Study on the spatial distribution patterns of histone modifications in Hippo pathway genes Article Swipe
 YOU?
·
· 2021
· Open Access
·
· DOI: https://doi.org/10.52601/bpr.2021.200042
YOU?
·
· 2021
· Open Access
·
· DOI: https://doi.org/10.52601/bpr.2021.200042
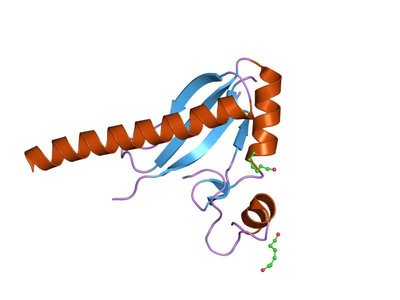
Hippo pathway can regulate cell division, differentiation and apoptosis, and control the shape and size of organs. To study the distribution patterns of histone modifications of Hippo pathway genes in embryonic stem cells is helpful to understand the molecular regulation mechanism of histone modification and Hippo pathway on stem cell self-renewal. In this study, 19 genes of Hippo pathway including YAP, TAZ, LATS1/2, MST1 and SAV1, and eight histone modifications in embryonic stem cells were chosen to study the spatial distribution patterns of histone modifications. It was found that there were obvious type specificity and the location preference of target regions in the distributions of histone modifications, and H3K4me3 and H3K36me3 played the most important regulatory roles. Through the correlation analysis of histone modifications, a histone modification functional cluster composed of H3K4ac, H3K4me3, H3K9ac and H3K27ac was detected in YAP. In addition, the spatial distribution patterns of histone modifications in Hippo pathway genes were obtained, which provided a new theoretical reference for elucidating the mechanism of histone modifications regulating the gene expression of Hippo pathway, and for revealing the molecular regulatory mechanism of histone modifications affecting the self-renewal of embryonic stem cells by regulating the Hippo pathway.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.52601/bpr.2021.200042
- OA Status
- diamond
- Cited By
- 4
- References
- 24
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W3153896515
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W3153896515Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.52601/bpr.2021.200042Digital Object Identifier
- Title
-
Study on the spatial distribution patterns of histone modifications in Hippo pathway genesWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2021Year of publication
- Publication date
-
2021-01-01Full publication date if available
- Authors
-
Wenxia Su, D Zhang, Zhang Qiang, Qian‐Zhong LiList of authors in order
- Landing page
-
https://doi.org/10.52601/bpr.2021.200042Publisher landing page
- Open access
-
YesWhether a free full text is available
- OA status
-
diamondOpen access status per OpenAlex
- OA URL
-
https://doi.org/10.52601/bpr.2021.200042Direct OA link when available
- Concepts
-
H3K4me3, Hippo signaling pathway, Histone, Biology, Cell biology, Histone H3, Histone code, Histone methylation, Histone methyltransferase, Genetics, Gene, Gene expression, Signal transduction, Nucleosome, Promoter, DNA methylationTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
4Total citation count in OpenAlex
- Citations by year (recent)
-
2025: 1, 2024: 1, 2023: 2Per-year citation counts (last 5 years)
- References (count)
-
24Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W3153896515 |
|---|---|
| doi | https://doi.org/10.52601/bpr.2021.200042 |
| ids.doi | https://doi.org/10.52601/bpr.2021.200042 |
| ids.mag | 3153896515 |
| ids.pmid | https://pubmed.ncbi.nlm.nih.gov/37288084 |
| ids.openalex | https://openalex.org/W3153896515 |
| fwci | 0.3911391 |
| type | article |
| title | Study on the spatial distribution patterns of histone modifications in Hippo pathway genes |
| biblio.issue | 1 |
| biblio.volume | 7 |
| biblio.last_page | 79 |
| biblio.first_page | 71 |
| topics[0].id | https://openalex.org/T12065 |
| topics[0].field.id | https://openalex.org/fields/13 |
| topics[0].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[0].score | 1.0 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1307 |
| topics[0].subfield.display_name | Cell Biology |
| topics[0].display_name | Hippo pathway signaling and YAP/TAZ |
| topics[1].id | https://openalex.org/T10604 |
| topics[1].field.id | https://openalex.org/fields/13 |
| topics[1].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[1].score | 0.9488999843597412 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1312 |
| topics[1].subfield.display_name | Molecular Biology |
| topics[1].display_name | RNA Research and Splicing |
| topics[2].id | https://openalex.org/T10929 |
| topics[2].field.id | https://openalex.org/fields/13 |
| topics[2].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[2].score | 0.9322999715805054 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1312 |
| topics[2].subfield.display_name | Molecular Biology |
| topics[2].display_name | Wnt/β-catenin signaling in development and cancer |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C2780914512 |
| concepts[0].level | 5 |
| concepts[0].score | 0.7592811584472656 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q25323943 |
| concepts[0].display_name | H3K4me3 |
| concepts[1].id | https://openalex.org/C172512520 |
| concepts[1].level | 3 |
| concepts[1].score | 0.7583656311035156 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q5768379 |
| concepts[1].display_name | Hippo signaling pathway |
| concepts[2].id | https://openalex.org/C64927066 |
| concepts[2].level | 3 |
| concepts[2].score | 0.709576427936554 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q36293 |
| concepts[2].display_name | Histone |
| concepts[3].id | https://openalex.org/C86803240 |
| concepts[3].level | 0 |
| concepts[3].score | 0.6490663290023804 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[3].display_name | Biology |
| concepts[4].id | https://openalex.org/C95444343 |
| concepts[4].level | 1 |
| concepts[4].score | 0.6354193091392517 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q7141 |
| concepts[4].display_name | Cell biology |
| concepts[5].id | https://openalex.org/C2776745794 |
| concepts[5].level | 4 |
| concepts[5].score | 0.5289364457130432 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q24777451 |
| concepts[5].display_name | Histone H3 |
| concepts[6].id | https://openalex.org/C10058791 |
| concepts[6].level | 5 |
| concepts[6].score | 0.5017390251159668 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q910897 |
| concepts[6].display_name | Histone code |
| concepts[7].id | https://openalex.org/C22709980 |
| concepts[7].level | 5 |
| concepts[7].score | 0.5006923675537109 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q4117777 |
| concepts[7].display_name | Histone methylation |
| concepts[8].id | https://openalex.org/C150425827 |
| concepts[8].level | 4 |
| concepts[8].score | 0.4490509629249573 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q2560317 |
| concepts[8].display_name | Histone methyltransferase |
| concepts[9].id | https://openalex.org/C54355233 |
| concepts[9].level | 1 |
| concepts[9].score | 0.38138043880462646 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[9].display_name | Genetics |
| concepts[10].id | https://openalex.org/C104317684 |
| concepts[10].level | 2 |
| concepts[10].score | 0.235899418592453 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[10].display_name | Gene |
| concepts[11].id | https://openalex.org/C150194340 |
| concepts[11].level | 3 |
| concepts[11].score | 0.17375102639198303 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q26972 |
| concepts[11].display_name | Gene expression |
| concepts[12].id | https://openalex.org/C62478195 |
| concepts[12].level | 2 |
| concepts[12].score | 0.17256596684455872 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q828130 |
| concepts[12].display_name | Signal transduction |
| concepts[13].id | https://openalex.org/C84772758 |
| concepts[13].level | 4 |
| concepts[13].score | 0.14750754833221436 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q271794 |
| concepts[13].display_name | Nucleosome |
| concepts[14].id | https://openalex.org/C101762097 |
| concepts[14].level | 4 |
| concepts[14].score | 0.1036422848701477 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q224093 |
| concepts[14].display_name | Promoter |
| concepts[15].id | https://openalex.org/C190727270 |
| concepts[15].level | 4 |
| concepts[15].score | 0.06431648135185242 |
| concepts[15].wikidata | https://www.wikidata.org/wiki/Q874745 |
| concepts[15].display_name | DNA methylation |
| keywords[0].id | https://openalex.org/keywords/h3k4me3 |
| keywords[0].score | 0.7592811584472656 |
| keywords[0].display_name | H3K4me3 |
| keywords[1].id | https://openalex.org/keywords/hippo-signaling-pathway |
| keywords[1].score | 0.7583656311035156 |
| keywords[1].display_name | Hippo signaling pathway |
| keywords[2].id | https://openalex.org/keywords/histone |
| keywords[2].score | 0.709576427936554 |
| keywords[2].display_name | Histone |
| keywords[3].id | https://openalex.org/keywords/biology |
| keywords[3].score | 0.6490663290023804 |
| keywords[3].display_name | Biology |
| keywords[4].id | https://openalex.org/keywords/cell-biology |
| keywords[4].score | 0.6354193091392517 |
| keywords[4].display_name | Cell biology |
| keywords[5].id | https://openalex.org/keywords/histone-h3 |
| keywords[5].score | 0.5289364457130432 |
| keywords[5].display_name | Histone H3 |
| keywords[6].id | https://openalex.org/keywords/histone-code |
| keywords[6].score | 0.5017390251159668 |
| keywords[6].display_name | Histone code |
| keywords[7].id | https://openalex.org/keywords/histone-methylation |
| keywords[7].score | 0.5006923675537109 |
| keywords[7].display_name | Histone methylation |
| keywords[8].id | https://openalex.org/keywords/histone-methyltransferase |
| keywords[8].score | 0.4490509629249573 |
| keywords[8].display_name | Histone methyltransferase |
| keywords[9].id | https://openalex.org/keywords/genetics |
| keywords[9].score | 0.38138043880462646 |
| keywords[9].display_name | Genetics |
| keywords[10].id | https://openalex.org/keywords/gene |
| keywords[10].score | 0.235899418592453 |
| keywords[10].display_name | Gene |
| keywords[11].id | https://openalex.org/keywords/gene-expression |
| keywords[11].score | 0.17375102639198303 |
| keywords[11].display_name | Gene expression |
| keywords[12].id | https://openalex.org/keywords/signal-transduction |
| keywords[12].score | 0.17256596684455872 |
| keywords[12].display_name | Signal transduction |
| keywords[13].id | https://openalex.org/keywords/nucleosome |
| keywords[13].score | 0.14750754833221436 |
| keywords[13].display_name | Nucleosome |
| keywords[14].id | https://openalex.org/keywords/promoter |
| keywords[14].score | 0.1036422848701477 |
| keywords[14].display_name | Promoter |
| keywords[15].id | https://openalex.org/keywords/dna-methylation |
| keywords[15].score | 0.06431648135185242 |
| keywords[15].display_name | DNA methylation |
| language | en |
| locations[0].id | doi:10.52601/bpr.2021.200042 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4210200332 |
| locations[0].source.issn | 2364-3420, 2364-3439 |
| locations[0].source.type | journal |
| locations[0].source.is_oa | True |
| locations[0].source.issn_l | 2364-3420 |
| locations[0].source.is_core | True |
| locations[0].source.is_in_doaj | True |
| locations[0].source.display_name | Biophysics Reports |
| locations[0].source.host_organization | https://openalex.org/P4310319900 |
| locations[0].source.host_organization_name | Springer Science+Business Media |
| locations[0].source.host_organization_lineage | https://openalex.org/P4310319900, https://openalex.org/P4310319965 |
| locations[0].source.host_organization_lineage_names | Springer Science+Business Media, Springer Nature |
| locations[0].license | cc-by |
| locations[0].pdf_url | |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | Biophysics Reports |
| locations[0].landing_page_url | https://doi.org/10.52601/bpr.2021.200042 |
| locations[1].id | pmid:37288084 |
| locations[1].is_oa | False |
| locations[1].source.id | https://openalex.org/S4306525036 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | PubMed |
| locations[1].source.host_organization | https://openalex.org/I1299303238 |
| locations[1].source.host_organization_name | National Institutes of Health |
| locations[1].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[1].license | |
| locations[1].pdf_url | |
| locations[1].version | publishedVersion |
| locations[1].raw_type | |
| locations[1].license_id | |
| locations[1].is_accepted | True |
| locations[1].is_published | True |
| locations[1].raw_source_name | Biophysics reports |
| locations[1].landing_page_url | https://pubmed.ncbi.nlm.nih.gov/37288084 |
| locations[2].id | pmh:oai:pubmedcentral.nih.gov:10240540 |
| locations[2].is_oa | True |
| locations[2].source.id | https://openalex.org/S2764455111 |
| locations[2].source.issn | |
| locations[2].source.type | repository |
| locations[2].source.is_oa | False |
| locations[2].source.issn_l | |
| locations[2].source.is_core | False |
| locations[2].source.is_in_doaj | False |
| locations[2].source.display_name | PubMed Central |
| locations[2].source.host_organization | https://openalex.org/I1299303238 |
| locations[2].source.host_organization_name | National Institutes of Health |
| locations[2].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[2].license | cc-by |
| locations[2].pdf_url | https://pmc.ncbi.nlm.nih.gov/articles/PMC10240540/pdf/br-7-1-71.pdf |
| locations[2].version | submittedVersion |
| locations[2].raw_type | Text |
| locations[2].license_id | https://openalex.org/licenses/cc-by |
| locations[2].is_accepted | False |
| locations[2].is_published | False |
| locations[2].raw_source_name | Biophys Rep |
| locations[2].landing_page_url | https://www.ncbi.nlm.nih.gov/pmc/articles/10240540 |
| indexed_in | crossref, doaj, pubmed |
| authorships[0].author.id | https://openalex.org/A5040879046 |
| authorships[0].author.orcid | https://orcid.org/0000-0001-8850-4614 |
| authorships[0].author.display_name | Wenxia Su |
| authorships[0].countries | CN |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I120379545 |
| authorships[0].affiliations[0].raw_affiliation_string | College of Science, Inner Mongolia Agriculture University, Hohhot 010018, China. |
| authorships[0].institutions[0].id | https://openalex.org/I120379545 |
| authorships[0].institutions[0].ror | https://ror.org/015d0jq83 |
| authorships[0].institutions[0].type | education |
| authorships[0].institutions[0].lineage | https://openalex.org/I120379545 |
| authorships[0].institutions[0].country_code | CN |
| authorships[0].institutions[0].display_name | Inner Mongolia Agricultural University |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Wenxia Su |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | College of Science, Inner Mongolia Agriculture University, Hohhot 010018, China. |
| authorships[1].author.id | https://openalex.org/A5025194184 |
| authorships[1].author.orcid | |
| authorships[1].author.display_name | D Zhang |
| authorships[1].countries | CN |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I120379545 |
| authorships[1].affiliations[0].raw_affiliation_string | College of Science, Inner Mongolia Agriculture University, Hohhot 010018, China. |
| authorships[1].institutions[0].id | https://openalex.org/I120379545 |
| authorships[1].institutions[0].ror | https://ror.org/015d0jq83 |
| authorships[1].institutions[0].type | education |
| authorships[1].institutions[0].lineage | https://openalex.org/I120379545 |
| authorships[1].institutions[0].country_code | CN |
| authorships[1].institutions[0].display_name | Inner Mongolia Agricultural University |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Dimeng Zhang |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | College of Science, Inner Mongolia Agriculture University, Hohhot 010018, China. |
| authorships[2].author.id | https://openalex.org/A5100786373 |
| authorships[2].author.orcid | https://orcid.org/0000-0003-2996-8093 |
| authorships[2].author.display_name | Zhang Qiang |
| authorships[2].countries | CN |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I120379545 |
| authorships[2].affiliations[0].raw_affiliation_string | College of Science, Inner Mongolia Agriculture University, Hohhot 010018, China. |
| authorships[2].institutions[0].id | https://openalex.org/I120379545 |
| authorships[2].institutions[0].ror | https://ror.org/015d0jq83 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I120379545 |
| authorships[2].institutions[0].country_code | CN |
| authorships[2].institutions[0].display_name | Inner Mongolia Agricultural University |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Qiang Zhang |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | College of Science, Inner Mongolia Agriculture University, Hohhot 010018, China. |
| authorships[3].author.id | https://openalex.org/A5054199900 |
| authorships[3].author.orcid | |
| authorships[3].author.display_name | Qian‐Zhong Li |
| authorships[3].countries | CN |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I2722730 |
| authorships[3].affiliations[0].raw_affiliation_string | The State Key Laboratory of Reproductive Regulation and Breeding of Grassland Livestock, Inner Mongolia University, Hohhot 010070, China. |
| authorships[3].affiliations[1].institution_ids | https://openalex.org/I2722730 |
| authorships[3].affiliations[1].raw_affiliation_string | Laboratory of Theoretical Biophysics, School of Physical Science and Technology, Inner Mongolia University, Hohhot 010021, China. |
| authorships[3].institutions[0].id | https://openalex.org/I2722730 |
| authorships[3].institutions[0].ror | https://ror.org/0106qb496 |
| authorships[3].institutions[0].type | education |
| authorships[3].institutions[0].lineage | https://openalex.org/I2722730 |
| authorships[3].institutions[0].country_code | CN |
| authorships[3].institutions[0].display_name | Inner Mongolia University |
| authorships[3].author_position | last |
| authorships[3].raw_author_name | Qianzhong Li |
| authorships[3].is_corresponding | False |
| authorships[3].raw_affiliation_strings | Laboratory of Theoretical Biophysics, School of Physical Science and Technology, Inner Mongolia University, Hohhot 010021, China., The State Key Laboratory of Reproductive Regulation and Breeding of Grassland Livestock, Inner Mongolia University, Hohhot 010070, China. |
| has_content.pdf | False |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://doi.org/10.52601/bpr.2021.200042 |
| open_access.oa_status | diamond |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Study on the spatial distribution patterns of histone modifications in Hippo pathway genes |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T12065 |
| primary_topic.field.id | https://openalex.org/fields/13 |
| primary_topic.field.display_name | Biochemistry, Genetics and Molecular Biology |
| primary_topic.score | 1.0 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1307 |
| primary_topic.subfield.display_name | Cell Biology |
| primary_topic.display_name | Hippo pathway signaling and YAP/TAZ |
| related_works | https://openalex.org/W2056228735, https://openalex.org/W2772381364, https://openalex.org/W2033441226, https://openalex.org/W1982738044, https://openalex.org/W3168171974, https://openalex.org/W3082608639, https://openalex.org/W1998449423, https://openalex.org/W2782621646, https://openalex.org/W3091707841, https://openalex.org/W2059180978 |
| cited_by_count | 4 |
| counts_by_year[0].year | 2025 |
| counts_by_year[0].cited_by_count | 1 |
| counts_by_year[1].year | 2024 |
| counts_by_year[1].cited_by_count | 1 |
| counts_by_year[2].year | 2023 |
| counts_by_year[2].cited_by_count | 2 |
| locations_count | 3 |
| best_oa_location.id | doi:10.52601/bpr.2021.200042 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4210200332 |
| best_oa_location.source.issn | 2364-3420, 2364-3439 |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | True |
| best_oa_location.source.issn_l | 2364-3420 |
| best_oa_location.source.is_core | True |
| best_oa_location.source.is_in_doaj | True |
| best_oa_location.source.display_name | Biophysics Reports |
| best_oa_location.source.host_organization | https://openalex.org/P4310319900 |
| best_oa_location.source.host_organization_name | Springer Science+Business Media |
| best_oa_location.source.host_organization_lineage | https://openalex.org/P4310319900, https://openalex.org/P4310319965 |
| best_oa_location.source.host_organization_lineage_names | Springer Science+Business Media, Springer Nature |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | journal-article |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | Biophysics Reports |
| best_oa_location.landing_page_url | https://doi.org/10.52601/bpr.2021.200042 |
| primary_location.id | doi:10.52601/bpr.2021.200042 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4210200332 |
| primary_location.source.issn | 2364-3420, 2364-3439 |
| primary_location.source.type | journal |
| primary_location.source.is_oa | True |
| primary_location.source.issn_l | 2364-3420 |
| primary_location.source.is_core | True |
| primary_location.source.is_in_doaj | True |
| primary_location.source.display_name | Biophysics Reports |
| primary_location.source.host_organization | https://openalex.org/P4310319900 |
| primary_location.source.host_organization_name | Springer Science+Business Media |
| primary_location.source.host_organization_lineage | https://openalex.org/P4310319900, https://openalex.org/P4310319965 |
| primary_location.source.host_organization_lineage_names | Springer Science+Business Media, Springer Nature |
| primary_location.license | cc-by |
| primary_location.pdf_url | |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | Biophysics Reports |
| primary_location.landing_page_url | https://doi.org/10.52601/bpr.2021.200042 |
| publication_date | 2021-01-01 |
| publication_year | 2021 |
| referenced_works | https://openalex.org/W1967364632, https://openalex.org/W2914151360, https://openalex.org/W2495778796, https://openalex.org/W2166840595, https://openalex.org/W2782258386, https://openalex.org/W2050666087, https://openalex.org/W2064553485, https://openalex.org/W1732693222, https://openalex.org/W2064906046, https://openalex.org/W2135726634, https://openalex.org/W2582384997, https://openalex.org/W2150163671, https://openalex.org/W2099709490, https://openalex.org/W2171501785, https://openalex.org/W2082019843, https://openalex.org/W2103274342, https://openalex.org/W2114525085, https://openalex.org/W2132264180, https://openalex.org/W2119000216, https://openalex.org/W2006778392, https://openalex.org/W2085197940, https://openalex.org/W2187077810, https://openalex.org/W2125987139, https://openalex.org/W2159376228 |
| referenced_works_count | 24 |
| abstract_inverted_index.a | 124, 157 |
| abstract_inverted_index.19 | 54 |
| abstract_inverted_index.In | 51, 140 |
| abstract_inverted_index.It | 85 |
| abstract_inverted_index.To | 17 |
| abstract_inverted_index.by | 192 |
| abstract_inverted_index.in | 29, 70, 101, 138, 149 |
| abstract_inverted_index.is | 33 |
| abstract_inverted_index.of | 15, 22, 25, 41, 56, 82, 98, 104, 121, 130, 146, 165, 172, 182, 188 |
| abstract_inverted_index.on | 47 |
| abstract_inverted_index.to | 35, 76 |
| abstract_inverted_index.and | 7, 9, 13, 44, 64, 66, 94, 107, 109, 134, 175 |
| abstract_inverted_index.can | 2 |
| abstract_inverted_index.for | 161, 176 |
| abstract_inverted_index.new | 158 |
| abstract_inverted_index.the | 11, 19, 37, 78, 95, 102, 112, 118, 142, 163, 169, 178, 186, 194 |
| abstract_inverted_index.was | 86, 136 |
| abstract_inverted_index.MST1 | 63 |
| abstract_inverted_index.TAZ, | 61 |
| abstract_inverted_index.YAP, | 60 |
| abstract_inverted_index.YAP. | 139 |
| abstract_inverted_index.cell | 4, 49 |
| abstract_inverted_index.gene | 170 |
| abstract_inverted_index.most | 113 |
| abstract_inverted_index.size | 14 |
| abstract_inverted_index.stem | 31, 48, 72, 190 |
| abstract_inverted_index.that | 88 |
| abstract_inverted_index.this | 52 |
| abstract_inverted_index.type | 92 |
| abstract_inverted_index.were | 74, 90, 153 |
| abstract_inverted_index.Hippo | 0, 26, 45, 57, 150, 173, 195 |
| abstract_inverted_index.SAV1, | 65 |
| abstract_inverted_index.cells | 32, 73, 191 |
| abstract_inverted_index.eight | 67 |
| abstract_inverted_index.found | 87 |
| abstract_inverted_index.genes | 28, 55, 152 |
| abstract_inverted_index.shape | 12 |
| abstract_inverted_index.study | 18, 77 |
| abstract_inverted_index.there | 89 |
| abstract_inverted_index.which | 155 |
| abstract_inverted_index.H3K9ac | 133 |
| abstract_inverted_index.chosen | 75 |
| abstract_inverted_index.played | 111 |
| abstract_inverted_index.roles. | 116 |
| abstract_inverted_index.study, | 53 |
| abstract_inverted_index.target | 99 |
| abstract_inverted_index.H3K27ac | 135 |
| abstract_inverted_index.H3K4ac, | 131 |
| abstract_inverted_index.H3K4me3 | 108 |
| abstract_inverted_index.Through | 117 |
| abstract_inverted_index.cluster | 128 |
| abstract_inverted_index.control | 10 |
| abstract_inverted_index.helpful | 34 |
| abstract_inverted_index.histone | 23, 42, 68, 83, 105, 122, 125, 147, 166, 183 |
| abstract_inverted_index.obvious | 91 |
| abstract_inverted_index.organs. | 16 |
| abstract_inverted_index.pathway | 1, 27, 46, 58, 151 |
| abstract_inverted_index.regions | 100 |
| abstract_inverted_index.spatial | 79, 143 |
| abstract_inverted_index.H3K36me3 | 110 |
| abstract_inverted_index.H3K4me3, | 132 |
| abstract_inverted_index.LATS1/2, | 62 |
| abstract_inverted_index.analysis | 120 |
| abstract_inverted_index.composed | 129 |
| abstract_inverted_index.detected | 137 |
| abstract_inverted_index.location | 96 |
| abstract_inverted_index.pathway, | 174 |
| abstract_inverted_index.pathway. | 196 |
| abstract_inverted_index.patterns | 21, 81, 145 |
| abstract_inverted_index.provided | 156 |
| abstract_inverted_index.regulate | 3 |
| abstract_inverted_index.addition, | 141 |
| abstract_inverted_index.affecting | 185 |
| abstract_inverted_index.division, | 5 |
| abstract_inverted_index.embryonic | 30, 71, 189 |
| abstract_inverted_index.important | 114 |
| abstract_inverted_index.including | 59 |
| abstract_inverted_index.mechanism | 40, 164, 181 |
| abstract_inverted_index.molecular | 38, 179 |
| abstract_inverted_index.obtained, | 154 |
| abstract_inverted_index.reference | 160 |
| abstract_inverted_index.revealing | 177 |
| abstract_inverted_index.apoptosis, | 8 |
| abstract_inverted_index.expression | 171 |
| abstract_inverted_index.functional | 127 |
| abstract_inverted_index.preference | 97 |
| abstract_inverted_index.regulating | 168, 193 |
| abstract_inverted_index.regulation | 39 |
| abstract_inverted_index.regulatory | 115, 180 |
| abstract_inverted_index.understand | 36 |
| abstract_inverted_index.correlation | 119 |
| abstract_inverted_index.elucidating | 162 |
| abstract_inverted_index.specificity | 93 |
| abstract_inverted_index.theoretical | 159 |
| abstract_inverted_index.distribution | 20, 80, 144 |
| abstract_inverted_index.modification | 43, 126 |
| abstract_inverted_index.self-renewal | 187 |
| abstract_inverted_index.distributions | 103 |
| abstract_inverted_index.modifications | 24, 69, 148, 167, 184 |
| abstract_inverted_index.self-renewal. | 50 |
| abstract_inverted_index.modifications, | 106, 123 |
| abstract_inverted_index.modifications. | 84 |
| abstract_inverted_index.differentiation | 6 |
| cited_by_percentile_year.max | 96 |
| cited_by_percentile_year.min | 90 |
| countries_distinct_count | 1 |
| institutions_distinct_count | 4 |
| citation_normalized_percentile.value | 0.53048638 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |