SynActJ: Easy-to-Use Automated Analysis of Synaptic Activity Article Swipe
 YOU?
·
· 2021
· Open Access
·
· DOI: https://doi.org/10.3389/fcomp.2021.777837
YOU?
·
· 2021
· Open Access
·
· DOI: https://doi.org/10.3389/fcomp.2021.777837
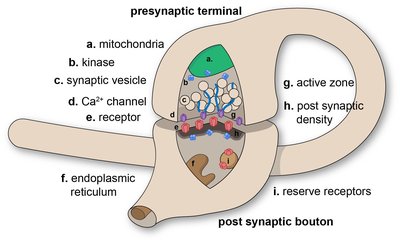
Neuronal synapses are highly dynamic communication hubs that mediate chemical neurotransmission via the exocytic fusion and subsequent endocytic recycling of neurotransmitter-containing synaptic vesicles (SVs). Functional imaging tools allow for the direct visualization of synaptic activity by detecting action potentials, pre- or postsynaptic calcium influx, SV exo- and endocytosis, and glutamate release. Fluorescent organic dyes or synapse-targeted genetic molecular reporters, such as calcium, voltage or neurotransmitter sensors and synapto-pHluorins reveal synaptic activity by undergoing rapid changes in their fluorescence intensity upon neuronal activity on timescales of milliseconds to seconds, which typically are recorded by fast and sensitive widefield live cell microscopy. The analysis of the resulting time-lapse movies in the past has been performed by either manually picking individual structures, custom scripts that have not been made widely available to the scientific community, or advanced software toolboxes that are complicated to use. For the precise, unbiased and reproducible measurement of synaptic activity, it is key that the research community has access to bio-image analysis tools that are easy-to-apply and allow the automated detection of fluorescent intensity changes in active synapses. Here we present SynActJ (Synaptic Activity in ImageJ), an easy-to-use fully open-source workflow that enables automated image and data analysis of synaptic activity. The workflow consists of a Fiji plugin performing the automated image analysis of active synapses in time-lapse movies via an interactive seeded watershed segmentation that can be easily adjusted and applied to a dataset in batch mode. The extracted intensity traces of each synaptic bouton are automatically processed, analyzed, and plotted using an R Shiny workflow. We validate the workflow on time-lapse images of stimulated synapses expressing the SV exo-/endocytosis reporter Synaptophysin-pHluorin or a synapse-targeted calcium sensor, Synaptophysin-RGECO. We compare the automatic workflow to manual analysis and compute calcium-influx and SV exo-/endocytosis kinetics and other parameters for synaptic vesicle recycling under different conditions. We predict SynActJ to become an important tool for the analysis of synaptic activity and synapse properties.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.3389/fcomp.2021.777837
- OA Status
- gold
- Cited By
- 18
- References
- 25
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4200484069
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4200484069Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.3389/fcomp.2021.777837Digital Object Identifier
- Title
-
SynActJ: Easy-to-Use Automated Analysis of Synaptic ActivityWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2021Year of publication
- Publication date
-
2021-12-09Full publication date if available
- Authors
-
Christopher Schmied, Tolga Soykan, Svenja Bolz, Volker Haucke, Martin LehmannList of authors in order
- Landing page
-
https://doi.org/10.3389/fcomp.2021.777837Publisher landing page
- Open access
-
YesWhether a free full text is available
- OA status
-
goldOpen access status per OpenAlex
- OA URL
-
https://doi.org/10.3389/fcomp.2021.777837Direct OA link when available
- Concepts
-
Calcium imaging, Neurotransmission, Synapse, Synaptic vesicle, Computer science, Calcium signaling, Neurotransmitter, Postsynaptic potential, Neuroscience, Chemistry, Biology, Calcium, Vesicle, Receptor, Central nervous system, Membrane, Organic chemistry, BiochemistryTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
18Total citation count in OpenAlex
- Citations by year (recent)
-
2025: 3, 2024: 7, 2023: 8Per-year citation counts (last 5 years)
- References (count)
-
25Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4200484069 |
|---|---|
| doi | https://doi.org/10.3389/fcomp.2021.777837 |
| ids.doi | https://doi.org/10.3389/fcomp.2021.777837 |
| ids.openalex | https://openalex.org/W4200484069 |
| fwci | 2.18433303 |
| type | article |
| title | SynActJ: Easy-to-Use Automated Analysis of Synaptic Activity |
| awards[0].id | https://openalex.org/G5570323996 |
| awards[0].funder_id | https://openalex.org/F4320320879 |
| awards[0].display_name | |
| awards[0].funder_award_id | HA2686/20-1 to V.H. |
| awards[0].funder_display_name | Deutsche Forschungsgemeinschaft |
| biblio.issue | |
| biblio.volume | 3 |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T12859 |
| topics[0].field.id | https://openalex.org/fields/13 |
| topics[0].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[0].score | 0.9987999796867371 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1304 |
| topics[0].subfield.display_name | Biophysics |
| topics[0].display_name | Cell Image Analysis Techniques |
| topics[1].id | https://openalex.org/T10540 |
| topics[1].field.id | https://openalex.org/fields/13 |
| topics[1].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[1].score | 0.9983000159263611 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1304 |
| topics[1].subfield.display_name | Biophysics |
| topics[1].display_name | Advanced Fluorescence Microscopy Techniques |
| topics[2].id | https://openalex.org/T10407 |
| topics[2].field.id | https://openalex.org/fields/13 |
| topics[2].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[2].score | 0.9918000102043152 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1312 |
| topics[2].subfield.display_name | Molecular Biology |
| topics[2].display_name | Lipid Membrane Structure and Behavior |
| funders[0].id | https://openalex.org/F4320320879 |
| funders[0].ror | https://ror.org/018mejw64 |
| funders[0].display_name | Deutsche Forschungsgemeinschaft |
| is_xpac | False |
| apc_list.value | 1150 |
| apc_list.currency | USD |
| apc_list.value_usd | 1150 |
| apc_paid.value | 1150 |
| apc_paid.currency | USD |
| apc_paid.value_usd | 1150 |
| concepts[0].id | https://openalex.org/C2776032975 |
| concepts[0].level | 3 |
| concepts[0].score | 0.5625597834587097 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q5018833 |
| concepts[0].display_name | Calcium imaging |
| concepts[1].id | https://openalex.org/C200170125 |
| concepts[1].level | 3 |
| concepts[1].score | 0.5466105341911316 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q969316 |
| concepts[1].display_name | Neurotransmission |
| concepts[2].id | https://openalex.org/C127445978 |
| concepts[2].level | 2 |
| concepts[2].score | 0.5079205632209778 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q187181 |
| concepts[2].display_name | Synapse |
| concepts[3].id | https://openalex.org/C148785051 |
| concepts[3].level | 4 |
| concepts[3].score | 0.48312780261039734 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q671136 |
| concepts[3].display_name | Synaptic vesicle |
| concepts[4].id | https://openalex.org/C41008148 |
| concepts[4].level | 0 |
| concepts[4].score | 0.4684583842754364 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q21198 |
| concepts[4].display_name | Computer science |
| concepts[5].id | https://openalex.org/C201571599 |
| concepts[5].level | 3 |
| concepts[5].score | 0.4222016930580139 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q5018840 |
| concepts[5].display_name | Calcium signaling |
| concepts[6].id | https://openalex.org/C2776219046 |
| concepts[6].level | 3 |
| concepts[6].score | 0.4210626184940338 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q162657 |
| concepts[6].display_name | Neurotransmitter |
| concepts[7].id | https://openalex.org/C197341189 |
| concepts[7].level | 3 |
| concepts[7].score | 0.41334670782089233 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q863533 |
| concepts[7].display_name | Postsynaptic potential |
| concepts[8].id | https://openalex.org/C169760540 |
| concepts[8].level | 1 |
| concepts[8].score | 0.4117390215396881 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q207011 |
| concepts[8].display_name | Neuroscience |
| concepts[9].id | https://openalex.org/C185592680 |
| concepts[9].level | 0 |
| concepts[9].score | 0.27440303564071655 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q2329 |
| concepts[9].display_name | Chemistry |
| concepts[10].id | https://openalex.org/C86803240 |
| concepts[10].level | 0 |
| concepts[10].score | 0.2701937258243561 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[10].display_name | Biology |
| concepts[11].id | https://openalex.org/C519063684 |
| concepts[11].level | 2 |
| concepts[11].score | 0.23473966121673584 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q706 |
| concepts[11].display_name | Calcium |
| concepts[12].id | https://openalex.org/C130316041 |
| concepts[12].level | 3 |
| concepts[12].score | 0.09622368216514587 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q189206 |
| concepts[12].display_name | Vesicle |
| concepts[13].id | https://openalex.org/C170493617 |
| concepts[13].level | 2 |
| concepts[13].score | 0.0 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q208467 |
| concepts[13].display_name | Receptor |
| concepts[14].id | https://openalex.org/C529278444 |
| concepts[14].level | 2 |
| concepts[14].score | 0.0 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q47273 |
| concepts[14].display_name | Central nervous system |
| concepts[15].id | https://openalex.org/C41625074 |
| concepts[15].level | 2 |
| concepts[15].score | 0.0 |
| concepts[15].wikidata | https://www.wikidata.org/wiki/Q176088 |
| concepts[15].display_name | Membrane |
| concepts[16].id | https://openalex.org/C178790620 |
| concepts[16].level | 1 |
| concepts[16].score | 0.0 |
| concepts[16].wikidata | https://www.wikidata.org/wiki/Q11351 |
| concepts[16].display_name | Organic chemistry |
| concepts[17].id | https://openalex.org/C55493867 |
| concepts[17].level | 1 |
| concepts[17].score | 0.0 |
| concepts[17].wikidata | https://www.wikidata.org/wiki/Q7094 |
| concepts[17].display_name | Biochemistry |
| keywords[0].id | https://openalex.org/keywords/calcium-imaging |
| keywords[0].score | 0.5625597834587097 |
| keywords[0].display_name | Calcium imaging |
| keywords[1].id | https://openalex.org/keywords/neurotransmission |
| keywords[1].score | 0.5466105341911316 |
| keywords[1].display_name | Neurotransmission |
| keywords[2].id | https://openalex.org/keywords/synapse |
| keywords[2].score | 0.5079205632209778 |
| keywords[2].display_name | Synapse |
| keywords[3].id | https://openalex.org/keywords/synaptic-vesicle |
| keywords[3].score | 0.48312780261039734 |
| keywords[3].display_name | Synaptic vesicle |
| keywords[4].id | https://openalex.org/keywords/computer-science |
| keywords[4].score | 0.4684583842754364 |
| keywords[4].display_name | Computer science |
| keywords[5].id | https://openalex.org/keywords/calcium-signaling |
| keywords[5].score | 0.4222016930580139 |
| keywords[5].display_name | Calcium signaling |
| keywords[6].id | https://openalex.org/keywords/neurotransmitter |
| keywords[6].score | 0.4210626184940338 |
| keywords[6].display_name | Neurotransmitter |
| keywords[7].id | https://openalex.org/keywords/postsynaptic-potential |
| keywords[7].score | 0.41334670782089233 |
| keywords[7].display_name | Postsynaptic potential |
| keywords[8].id | https://openalex.org/keywords/neuroscience |
| keywords[8].score | 0.4117390215396881 |
| keywords[8].display_name | Neuroscience |
| keywords[9].id | https://openalex.org/keywords/chemistry |
| keywords[9].score | 0.27440303564071655 |
| keywords[9].display_name | Chemistry |
| keywords[10].id | https://openalex.org/keywords/biology |
| keywords[10].score | 0.2701937258243561 |
| keywords[10].display_name | Biology |
| keywords[11].id | https://openalex.org/keywords/calcium |
| keywords[11].score | 0.23473966121673584 |
| keywords[11].display_name | Calcium |
| keywords[12].id | https://openalex.org/keywords/vesicle |
| keywords[12].score | 0.09622368216514587 |
| keywords[12].display_name | Vesicle |
| language | en |
| locations[0].id | doi:10.3389/fcomp.2021.777837 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4210211086 |
| locations[0].source.issn | 2624-9898 |
| locations[0].source.type | journal |
| locations[0].source.is_oa | True |
| locations[0].source.issn_l | 2624-9898 |
| locations[0].source.is_core | True |
| locations[0].source.is_in_doaj | True |
| locations[0].source.display_name | Frontiers in Computer Science |
| locations[0].source.host_organization | https://openalex.org/P4310320527 |
| locations[0].source.host_organization_name | Frontiers Media |
| locations[0].source.host_organization_lineage | https://openalex.org/P4310320527 |
| locations[0].source.host_organization_lineage_names | Frontiers Media |
| locations[0].license | cc-by |
| locations[0].pdf_url | |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | Frontiers in Computer Science |
| locations[0].landing_page_url | https://doi.org/10.3389/fcomp.2021.777837 |
| locations[1].id | pmh:oai:doaj.org/article:23d32116dbec4ff281f4b8f137fb8936 |
| locations[1].is_oa | True |
| locations[1].source.id | https://openalex.org/S4306401280 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | DOAJ (DOAJ: Directory of Open Access Journals) |
| locations[1].source.host_organization | |
| locations[1].source.host_organization_name | |
| locations[1].license | cc-by-sa |
| locations[1].pdf_url | |
| locations[1].version | submittedVersion |
| locations[1].raw_type | article |
| locations[1].license_id | https://openalex.org/licenses/cc-by-sa |
| locations[1].is_accepted | False |
| locations[1].is_published | False |
| locations[1].raw_source_name | Frontiers in Computer Science, Vol 3 (2021) |
| locations[1].landing_page_url | https://doaj.org/article/23d32116dbec4ff281f4b8f137fb8936 |
| indexed_in | crossref, doaj |
| authorships[0].author.id | https://openalex.org/A5038375914 |
| authorships[0].author.orcid | https://orcid.org/0000-0003-2058-1124 |
| authorships[0].author.display_name | Christopher Schmied |
| authorships[0].countries | DE, IT |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I4210099563 |
| authorships[0].affiliations[0].raw_affiliation_string | Leibniz-Forschungsinstitut für Molekulare Pharmakologie (FMP), Berlin, Germany |
| authorships[0].affiliations[1].institution_ids | https://openalex.org/I4210115812 |
| authorships[0].affiliations[1].raw_affiliation_string | Fondazione Human Technopole (HT), Milano, Italy |
| authorships[0].institutions[0].id | https://openalex.org/I4210099563 |
| authorships[0].institutions[0].ror | https://ror.org/010s54n03 |
| authorships[0].institutions[0].type | facility |
| authorships[0].institutions[0].lineage | https://openalex.org/I315704651, https://openalex.org/I4210099563 |
| authorships[0].institutions[0].country_code | DE |
| authorships[0].institutions[0].display_name | Leibniz-Forschungsinstitut für Molekulare Pharmakologie |
| authorships[0].institutions[1].id | https://openalex.org/I4210115812 |
| authorships[0].institutions[1].ror | https://ror.org/029gmnc79 |
| authorships[0].institutions[1].type | facility |
| authorships[0].institutions[1].lineage | https://openalex.org/I4210115812 |
| authorships[0].institutions[1].country_code | IT |
| authorships[0].institutions[1].display_name | Human Technopole |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Christopher Schmied |
| authorships[0].is_corresponding | True |
| authorships[0].raw_affiliation_strings | Fondazione Human Technopole (HT), Milano, Italy, Leibniz-Forschungsinstitut für Molekulare Pharmakologie (FMP), Berlin, Germany |
| authorships[1].author.id | https://openalex.org/A5013452452 |
| authorships[1].author.orcid | https://orcid.org/0000-0002-1324-9601 |
| authorships[1].author.display_name | Tolga Soykan |
| authorships[1].countries | DE |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I4210099563 |
| authorships[1].affiliations[0].raw_affiliation_string | Leibniz-Forschungsinstitut für Molekulare Pharmakologie (FMP), Berlin, Germany |
| authorships[1].institutions[0].id | https://openalex.org/I4210099563 |
| authorships[1].institutions[0].ror | https://ror.org/010s54n03 |
| authorships[1].institutions[0].type | facility |
| authorships[1].institutions[0].lineage | https://openalex.org/I315704651, https://openalex.org/I4210099563 |
| authorships[1].institutions[0].country_code | DE |
| authorships[1].institutions[0].display_name | Leibniz-Forschungsinstitut für Molekulare Pharmakologie |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Tolga Soykan |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | Leibniz-Forschungsinstitut für Molekulare Pharmakologie (FMP), Berlin, Germany |
| authorships[2].author.id | https://openalex.org/A5088925673 |
| authorships[2].author.orcid | |
| authorships[2].author.display_name | Svenja Bolz |
| authorships[2].countries | DE |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I4210099563 |
| authorships[2].affiliations[0].raw_affiliation_string | Leibniz-Forschungsinstitut für Molekulare Pharmakologie (FMP), Berlin, Germany |
| authorships[2].institutions[0].id | https://openalex.org/I4210099563 |
| authorships[2].institutions[0].ror | https://ror.org/010s54n03 |
| authorships[2].institutions[0].type | facility |
| authorships[2].institutions[0].lineage | https://openalex.org/I315704651, https://openalex.org/I4210099563 |
| authorships[2].institutions[0].country_code | DE |
| authorships[2].institutions[0].display_name | Leibniz-Forschungsinstitut für Molekulare Pharmakologie |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Svenja Bolz |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | Leibniz-Forschungsinstitut für Molekulare Pharmakologie (FMP), Berlin, Germany |
| authorships[3].author.id | https://openalex.org/A5010631390 |
| authorships[3].author.orcid | https://orcid.org/0000-0003-3119-6993 |
| authorships[3].author.display_name | Volker Haucke |
| authorships[3].countries | DE |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I4210099563 |
| authorships[3].affiliations[0].raw_affiliation_string | Leibniz-Forschungsinstitut für Molekulare Pharmakologie (FMP), Berlin, Germany |
| authorships[3].institutions[0].id | https://openalex.org/I4210099563 |
| authorships[3].institutions[0].ror | https://ror.org/010s54n03 |
| authorships[3].institutions[0].type | facility |
| authorships[3].institutions[0].lineage | https://openalex.org/I315704651, https://openalex.org/I4210099563 |
| authorships[3].institutions[0].country_code | DE |
| authorships[3].institutions[0].display_name | Leibniz-Forschungsinstitut für Molekulare Pharmakologie |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Volker Haucke |
| authorships[3].is_corresponding | True |
| authorships[3].raw_affiliation_strings | Leibniz-Forschungsinstitut für Molekulare Pharmakologie (FMP), Berlin, Germany |
| authorships[4].author.id | https://openalex.org/A5068664764 |
| authorships[4].author.orcid | https://orcid.org/0000-0002-8370-6353 |
| authorships[4].author.display_name | Martin Lehmann |
| authorships[4].countries | DE |
| authorships[4].affiliations[0].institution_ids | https://openalex.org/I4210099563 |
| authorships[4].affiliations[0].raw_affiliation_string | Leibniz-Forschungsinstitut für Molekulare Pharmakologie (FMP), Berlin, Germany |
| authorships[4].institutions[0].id | https://openalex.org/I4210099563 |
| authorships[4].institutions[0].ror | https://ror.org/010s54n03 |
| authorships[4].institutions[0].type | facility |
| authorships[4].institutions[0].lineage | https://openalex.org/I315704651, https://openalex.org/I4210099563 |
| authorships[4].institutions[0].country_code | DE |
| authorships[4].institutions[0].display_name | Leibniz-Forschungsinstitut für Molekulare Pharmakologie |
| authorships[4].author_position | last |
| authorships[4].raw_author_name | Martin Lehmann |
| authorships[4].is_corresponding | True |
| authorships[4].raw_affiliation_strings | Leibniz-Forschungsinstitut für Molekulare Pharmakologie (FMP), Berlin, Germany |
| has_content.pdf | False |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://doi.org/10.3389/fcomp.2021.777837 |
| open_access.oa_status | gold |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | SynActJ: Easy-to-Use Automated Analysis of Synaptic Activity |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T12859 |
| primary_topic.field.id | https://openalex.org/fields/13 |
| primary_topic.field.display_name | Biochemistry, Genetics and Molecular Biology |
| primary_topic.score | 0.9987999796867371 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1304 |
| primary_topic.subfield.display_name | Biophysics |
| primary_topic.display_name | Cell Image Analysis Techniques |
| related_works | https://openalex.org/W2074830850, https://openalex.org/W2994371818, https://openalex.org/W1617322396, https://openalex.org/W4289975053, https://openalex.org/W2074688705, https://openalex.org/W2066247450, https://openalex.org/W1599287250, https://openalex.org/W2019644150, https://openalex.org/W2329602648, https://openalex.org/W2442189777 |
| cited_by_count | 18 |
| counts_by_year[0].year | 2025 |
| counts_by_year[0].cited_by_count | 3 |
| counts_by_year[1].year | 2024 |
| counts_by_year[1].cited_by_count | 7 |
| counts_by_year[2].year | 2023 |
| counts_by_year[2].cited_by_count | 8 |
| locations_count | 2 |
| best_oa_location.id | doi:10.3389/fcomp.2021.777837 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4210211086 |
| best_oa_location.source.issn | 2624-9898 |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | True |
| best_oa_location.source.issn_l | 2624-9898 |
| best_oa_location.source.is_core | True |
| best_oa_location.source.is_in_doaj | True |
| best_oa_location.source.display_name | Frontiers in Computer Science |
| best_oa_location.source.host_organization | https://openalex.org/P4310320527 |
| best_oa_location.source.host_organization_name | Frontiers Media |
| best_oa_location.source.host_organization_lineage | https://openalex.org/P4310320527 |
| best_oa_location.source.host_organization_lineage_names | Frontiers Media |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | journal-article |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | Frontiers in Computer Science |
| best_oa_location.landing_page_url | https://doi.org/10.3389/fcomp.2021.777837 |
| primary_location.id | doi:10.3389/fcomp.2021.777837 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4210211086 |
| primary_location.source.issn | 2624-9898 |
| primary_location.source.type | journal |
| primary_location.source.is_oa | True |
| primary_location.source.issn_l | 2624-9898 |
| primary_location.source.is_core | True |
| primary_location.source.is_in_doaj | True |
| primary_location.source.display_name | Frontiers in Computer Science |
| primary_location.source.host_organization | https://openalex.org/P4310320527 |
| primary_location.source.host_organization_name | Frontiers Media |
| primary_location.source.host_organization_lineage | https://openalex.org/P4310320527 |
| primary_location.source.host_organization_lineage_names | Frontiers Media |
| primary_location.license | cc-by |
| primary_location.pdf_url | |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | Frontiers in Computer Science |
| primary_location.landing_page_url | https://doi.org/10.3389/fcomp.2021.777837 |
| publication_date | 2021-12-09 |
| publication_year | 2021 |
| referenced_works | https://openalex.org/W3025414101, https://openalex.org/W2799552138, https://openalex.org/W2066501531, https://openalex.org/W2040360409, https://openalex.org/W2805271873, https://openalex.org/W2506916679, https://openalex.org/W1993355218, https://openalex.org/W2047562589, https://openalex.org/W2089227939, https://openalex.org/W2467628122, https://openalex.org/W2513986326, https://openalex.org/W2016166286, https://openalex.org/W2469581795, https://openalex.org/W1964713902, https://openalex.org/W3149302371, https://openalex.org/W2143623375, https://openalex.org/W2167279371, https://openalex.org/W2099540110, https://openalex.org/W2305957222, https://openalex.org/W2590662729, https://openalex.org/W2060384627, https://openalex.org/W1990646890, https://openalex.org/W1966782694, https://openalex.org/W1990076574, https://openalex.org/W4399638148 |
| referenced_works_count | 25 |
| abstract_inverted_index.R | 255 |
| abstract_inverted_index.a | 206, 234, 275 |
| abstract_inverted_index.SV | 44, 270, 292 |
| abstract_inverted_index.We | 258, 280, 305 |
| abstract_inverted_index.an | 187, 221, 254, 310 |
| abstract_inverted_index.as | 60 |
| abstract_inverted_index.be | 228 |
| abstract_inverted_index.by | 35, 71, 92, 113 |
| abstract_inverted_index.in | 75, 107, 176, 185, 217, 236 |
| abstract_inverted_index.is | 152 |
| abstract_inverted_index.it | 151 |
| abstract_inverted_index.of | 19, 32, 84, 102, 148, 172, 199, 205, 214, 243, 265, 316 |
| abstract_inverted_index.on | 82, 262 |
| abstract_inverted_index.or | 40, 54, 63, 132, 274 |
| abstract_inverted_index.to | 86, 128, 139, 160, 233, 285, 308 |
| abstract_inverted_index.we | 180 |
| abstract_inverted_index.For | 141 |
| abstract_inverted_index.The | 100, 202, 239 |
| abstract_inverted_index.and | 15, 46, 48, 66, 94, 145, 167, 196, 231, 251, 288, 291, 295, 319 |
| abstract_inverted_index.are | 2, 90, 137, 165, 247 |
| abstract_inverted_index.can | 227 |
| abstract_inverted_index.for | 28, 298, 313 |
| abstract_inverted_index.has | 110, 158 |
| abstract_inverted_index.key | 153 |
| abstract_inverted_index.not | 123 |
| abstract_inverted_index.the | 12, 29, 103, 108, 129, 142, 155, 169, 210, 260, 269, 282, 314 |
| abstract_inverted_index.via | 11, 220 |
| abstract_inverted_index.Fiji | 207 |
| abstract_inverted_index.Here | 179 |
| abstract_inverted_index.been | 111, 124 |
| abstract_inverted_index.cell | 98 |
| abstract_inverted_index.data | 197 |
| abstract_inverted_index.dyes | 53 |
| abstract_inverted_index.each | 244 |
| abstract_inverted_index.exo- | 45 |
| abstract_inverted_index.fast | 93 |
| abstract_inverted_index.have | 122 |
| abstract_inverted_index.hubs | 6 |
| abstract_inverted_index.live | 97 |
| abstract_inverted_index.made | 125 |
| abstract_inverted_index.past | 109 |
| abstract_inverted_index.pre- | 39 |
| abstract_inverted_index.such | 59 |
| abstract_inverted_index.that | 7, 121, 136, 154, 164, 192, 226 |
| abstract_inverted_index.tool | 312 |
| abstract_inverted_index.upon | 79 |
| abstract_inverted_index.use. | 140 |
| abstract_inverted_index.Shiny | 256 |
| abstract_inverted_index.allow | 27, 168 |
| abstract_inverted_index.batch | 237 |
| abstract_inverted_index.fully | 189 |
| abstract_inverted_index.image | 195, 212 |
| abstract_inverted_index.mode. | 238 |
| abstract_inverted_index.other | 296 |
| abstract_inverted_index.rapid | 73 |
| abstract_inverted_index.their | 76 |
| abstract_inverted_index.tools | 26, 163 |
| abstract_inverted_index.under | 302 |
| abstract_inverted_index.using | 253 |
| abstract_inverted_index.which | 88 |
| abstract_inverted_index.(SVs). | 23 |
| abstract_inverted_index.access | 159 |
| abstract_inverted_index.action | 37 |
| abstract_inverted_index.active | 177, 215 |
| abstract_inverted_index.become | 309 |
| abstract_inverted_index.bouton | 246 |
| abstract_inverted_index.custom | 119 |
| abstract_inverted_index.direct | 30 |
| abstract_inverted_index.easily | 229 |
| abstract_inverted_index.either | 114 |
| abstract_inverted_index.fusion | 14 |
| abstract_inverted_index.highly | 3 |
| abstract_inverted_index.images | 264 |
| abstract_inverted_index.manual | 286 |
| abstract_inverted_index.movies | 106, 219 |
| abstract_inverted_index.plugin | 208 |
| abstract_inverted_index.reveal | 68 |
| abstract_inverted_index.seeded | 223 |
| abstract_inverted_index.traces | 242 |
| abstract_inverted_index.widely | 126 |
| abstract_inverted_index.SynActJ | 182, 307 |
| abstract_inverted_index.applied | 232 |
| abstract_inverted_index.calcium | 42, 277 |
| abstract_inverted_index.changes | 74, 175 |
| abstract_inverted_index.compare | 281 |
| abstract_inverted_index.compute | 289 |
| abstract_inverted_index.dataset | 235 |
| abstract_inverted_index.dynamic | 4 |
| abstract_inverted_index.enables | 193 |
| abstract_inverted_index.genetic | 56 |
| abstract_inverted_index.imaging | 25 |
| abstract_inverted_index.influx, | 43 |
| abstract_inverted_index.mediate | 8 |
| abstract_inverted_index.organic | 52 |
| abstract_inverted_index.picking | 116 |
| abstract_inverted_index.plotted | 252 |
| abstract_inverted_index.predict | 306 |
| abstract_inverted_index.present | 181 |
| abstract_inverted_index.scripts | 120 |
| abstract_inverted_index.sensor, | 278 |
| abstract_inverted_index.sensors | 65 |
| abstract_inverted_index.synapse | 320 |
| abstract_inverted_index.vesicle | 300 |
| abstract_inverted_index.voltage | 62 |
| abstract_inverted_index.Activity | 184 |
| abstract_inverted_index.ImageJ), | 186 |
| abstract_inverted_index.Neuronal | 0 |
| abstract_inverted_index.activity | 34, 70, 81, 318 |
| abstract_inverted_index.adjusted | 230 |
| abstract_inverted_index.advanced | 133 |
| abstract_inverted_index.analysis | 101, 162, 198, 213, 287, 315 |
| abstract_inverted_index.calcium, | 61 |
| abstract_inverted_index.chemical | 9 |
| abstract_inverted_index.consists | 204 |
| abstract_inverted_index.exocytic | 13 |
| abstract_inverted_index.kinetics | 294 |
| abstract_inverted_index.manually | 115 |
| abstract_inverted_index.neuronal | 80 |
| abstract_inverted_index.precise, | 143 |
| abstract_inverted_index.recorded | 91 |
| abstract_inverted_index.release. | 50 |
| abstract_inverted_index.reporter | 272 |
| abstract_inverted_index.research | 156 |
| abstract_inverted_index.seconds, | 87 |
| abstract_inverted_index.software | 134 |
| abstract_inverted_index.synapses | 1, 216, 267 |
| abstract_inverted_index.synaptic | 21, 33, 69, 149, 200, 245, 299, 317 |
| abstract_inverted_index.unbiased | 144 |
| abstract_inverted_index.validate | 259 |
| abstract_inverted_index.vesicles | 22 |
| abstract_inverted_index.workflow | 191, 203, 261, 284 |
| abstract_inverted_index.(Synaptic | 183 |
| abstract_inverted_index.activity, | 150 |
| abstract_inverted_index.activity. | 201 |
| abstract_inverted_index.analyzed, | 250 |
| abstract_inverted_index.automated | 170, 194, 211 |
| abstract_inverted_index.automatic | 283 |
| abstract_inverted_index.available | 127 |
| abstract_inverted_index.bio-image | 161 |
| abstract_inverted_index.community | 157 |
| abstract_inverted_index.detecting | 36 |
| abstract_inverted_index.detection | 171 |
| abstract_inverted_index.different | 303 |
| abstract_inverted_index.endocytic | 17 |
| abstract_inverted_index.extracted | 240 |
| abstract_inverted_index.glutamate | 49 |
| abstract_inverted_index.important | 311 |
| abstract_inverted_index.intensity | 78, 174, 241 |
| abstract_inverted_index.molecular | 57 |
| abstract_inverted_index.performed | 112 |
| abstract_inverted_index.recycling | 18, 301 |
| abstract_inverted_index.resulting | 104 |
| abstract_inverted_index.sensitive | 95 |
| abstract_inverted_index.synapses. | 178 |
| abstract_inverted_index.toolboxes | 135 |
| abstract_inverted_index.typically | 89 |
| abstract_inverted_index.watershed | 224 |
| abstract_inverted_index.widefield | 96 |
| abstract_inverted_index.workflow. | 257 |
| abstract_inverted_index.Functional | 24 |
| abstract_inverted_index.community, | 131 |
| abstract_inverted_index.expressing | 268 |
| abstract_inverted_index.individual | 117 |
| abstract_inverted_index.parameters | 297 |
| abstract_inverted_index.performing | 209 |
| abstract_inverted_index.processed, | 249 |
| abstract_inverted_index.reporters, | 58 |
| abstract_inverted_index.scientific | 130 |
| abstract_inverted_index.stimulated | 266 |
| abstract_inverted_index.subsequent | 16 |
| abstract_inverted_index.time-lapse | 105, 218, 263 |
| abstract_inverted_index.timescales | 83 |
| abstract_inverted_index.undergoing | 72 |
| abstract_inverted_index.Fluorescent | 51 |
| abstract_inverted_index.complicated | 138 |
| abstract_inverted_index.conditions. | 304 |
| abstract_inverted_index.easy-to-use | 188 |
| abstract_inverted_index.fluorescent | 173 |
| abstract_inverted_index.interactive | 222 |
| abstract_inverted_index.measurement | 147 |
| abstract_inverted_index.microscopy. | 99 |
| abstract_inverted_index.open-source | 190 |
| abstract_inverted_index.potentials, | 38 |
| abstract_inverted_index.properties. | 321 |
| abstract_inverted_index.structures, | 118 |
| abstract_inverted_index.endocytosis, | 47 |
| abstract_inverted_index.fluorescence | 77 |
| abstract_inverted_index.milliseconds | 85 |
| abstract_inverted_index.postsynaptic | 41 |
| abstract_inverted_index.reproducible | 146 |
| abstract_inverted_index.segmentation | 225 |
| abstract_inverted_index.automatically | 248 |
| abstract_inverted_index.communication | 5 |
| abstract_inverted_index.easy-to-apply | 166 |
| abstract_inverted_index.visualization | 31 |
| abstract_inverted_index.calcium-influx | 290 |
| abstract_inverted_index.exo-/endocytosis | 271, 293 |
| abstract_inverted_index.neurotransmitter | 64 |
| abstract_inverted_index.synapse-targeted | 55, 276 |
| abstract_inverted_index.neurotransmission | 10 |
| abstract_inverted_index.synapto-pHluorins | 67 |
| abstract_inverted_index.Synaptophysin-RGECO. | 279 |
| abstract_inverted_index.Synaptophysin-pHluorin | 273 |
| abstract_inverted_index.neurotransmitter-containing | 20 |
| cited_by_percentile_year.max | 99 |
| cited_by_percentile_year.min | 97 |
| corresponding_author_ids | https://openalex.org/A5068664764, https://openalex.org/A5038375914, https://openalex.org/A5010631390 |
| countries_distinct_count | 2 |
| institutions_distinct_count | 5 |
| corresponding_institution_ids | https://openalex.org/I4210099563, https://openalex.org/I4210115812 |
| citation_normalized_percentile.value | 0.93500526 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |