Supplementary Figure 6 from Complex Tumor Genomes Inferred from Single Circulating Tumor Cells by Array-CGH and Next-Generation Sequencing Article Swipe
Ellen Heitzer
,
Martina Auer
,
Christin Gasch
,
Martin Pichler
,
Peter Ulz
,
Eva Maria Hoffmann
,
Sigurd Lax
,
Julie Waldispuehl‐Geigl
,
Oliver Mauermann
,
Carolin Lackner
,
Gerald Höfler
,
Florian Eisner
,
Heinz Sill
,
Hellmut Samonigg
,
Klaus Pantel
,
Sabine Riethdorf
,
Thomas Bauernhofer
,
Jochen B. Geigl
,
Michael R. Speicher
·
 YOU?
·
· 2023
· Open Access
·
· DOI: https://doi.org/10.1158/0008-5472.22398303.v1
YOU?
·
· 2023
· Open Access
·
· DOI: https://doi.org/10.1158/0008-5472.22398303.v1
 YOU?
·
· 2023
· Open Access
·
· DOI: https://doi.org/10.1158/0008-5472.22398303.v1
YOU?
·
· 2023
· Open Access
·
· DOI: https://doi.org/10.1158/0008-5472.22398303.v1
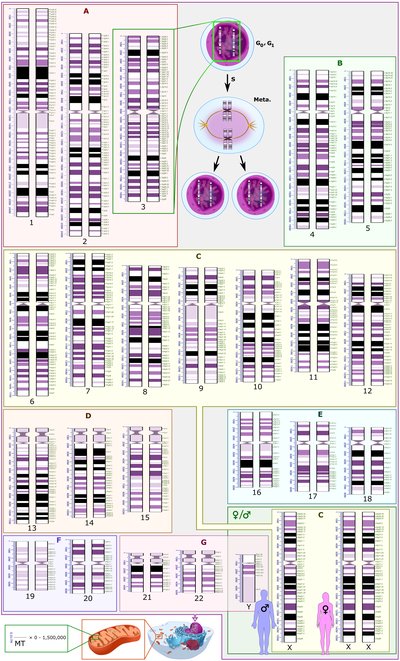
PDF file - 188K, Sequencing of UCR41 and flanking regions: UCR41 is a 217-bp-long ultra-conserved region depicted in dark green. The gray bar indicates the extremely conserved sequence. The region is flanked by two SNPs that are frequent in the European population (blue arrows). The black curve indicates the mutability, which is higher in non-conserved regions compared with conserved segments. The black bars illustrate the 5 overlapping amplicons used for amplification. Mutations identified in our samples are indicated with red arrows. The genomic coordinates refer to the hg18 assembly of the human genome. (The graph is reproduced with permission from De Grassi et al.)
Related Topics
Concepts
Amplicon
Biology
Genetics
Genome
Conserved sequence
DNA sequencing
Human genome
Computational biology
Evolutionary biology
DNA
Gene
Base sequence
Polymerase chain reaction
Metadata
- Type
- preprint
- Language
- en
- Landing Page
- https://doi.org/10.1158/0008-5472.22398303.v1
- https://aacr.figshare.com/articles/journal_contribution/Supplementary_Figure_6_from_Complex_Tumor_Genomes_Inferred_from_Single_Circulating_Tumor_Cells_by_Array-CGH_and_Next-Generation_Sequencing/22398303/1/files/39844017.pdf
- OA Status
- gold
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4362164172
All OpenAlex metadata
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4362164172Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1158/0008-5472.22398303.v1Digital Object Identifier
- Title
-
Supplementary Figure 6 from Complex Tumor Genomes Inferred from Single Circulating Tumor Cells by Array-CGH and Next-Generation SequencingWork title
- Type
-
preprintOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2023Year of publication
- Publication date
-
2023-03-30Full publication date if available
- Authors
-
Ellen Heitzer, Martina Auer, Christin Gasch, Martin Pichler, Peter Ulz, Eva Maria Hoffmann, Sigurd Lax, Julie Waldispuehl‐Geigl, Oliver Mauermann, Carolin Lackner, Gerald Höfler, Florian Eisner, Heinz Sill, Hellmut Samonigg, Klaus Pantel, Sabine Riethdorf, Thomas Bauernhofer, Jochen B. Geigl, Michael R. SpeicherList of authors in order
- Landing page
-
https://doi.org/10.1158/0008-5472.22398303.v1Publisher landing page
- PDF URL
-
https://aacr.figshare.com/articles/journal_contribution/Supplementary_Figure_6_from_Complex_Tumor_Genomes_Inferred_from_Single_Circulating_Tumor_Cells_by_Array-CGH_and_Next-Generation_Sequencing/22398303/1/files/39844017.pdfDirect link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
goldOpen access status per OpenAlex
- OA URL
-
https://aacr.figshare.com/articles/journal_contribution/Supplementary_Figure_6_from_Complex_Tumor_Genomes_Inferred_from_Single_Circulating_Tumor_Cells_by_Array-CGH_and_Next-Generation_Sequencing/22398303/1/files/39844017.pdfDirect OA link when available
- Concepts
-
Amplicon, Biology, Genetics, Genome, Conserved sequence, DNA sequencing, Human genome, Computational biology, Evolutionary biology, DNA, Gene, Base sequence, Polymerase chain reactionTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
0Total citation count in OpenAlex
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4362164172 |
|---|---|
| doi | https://doi.org/10.1158/0008-5472.22398303.v1 |
| ids.doi | https://doi.org/10.1158/0008-5472.22398303.v1 |
| ids.openalex | https://openalex.org/W4362164172 |
| fwci | 0.0 |
| type | preprint |
| title | Supplementary Figure 6 from Complex Tumor Genomes Inferred from Single Circulating Tumor Cells by Array-CGH and Next-Generation Sequencing |
| biblio.issue | |
| biblio.volume | |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T11287 |
| topics[0].field.id | https://openalex.org/fields/13 |
| topics[0].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[0].score | 0.9624999761581421 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1306 |
| topics[0].subfield.display_name | Cancer Research |
| topics[0].display_name | Cancer Genomics and Diagnostics |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C8185291 |
| concepts[0].level | 4 |
| concepts[0].score | 0.6223672032356262 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q2844457 |
| concepts[0].display_name | Amplicon |
| concepts[1].id | https://openalex.org/C86803240 |
| concepts[1].level | 0 |
| concepts[1].score | 0.5835211277008057 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[1].display_name | Biology |
| concepts[2].id | https://openalex.org/C54355233 |
| concepts[2].level | 1 |
| concepts[2].score | 0.5660140514373779 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[2].display_name | Genetics |
| concepts[3].id | https://openalex.org/C141231307 |
| concepts[3].level | 3 |
| concepts[3].score | 0.5611206293106079 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q7020 |
| concepts[3].display_name | Genome |
| concepts[4].id | https://openalex.org/C199216141 |
| concepts[4].level | 4 |
| concepts[4].score | 0.5257809162139893 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q4995178 |
| concepts[4].display_name | Conserved sequence |
| concepts[5].id | https://openalex.org/C51679486 |
| concepts[5].level | 3 |
| concepts[5].score | 0.4947035014629364 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q380546 |
| concepts[5].display_name | DNA sequencing |
| concepts[6].id | https://openalex.org/C197077220 |
| concepts[6].level | 4 |
| concepts[6].score | 0.4581114649772644 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q720988 |
| concepts[6].display_name | Human genome |
| concepts[7].id | https://openalex.org/C70721500 |
| concepts[7].level | 1 |
| concepts[7].score | 0.42319613695144653 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q177005 |
| concepts[7].display_name | Computational biology |
| concepts[8].id | https://openalex.org/C78458016 |
| concepts[8].level | 1 |
| concepts[8].score | 0.35422319173812866 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q840400 |
| concepts[8].display_name | Evolutionary biology |
| concepts[9].id | https://openalex.org/C552990157 |
| concepts[9].level | 2 |
| concepts[9].score | 0.2948629856109619 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q7430 |
| concepts[9].display_name | DNA |
| concepts[10].id | https://openalex.org/C104317684 |
| concepts[10].level | 2 |
| concepts[10].score | 0.2826291024684906 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[10].display_name | Gene |
| concepts[11].id | https://openalex.org/C3017666073 |
| concepts[11].level | 3 |
| concepts[11].score | 0.1027260422706604 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q1764062 |
| concepts[11].display_name | Base sequence |
| concepts[12].id | https://openalex.org/C49105822 |
| concepts[12].level | 3 |
| concepts[12].score | 0.099831223487854 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q176996 |
| concepts[12].display_name | Polymerase chain reaction |
| keywords[0].id | https://openalex.org/keywords/amplicon |
| keywords[0].score | 0.6223672032356262 |
| keywords[0].display_name | Amplicon |
| keywords[1].id | https://openalex.org/keywords/biology |
| keywords[1].score | 0.5835211277008057 |
| keywords[1].display_name | Biology |
| keywords[2].id | https://openalex.org/keywords/genetics |
| keywords[2].score | 0.5660140514373779 |
| keywords[2].display_name | Genetics |
| keywords[3].id | https://openalex.org/keywords/genome |
| keywords[3].score | 0.5611206293106079 |
| keywords[3].display_name | Genome |
| keywords[4].id | https://openalex.org/keywords/conserved-sequence |
| keywords[4].score | 0.5257809162139893 |
| keywords[4].display_name | Conserved sequence |
| keywords[5].id | https://openalex.org/keywords/dna-sequencing |
| keywords[5].score | 0.4947035014629364 |
| keywords[5].display_name | DNA sequencing |
| keywords[6].id | https://openalex.org/keywords/human-genome |
| keywords[6].score | 0.4581114649772644 |
| keywords[6].display_name | Human genome |
| keywords[7].id | https://openalex.org/keywords/computational-biology |
| keywords[7].score | 0.42319613695144653 |
| keywords[7].display_name | Computational biology |
| keywords[8].id | https://openalex.org/keywords/evolutionary-biology |
| keywords[8].score | 0.35422319173812866 |
| keywords[8].display_name | Evolutionary biology |
| keywords[9].id | https://openalex.org/keywords/dna |
| keywords[9].score | 0.2948629856109619 |
| keywords[9].display_name | DNA |
| keywords[10].id | https://openalex.org/keywords/gene |
| keywords[10].score | 0.2826291024684906 |
| keywords[10].display_name | Gene |
| keywords[11].id | https://openalex.org/keywords/base-sequence |
| keywords[11].score | 0.1027260422706604 |
| keywords[11].display_name | Base sequence |
| keywords[12].id | https://openalex.org/keywords/polymerase-chain-reaction |
| keywords[12].score | 0.099831223487854 |
| keywords[12].display_name | Polymerase chain reaction |
| language | en |
| locations[0].id | doi:10.1158/0008-5472.22398303.v1 |
| locations[0].is_oa | True |
| locations[0].source | |
| locations[0].license | cc-by |
| locations[0].pdf_url | https://aacr.figshare.com/articles/journal_contribution/Supplementary_Figure_6_from_Complex_Tumor_Genomes_Inferred_from_Single_Circulating_Tumor_Cells_by_Array-CGH_and_Next-Generation_Sequencing/22398303/1/files/39844017.pdf |
| locations[0].version | acceptedVersion |
| locations[0].raw_type | posted-content |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | True |
| locations[0].is_published | False |
| locations[0].raw_source_name | |
| locations[0].landing_page_url | https://doi.org/10.1158/0008-5472.22398303.v1 |
| locations[1].id | pmh:oai:figshare.com:article/22398303 |
| locations[1].is_oa | True |
| locations[1].source.id | https://openalex.org/S4306400572 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | OPAL (Open@LaTrobe) (La Trobe University) |
| locations[1].source.host_organization | https://openalex.org/I196829312 |
| locations[1].source.host_organization_name | La Trobe University |
| locations[1].source.host_organization_lineage | https://openalex.org/I196829312 |
| locations[1].license | cc-by |
| locations[1].pdf_url | |
| locations[1].version | submittedVersion |
| locations[1].raw_type | Text |
| locations[1].license_id | https://openalex.org/licenses/cc-by |
| locations[1].is_accepted | False |
| locations[1].is_published | False |
| locations[1].raw_source_name | |
| locations[1].landing_page_url | https://figshare.com/articles/journal_contribution/Supplementary_Figure_6_from_Complex_Tumor_Genomes_Inferred_from_Single_Circulating_Tumor_Cells_by_Array-CGH_and_Next-Generation_Sequencing/22398303 |
| indexed_in | crossref |
| authorships[0].author.id | https://openalex.org/A5069144652 |
| authorships[0].author.orcid | https://orcid.org/0000-0002-8815-7859 |
| authorships[0].author.display_name | Ellen Heitzer |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Ellen Heitzer |
| authorships[0].is_corresponding | False |
| authorships[1].author.id | https://openalex.org/A5060340775 |
| authorships[1].author.orcid | |
| authorships[1].author.display_name | Martina Auer |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Martina Auer |
| authorships[1].is_corresponding | False |
| authorships[2].author.id | https://openalex.org/A5030787310 |
| authorships[2].author.orcid | |
| authorships[2].author.display_name | Christin Gasch |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Christin Gasch |
| authorships[2].is_corresponding | False |
| authorships[3].author.id | https://openalex.org/A5006483196 |
| authorships[3].author.orcid | https://orcid.org/0000-0002-8701-9462 |
| authorships[3].author.display_name | Martin Pichler |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Martin Pichler |
| authorships[3].is_corresponding | False |
| authorships[4].author.id | https://openalex.org/A5018666322 |
| authorships[4].author.orcid | https://orcid.org/0000-0001-5941-1160 |
| authorships[4].author.display_name | Peter Ulz |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Peter Ulz |
| authorships[4].is_corresponding | False |
| authorships[5].author.id | https://openalex.org/A5043122569 |
| authorships[5].author.orcid | |
| authorships[5].author.display_name | Eva Maria Hoffmann |
| authorships[5].author_position | middle |
| authorships[5].raw_author_name | Eva Maria Hoffmann |
| authorships[5].is_corresponding | False |
| authorships[6].author.id | https://openalex.org/A5054024247 |
| authorships[6].author.orcid | https://orcid.org/0000-0002-3576-601X |
| authorships[6].author.display_name | Sigurd Lax |
| authorships[6].author_position | middle |
| authorships[6].raw_author_name | Sigurd Lax |
| authorships[6].is_corresponding | False |
| authorships[7].author.id | https://openalex.org/A5035464450 |
| authorships[7].author.orcid | |
| authorships[7].author.display_name | Julie Waldispuehl‐Geigl |
| authorships[7].author_position | middle |
| authorships[7].raw_author_name | Julie Waldispuehl-Geigl |
| authorships[7].is_corresponding | False |
| authorships[8].author.id | https://openalex.org/A5079558854 |
| authorships[8].author.orcid | |
| authorships[8].author.display_name | Oliver Mauermann |
| authorships[8].author_position | middle |
| authorships[8].raw_author_name | Oliver Mauermann |
| authorships[8].is_corresponding | False |
| authorships[9].author.id | https://openalex.org/A5062226170 |
| authorships[9].author.orcid | https://orcid.org/0000-0002-9809-4388 |
| authorships[9].author.display_name | Carolin Lackner |
| authorships[9].author_position | middle |
| authorships[9].raw_author_name | Carolin Lackner |
| authorships[9].is_corresponding | False |
| authorships[10].author.id | https://openalex.org/A5071820041 |
| authorships[10].author.orcid | |
| authorships[10].author.display_name | Gerald Höfler |
| authorships[10].author_position | middle |
| authorships[10].raw_author_name | Gerald Höfler |
| authorships[10].is_corresponding | False |
| authorships[11].author.id | https://openalex.org/A5068348685 |
| authorships[11].author.orcid | |
| authorships[11].author.display_name | Florian Eisner |
| authorships[11].author_position | middle |
| authorships[11].raw_author_name | Florian Eisner |
| authorships[11].is_corresponding | False |
| authorships[12].author.id | https://openalex.org/A5073135548 |
| authorships[12].author.orcid | https://orcid.org/0000-0003-0993-4371 |
| authorships[12].author.display_name | Heinz Sill |
| authorships[12].author_position | middle |
| authorships[12].raw_author_name | Heinz Sill |
| authorships[12].is_corresponding | False |
| authorships[13].author.id | https://openalex.org/A5105193795 |
| authorships[13].author.orcid | |
| authorships[13].author.display_name | Hellmut Samonigg |
| authorships[13].author_position | middle |
| authorships[13].raw_author_name | Hellmut Samonigg |
| authorships[13].is_corresponding | False |
| authorships[14].author.id | https://openalex.org/A5048105827 |
| authorships[14].author.orcid | https://orcid.org/0000-0001-5736-2772 |
| authorships[14].author.display_name | Klaus Pantel |
| authorships[14].author_position | middle |
| authorships[14].raw_author_name | Klaus Pantel |
| authorships[14].is_corresponding | False |
| authorships[15].author.id | https://openalex.org/A5078288046 |
| authorships[15].author.orcid | https://orcid.org/0000-0003-0028-5643 |
| authorships[15].author.display_name | Sabine Riethdorf |
| authorships[15].author_position | middle |
| authorships[15].raw_author_name | Sabine Riethdorf |
| authorships[15].is_corresponding | False |
| authorships[16].author.id | https://openalex.org/A5046917464 |
| authorships[16].author.orcid | https://orcid.org/0000-0001-9124-4876 |
| authorships[16].author.display_name | Thomas Bauernhofer |
| authorships[16].author_position | middle |
| authorships[16].raw_author_name | Thomas Bauernhofer |
| authorships[16].is_corresponding | False |
| authorships[17].author.id | https://openalex.org/A5060464938 |
| authorships[17].author.orcid | https://orcid.org/0000-0001-9160-0682 |
| authorships[17].author.display_name | Jochen B. Geigl |
| authorships[17].author_position | middle |
| authorships[17].raw_author_name | Jochen B. Geigl |
| authorships[17].is_corresponding | False |
| authorships[18].author.id | https://openalex.org/A5062402346 |
| authorships[18].author.orcid | https://orcid.org/0000-0003-0105-955X |
| authorships[18].author.display_name | Michael R. Speicher |
| authorships[18].author_position | last |
| authorships[18].raw_author_name | Michael R. Speicher |
| authorships[18].is_corresponding | False |
| has_content.pdf | True |
| has_content.grobid_xml | True |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://aacr.figshare.com/articles/journal_contribution/Supplementary_Figure_6_from_Complex_Tumor_Genomes_Inferred_from_Single_Circulating_Tumor_Cells_by_Array-CGH_and_Next-Generation_Sequencing/22398303/1/files/39844017.pdf |
| open_access.oa_status | gold |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Supplementary Figure 6 from Complex Tumor Genomes Inferred from Single Circulating Tumor Cells by Array-CGH and Next-Generation Sequencing |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T11287 |
| primary_topic.field.id | https://openalex.org/fields/13 |
| primary_topic.field.display_name | Biochemistry, Genetics and Molecular Biology |
| primary_topic.score | 0.9624999761581421 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1306 |
| primary_topic.subfield.display_name | Cancer Research |
| primary_topic.display_name | Cancer Genomics and Diagnostics |
| related_works | https://openalex.org/W2995118009, https://openalex.org/W2786748503, https://openalex.org/W2011298649, https://openalex.org/W2036617847, https://openalex.org/W3088281179, https://openalex.org/W1966203981, https://openalex.org/W2898640786, https://openalex.org/W1969895229, https://openalex.org/W2108497208, https://openalex.org/W2122609286 |
| cited_by_count | 0 |
| locations_count | 2 |
| best_oa_location.id | doi:10.1158/0008-5472.22398303.v1 |
| best_oa_location.is_oa | True |
| best_oa_location.source | |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | https://aacr.figshare.com/articles/journal_contribution/Supplementary_Figure_6_from_Complex_Tumor_Genomes_Inferred_from_Single_Circulating_Tumor_Cells_by_Array-CGH_and_Next-Generation_Sequencing/22398303/1/files/39844017.pdf |
| best_oa_location.version | acceptedVersion |
| best_oa_location.raw_type | posted-content |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | False |
| best_oa_location.raw_source_name | |
| best_oa_location.landing_page_url | https://doi.org/10.1158/0008-5472.22398303.v1 |
| primary_location.id | doi:10.1158/0008-5472.22398303.v1 |
| primary_location.is_oa | True |
| primary_location.source | |
| primary_location.license | cc-by |
| primary_location.pdf_url | https://aacr.figshare.com/articles/journal_contribution/Supplementary_Figure_6_from_Complex_Tumor_Genomes_Inferred_from_Single_Circulating_Tumor_Cells_by_Array-CGH_and_Next-Generation_Sequencing/22398303/1/files/39844017.pdf |
| primary_location.version | acceptedVersion |
| primary_location.raw_type | posted-content |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | True |
| primary_location.is_published | False |
| primary_location.raw_source_name | |
| primary_location.landing_page_url | https://doi.org/10.1158/0008-5472.22398303.v1 |
| publication_date | 2023-03-30 |
| publication_year | 2023 |
| referenced_works_count | 0 |
| abstract_inverted_index.- | 2 |
| abstract_inverted_index.5 | 65 |
| abstract_inverted_index.a | 12 |
| abstract_inverted_index.De | 100 |
| abstract_inverted_index.by | 32 |
| abstract_inverted_index.et | 102 |
| abstract_inverted_index.in | 17, 38, 53, 73 |
| abstract_inverted_index.is | 11, 30, 51, 95 |
| abstract_inverted_index.of | 5, 89 |
| abstract_inverted_index.to | 85 |
| abstract_inverted_index.The | 20, 28, 44, 60, 81 |
| abstract_inverted_index.and | 7 |
| abstract_inverted_index.are | 36, 76 |
| abstract_inverted_index.bar | 22 |
| abstract_inverted_index.for | 69 |
| abstract_inverted_index.our | 74 |
| abstract_inverted_index.red | 79 |
| abstract_inverted_index.the | 24, 39, 48, 64, 86, 90 |
| abstract_inverted_index.two | 33 |
| abstract_inverted_index.(The | 93 |
| abstract_inverted_index.SNPs | 34 |
| abstract_inverted_index.bars | 62 |
| abstract_inverted_index.dark | 18 |
| abstract_inverted_index.file | 1 |
| abstract_inverted_index.from | 99 |
| abstract_inverted_index.gray | 21 |
| abstract_inverted_index.hg18 | 87 |
| abstract_inverted_index.that | 35 |
| abstract_inverted_index.used | 68 |
| abstract_inverted_index.with | 57, 78, 97 |
| abstract_inverted_index.(blue | 42 |
| abstract_inverted_index.188K, | 3 |
| abstract_inverted_index.UCR41 | 6, 10 |
| abstract_inverted_index.black | 45, 61 |
| abstract_inverted_index.curve | 46 |
| abstract_inverted_index.graph | 94 |
| abstract_inverted_index.human | 91 |
| abstract_inverted_index.refer | 84 |
| abstract_inverted_index.which | 50 |
| abstract_inverted_index.Grassi | 101 |
| abstract_inverted_index.green. | 19 |
| abstract_inverted_index.higher | 52 |
| abstract_inverted_index.region | 15, 29 |
| abstract_inverted_index.arrows. | 80 |
| abstract_inverted_index.flanked | 31 |
| abstract_inverted_index.genome. | 92 |
| abstract_inverted_index.genomic | 82 |
| abstract_inverted_index.regions | 55 |
| abstract_inverted_index.samples | 75 |
| abstract_inverted_index.European | 40 |
| abstract_inverted_index.arrows). | 43 |
| abstract_inverted_index.assembly | 88 |
| abstract_inverted_index.compared | 56 |
| abstract_inverted_index.depicted | 16 |
| abstract_inverted_index.flanking | 8 |
| abstract_inverted_index.frequent | 37 |
| abstract_inverted_index.regions: | 9 |
| abstract_inverted_index.Mutations | 71 |
| abstract_inverted_index.amplicons | 67 |
| abstract_inverted_index.conserved | 26, 58 |
| abstract_inverted_index.extremely | 25 |
| abstract_inverted_index.indicated | 77 |
| abstract_inverted_index.indicates | 23, 47 |
| abstract_inverted_index.segments. | 59 |
| abstract_inverted_index.sequence. | 27 |
| abstract_inverted_index.Sequencing | 4 |
| abstract_inverted_index.identified | 72 |
| abstract_inverted_index.illustrate | 63 |
| abstract_inverted_index.permission | 98 |
| abstract_inverted_index.population | 41 |
| abstract_inverted_index.reproduced | 96 |
| abstract_inverted_index.217-bp-long | 13 |
| abstract_inverted_index.coordinates | 83 |
| abstract_inverted_index.mutability, | 49 |
| abstract_inverted_index.overlapping | 66 |
| abstract_inverted_index.<p>PDF | 0 |
| abstract_inverted_index.non-conserved | 54 |
| abstract_inverted_index.al.)</p> | 103 |
| abstract_inverted_index.amplification. | 70 |
| abstract_inverted_index.ultra-conserved | 14 |
| cited_by_percentile_year | |
| countries_distinct_count | 0 |
| institutions_distinct_count | 19 |
| citation_normalized_percentile.value | 0.24528755 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |