index.activity | 7, 113, 163 |
| abstract_inverted_index.analyses | 50 |
| abstract_inverted_index.bacteria | 65 |
| abstract_inverted_index.cholerae | 69 |
| abstract_inverted_index.critical | 227 |
| abstract_inverted_index.decrease | 110 |
| abstract_inverted_index.deletion | 103 |
| abstract_inverted_index.encoding | 26 |
| abstract_inverted_index.function | 76, 195, 217 |
| abstract_inverted_index.genomics | 41 |
| abstract_inverted_index.insights | 31 |
| abstract_inverted_index.permease | 6 |
| abstract_inverted_index.reduced. | 201 |
| abstract_inverted_index.residues | 123 |
| abstract_inverted_index.resulted | 129 |
| abstract_inverted_index.targeted | 232 |
| abstract_inverted_index.tertiary | 45 |
| abstract_inverted_index.analysis, | 42 |
| abstract_inverted_index.conserved | 121, 208 |
| abstract_inverted_index.decreased | 162, 177 |
| abstract_inverted_index.different | 62 |
| abstract_inverted_index.essential | 19, 212 |
| abstract_inverted_index.function, | 39, 134 |
| abstract_inverted_index.induction | 22 |
| abstract_inverted_index.modeling, | 47 |
| abstract_inverted_index.performed | 56 |
| abstract_inverted_index.periplasm | 13 |
| abstract_inverted_index.resulting | 135 |
| abstract_inverted_index.secondary | 43 |
| abstract_inverted_index.structure | 37, 46 |
| abstract_inverted_index.ampicillin | 86, 142, 159 |
| abstract_inverted_index.baumannii) | 72 |
| abstract_inverted_index.complement | 74 |
| abstract_inverted_index.cytoplasm, | 16 |
| abstract_inverted_index.inhibitory | 82 |
| abstract_inverted_index.mutational | 49 |
| abstract_inverted_index.resistance | 140, 157 |
| abstract_inverted_index.structural | 224 |
| abstract_inverted_index.transcript | 180, 191 |
| abstract_inverted_index.transports | 9 |
| abstract_inverted_index.PAO1ΔampG. | 144 |
| abstract_inverted_index.Pseudomonas | 78 |
| abstract_inverted_index.aeruginosa. | 79 |
| abstract_inverted_index.comparative | 40 |
| abstract_inverted_index.demonstrate | 205 |
| abstract_inverted_index.development | 235 |
| abstract_inverted_index.drastically | 200 |
| abstract_inverted_index.experiments | 54 |
| abstract_inverted_index.maintaining | 215 |
| abstract_inverted_index.mutagenesis | 119 |
| abstract_inverted_index.(Escherichia | 66 |
| abstract_inverted_index.(PAO1ΔampG) | 106 |
| abstract_inverted_index.ampC-encoded | 116 |
| abstract_inverted_index.experimental | 203 |
| abstract_inverted_index.information, | 225 |
| abstract_inverted_index.meuropeptide | 10 |
| abstract_inverted_index.relationship | 34 |
| abstract_inverted_index.Acinetobacter | 71 |
| abstract_inverted_index.Site-directed | 118 |
| abstract_inverted_index.concentration | 83 |
| abstract_inverted_index.corresponding | 109 |
| abstract_inverted_index.significantly | 161 |
| abstract_inverted_index.site-directed | 48 |
| abstract_inverted_index.transmembrane | 3 |
| abstract_inverted_index.β-lactamase. | 27, 117, 167 |
| abstract_inverted_index.anti-bacterial | 238 |
| abstract_inverted_index.complementation | 53 |
| cited_by_percentile_year.max | 97 |
| cited_by_percentile_year.min | 89 |
| corresponding_author_ids | https://openalex.org/A5087478830, https://openalex.org/A5101943568, https://openalex.org/A5003791054, https://openalex.org/A5040873566, https://openalex.org/A5101953347, https://openalex.org/A5036438615, https://openalex.org/A5087461678, https://openalex.org/A5102006839, https://openalex.org/A5102016185, https://openalex.org/A5100370474, https://openalex.org/A5009099198, https://openalex.org/A5006942289, https://openalex.org/A5101760487 |
| countries_distinct_count | 2 |
| institutions_distinct_count | 13 |
| corresponding_institution_ids | https://openalex.org/I27781120, https://openalex.org/I3129496460, https://openalex.org/I33213144, https://openalex.org/I4210144189 |
| sustainable_development_goals[0].id | https://metadata.un.org/sdg/15 |
| sustainable_development_goals[0].score | 0.41999998688697815 |
| sustainable_development_goals[0].display_name | Life in Land |
| citation_normalized_percentile.value | 0.71707748 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |