Systematic tissue-specific functional annotation of the human genome highlights immune-related DNA elements for late-onset Alzheimer’s disease Article Swipe
 YOU?
·
· 2016
· Open Access
·
· DOI: https://doi.org/10.1101/078865
YOU?
·
· 2016
· Open Access
·
· DOI: https://doi.org/10.1101/078865
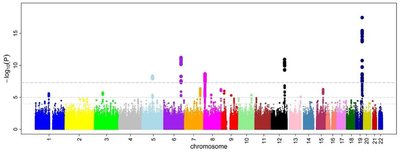
Continuing efforts from large international consortia have made genome-wide epigenomic and transcriptomic annotation data publicly available for a variety of cell and tissue types. However, synthesis of these datasets into effective summary metrics to characterize the functional non-coding genome remains a challenge. Here, we present GenoSkyline-Plus, an extension of our previous work through integration of an expanded set of epigenomic and transcriptomic annotations to produce high-resolution, single tissue annotations. After validating our annotations with a catalog of tissue-specific non-coding elements previously identified in the literature, we apply our method using data from 127 different cell and tissue types to present an atlas of heritability enrichment across 45 different GWAS traits. We show that broader organ system categories (e.g. immune system) increase statistical power in identifying biologically relevant tissue types for complex diseases while annotations of individual cell types (e.g. monocytes or B-cells) provide deeper insights into disease etiology. Additionally, we use our GenoSkyline-Plus annotations in an in-depth case study of late-onset Alzheimer’s disease (LOAD). Our analyses suggest a strong connection between LOAD heritability and genetic variants contained in regions of the genome functional in monocytes. Furthermore, we show that LOAD shares a similar localization of SNPs to monocyte-functional regions with Parkinson’s disease. Overall, we demonstrate that integrated genome annotations at the single tissue level provide a valuable tool for understanding the etiology of complex human diseases. Our GenoSkyline-Plus annotations are freely available at http://genocanyon.med.yale.edu/GenoSkyline . Author Summary After years of community efforts, many experimental and computational approaches have been developed and applied for functional annotation of the human genome, yet proper annotation still remains challenging, especially in non-coding regions. As complex disease research rapidly advances, increasing evidence suggests that non-coding regulatory DNA elements may be the primary regions harboring risk variants in human complex diseases. In this paper, we introduce GenoSkyline-Plus, a principled annotation framework to identify tissue and cell type-specific functional regions in the human genome through integration of diverse high-throughput epigenomic and transcriptomic data. Through validation of known non-coding tissue-specific regulatory regions, enrichment analyses on 45 complex traits, and an in-depth case study of neurodegenerative diseases, we demonstrate the ability of GenoSkyline-Plus to accurately identify tissue-specific functionality in the human genome and provide unbiased, genome-wide insights into the genetic basis of human complex diseases.
Related Topics
- Type
- preprint
- Language
- en
- Landing Page
- https://doi.org/10.1101/078865
- https://www.biorxiv.org/content/biorxiv/early/2017/05/28/078865.full.pdf
- OA Status
- green
- Cited By
- 8
- References
- 89
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W2529601773
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W2529601773Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1101/078865Digital Object Identifier
- Title
-
Systematic tissue-specific functional annotation of the human genome highlights immune-related DNA elements for late-onset Alzheimer’s diseaseWork title
- Type
-
preprintOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2016Year of publication
- Publication date
-
2016-10-02Full publication date if available
- Authors
-
Qiongshi Lu, Ryan L. Powles, Sarah B. Abdallah, Derek Ou, Qian Wang, Yiming Hu, Yisi Lu, Wei Liu, Boyang Li, Shubhabrata Mukherjee, Paul K. Crane, Hongyu ZhaoList of authors in order
- Landing page
-
https://doi.org/10.1101/078865Publisher landing page
- PDF URL
-
https://www.biorxiv.org/content/biorxiv/early/2017/05/28/078865.full.pdfDirect link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
greenOpen access status per OpenAlex
- OA URL
-
https://www.biorxiv.org/content/biorxiv/early/2017/05/28/078865.full.pdfDirect OA link when available
- Concepts
-
Epigenomics, Genome-wide association study, Biology, Computational biology, Genome, Annotation, Epigenome, Disease, Genetics, Single-nucleotide polymorphism, DNA methylation, Gene, Medicine, Pathology, Gene expression, GenotypeTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
8Total citation count in OpenAlex
- Citations by year (recent)
-
2025: 1, 2023: 2, 2021: 1, 2020: 3, 2017: 1Per-year citation counts (last 5 years)
- References (count)
-
89Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W2529601773 |
|---|---|
| doi | https://doi.org/10.1101/078865 |
| ids.doi | https://doi.org/10.1101/078865 |
| ids.mag | 2529601773 |
| ids.openalex | https://openalex.org/W2529601773 |
| fwci | 0.34665233 |
| type | preprint |
| title | Systematic tissue-specific functional annotation of the human genome highlights immune-related DNA elements for late-onset Alzheimer’s disease |
| biblio.issue | |
| biblio.volume | |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T10269 |
| topics[0].field.id | https://openalex.org/fields/13 |
| topics[0].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[0].score | 0.9987000226974487 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1312 |
| topics[0].subfield.display_name | Molecular Biology |
| topics[0].display_name | Epigenetics and DNA Methylation |
| topics[1].id | https://openalex.org/T10112 |
| topics[1].field.id | https://openalex.org/fields/24 |
| topics[1].field.display_name | Immunology and Microbiology |
| topics[1].score | 0.9915000200271606 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/2406 |
| topics[1].subfield.display_name | Virology |
| topics[1].display_name | HIV Research and Treatment |
| topics[2].id | https://openalex.org/T10604 |
| topics[2].field.id | https://openalex.org/fields/13 |
| topics[2].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[2].score | 0.9904999732971191 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1312 |
| topics[2].subfield.display_name | Molecular Biology |
| topics[2].display_name | RNA Research and Splicing |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C121912465 |
| concepts[0].level | 5 |
| concepts[0].score | 0.787238597869873 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q3589153 |
| concepts[0].display_name | Epigenomics |
| concepts[1].id | https://openalex.org/C106208931 |
| concepts[1].level | 5 |
| concepts[1].score | 0.6425719857215881 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q1098876 |
| concepts[1].display_name | Genome-wide association study |
| concepts[2].id | https://openalex.org/C86803240 |
| concepts[2].level | 0 |
| concepts[2].score | 0.5813406705856323 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[2].display_name | Biology |
| concepts[3].id | https://openalex.org/C70721500 |
| concepts[3].level | 1 |
| concepts[3].score | 0.5778810977935791 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q177005 |
| concepts[3].display_name | Computational biology |
| concepts[4].id | https://openalex.org/C141231307 |
| concepts[4].level | 3 |
| concepts[4].score | 0.5777226686477661 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q7020 |
| concepts[4].display_name | Genome |
| concepts[5].id | https://openalex.org/C2776321320 |
| concepts[5].level | 2 |
| concepts[5].score | 0.5439798831939697 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q857525 |
| concepts[5].display_name | Annotation |
| concepts[6].id | https://openalex.org/C117717151 |
| concepts[6].level | 5 |
| concepts[6].score | 0.5056545734405518 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q3589155 |
| concepts[6].display_name | Epigenome |
| concepts[7].id | https://openalex.org/C2779134260 |
| concepts[7].level | 2 |
| concepts[7].score | 0.46004819869995117 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q12136 |
| concepts[7].display_name | Disease |
| concepts[8].id | https://openalex.org/C54355233 |
| concepts[8].level | 1 |
| concepts[8].score | 0.3702659010887146 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[8].display_name | Genetics |
| concepts[9].id | https://openalex.org/C153209595 |
| concepts[9].level | 4 |
| concepts[9].score | 0.2853546738624573 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q501128 |
| concepts[9].display_name | Single-nucleotide polymorphism |
| concepts[10].id | https://openalex.org/C190727270 |
| concepts[10].level | 4 |
| concepts[10].score | 0.2639768719673157 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q874745 |
| concepts[10].display_name | DNA methylation |
| concepts[11].id | https://openalex.org/C104317684 |
| concepts[11].level | 2 |
| concepts[11].score | 0.17972150444984436 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[11].display_name | Gene |
| concepts[12].id | https://openalex.org/C71924100 |
| concepts[12].level | 0 |
| concepts[12].score | 0.11793252825737 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q11190 |
| concepts[12].display_name | Medicine |
| concepts[13].id | https://openalex.org/C142724271 |
| concepts[13].level | 1 |
| concepts[13].score | 0.08921915292739868 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q7208 |
| concepts[13].display_name | Pathology |
| concepts[14].id | https://openalex.org/C150194340 |
| concepts[14].level | 3 |
| concepts[14].score | 0.0 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q26972 |
| concepts[14].display_name | Gene expression |
| concepts[15].id | https://openalex.org/C135763542 |
| concepts[15].level | 3 |
| concepts[15].score | 0.0 |
| concepts[15].wikidata | https://www.wikidata.org/wiki/Q106016 |
| concepts[15].display_name | Genotype |
| keywords[0].id | https://openalex.org/keywords/epigenomics |
| keywords[0].score | 0.787238597869873 |
| keywords[0].display_name | Epigenomics |
| keywords[1].id | https://openalex.org/keywords/genome-wide-association-study |
| keywords[1].score | 0.6425719857215881 |
| keywords[1].display_name | Genome-wide association study |
| keywords[2].id | https://openalex.org/keywords/biology |
| keywords[2].score | 0.5813406705856323 |
| keywords[2].display_name | Biology |
| keywords[3].id | https://openalex.org/keywords/computational-biology |
| keywords[3].score | 0.5778810977935791 |
| keywords[3].display_name | Computational biology |
| keywords[4].id | https://openalex.org/keywords/genome |
| keywords[4].score | 0.5777226686477661 |
| keywords[4].display_name | Genome |
| keywords[5].id | https://openalex.org/keywords/annotation |
| keywords[5].score | 0.5439798831939697 |
| keywords[5].display_name | Annotation |
| keywords[6].id | https://openalex.org/keywords/epigenome |
| keywords[6].score | 0.5056545734405518 |
| keywords[6].display_name | Epigenome |
| keywords[7].id | https://openalex.org/keywords/disease |
| keywords[7].score | 0.46004819869995117 |
| keywords[7].display_name | Disease |
| keywords[8].id | https://openalex.org/keywords/genetics |
| keywords[8].score | 0.3702659010887146 |
| keywords[8].display_name | Genetics |
| keywords[9].id | https://openalex.org/keywords/single-nucleotide-polymorphism |
| keywords[9].score | 0.2853546738624573 |
| keywords[9].display_name | Single-nucleotide polymorphism |
| keywords[10].id | https://openalex.org/keywords/dna-methylation |
| keywords[10].score | 0.2639768719673157 |
| keywords[10].display_name | DNA methylation |
| keywords[11].id | https://openalex.org/keywords/gene |
| keywords[11].score | 0.17972150444984436 |
| keywords[11].display_name | Gene |
| keywords[12].id | https://openalex.org/keywords/medicine |
| keywords[12].score | 0.11793252825737 |
| keywords[12].display_name | Medicine |
| keywords[13].id | https://openalex.org/keywords/pathology |
| keywords[13].score | 0.08921915292739868 |
| keywords[13].display_name | Pathology |
| language | en |
| locations[0].id | doi:10.1101/078865 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4306402567 |
| locations[0].source.issn | |
| locations[0].source.type | repository |
| locations[0].source.is_oa | False |
| locations[0].source.issn_l | |
| locations[0].source.is_core | False |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| locations[0].source.host_organization | https://openalex.org/I2750212522 |
| locations[0].source.host_organization_name | Cold Spring Harbor Laboratory |
| locations[0].source.host_organization_lineage | https://openalex.org/I2750212522 |
| locations[0].license | cc-by-nc-nd |
| locations[0].pdf_url | https://www.biorxiv.org/content/biorxiv/early/2017/05/28/078865.full.pdf |
| locations[0].version | acceptedVersion |
| locations[0].raw_type | posted-content |
| locations[0].license_id | https://openalex.org/licenses/cc-by-nc-nd |
| locations[0].is_accepted | True |
| locations[0].is_published | False |
| locations[0].raw_source_name | |
| locations[0].landing_page_url | https://doi.org/10.1101/078865 |
| indexed_in | crossref |
| authorships[0].author.id | https://openalex.org/A5083375749 |
| authorships[0].author.orcid | https://orcid.org/0000-0002-4514-0969 |
| authorships[0].author.display_name | Qiongshi Lu |
| authorships[0].countries | US |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I32971472 |
| authorships[0].affiliations[0].raw_affiliation_string | Department of Biostatistics, Yale School of Public Health, New Haven, CT, USA |
| authorships[0].institutions[0].id | https://openalex.org/I32971472 |
| authorships[0].institutions[0].ror | https://ror.org/03v76x132 |
| authorships[0].institutions[0].type | education |
| authorships[0].institutions[0].lineage | https://openalex.org/I32971472 |
| authorships[0].institutions[0].country_code | US |
| authorships[0].institutions[0].display_name | Yale University |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Qiongshi Lu |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Department of Biostatistics, Yale School of Public Health, New Haven, CT, USA |
| authorships[1].author.id | https://openalex.org/A5000633263 |
| authorships[1].author.orcid | https://orcid.org/0009-0006-3706-9281 |
| authorships[1].author.display_name | Ryan L. Powles |
| authorships[1].countries | US |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I32971472 |
| authorships[1].affiliations[0].raw_affiliation_string | Program of Computational Biology and Bioinformatics, Yale University, New Haven, CT, USA |
| authorships[1].institutions[0].id | https://openalex.org/I32971472 |
| authorships[1].institutions[0].ror | https://ror.org/03v76x132 |
| authorships[1].institutions[0].type | education |
| authorships[1].institutions[0].lineage | https://openalex.org/I32971472 |
| authorships[1].institutions[0].country_code | US |
| authorships[1].institutions[0].display_name | Yale University |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Ryan L. Powles |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | Program of Computational Biology and Bioinformatics, Yale University, New Haven, CT, USA |
| authorships[2].author.id | https://openalex.org/A5112469916 |
| authorships[2].author.orcid | |
| authorships[2].author.display_name | Sarah B. Abdallah |
| authorships[2].countries | US |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I32971472 |
| authorships[2].affiliations[0].raw_affiliation_string | Yale University School of Medicine, New Haven, CT, USA |
| authorships[2].institutions[0].id | https://openalex.org/I32971472 |
| authorships[2].institutions[0].ror | https://ror.org/03v76x132 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I32971472 |
| authorships[2].institutions[0].country_code | US |
| authorships[2].institutions[0].display_name | Yale University |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Sarah Abdallah |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | Yale University School of Medicine, New Haven, CT, USA |
| authorships[3].author.id | https://openalex.org/A5039287462 |
| authorships[3].author.orcid | |
| authorships[3].author.display_name | Derek Ou |
| authorships[3].countries | US |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I32971472 |
| authorships[3].affiliations[0].raw_affiliation_string | Yale University School of Medicine, New Haven, CT, USA |
| authorships[3].institutions[0].id | https://openalex.org/I32971472 |
| authorships[3].institutions[0].ror | https://ror.org/03v76x132 |
| authorships[3].institutions[0].type | education |
| authorships[3].institutions[0].lineage | https://openalex.org/I32971472 |
| authorships[3].institutions[0].country_code | US |
| authorships[3].institutions[0].display_name | Yale University |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Derek Ou |
| authorships[3].is_corresponding | False |
| authorships[3].raw_affiliation_strings | Yale University School of Medicine, New Haven, CT, USA |
| authorships[4].author.id | https://openalex.org/A5100391090 |
| authorships[4].author.orcid | https://orcid.org/0000-0002-6514-3470 |
| authorships[4].author.display_name | Qian Wang |
| authorships[4].countries | US |
| authorships[4].affiliations[0].institution_ids | https://openalex.org/I32971472 |
| authorships[4].affiliations[0].raw_affiliation_string | Program of Computational Biology and Bioinformatics, Yale University, New Haven, CT, USA |
| authorships[4].institutions[0].id | https://openalex.org/I32971472 |
| authorships[4].institutions[0].ror | https://ror.org/03v76x132 |
| authorships[4].institutions[0].type | education |
| authorships[4].institutions[0].lineage | https://openalex.org/I32971472 |
| authorships[4].institutions[0].country_code | US |
| authorships[4].institutions[0].display_name | Yale University |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Qian Wang |
| authorships[4].is_corresponding | False |
| authorships[4].raw_affiliation_strings | Program of Computational Biology and Bioinformatics, Yale University, New Haven, CT, USA |
| authorships[5].author.id | https://openalex.org/A5034686090 |
| authorships[5].author.orcid | https://orcid.org/0000-0002-0988-1030 |
| authorships[5].author.display_name | Yiming Hu |
| authorships[5].countries | US |
| authorships[5].affiliations[0].institution_ids | https://openalex.org/I32971472 |
| authorships[5].affiliations[0].raw_affiliation_string | Department of Biostatistics, Yale School of Public Health, New Haven, CT, USA |
| authorships[5].institutions[0].id | https://openalex.org/I32971472 |
| authorships[5].institutions[0].ror | https://ror.org/03v76x132 |
| authorships[5].institutions[0].type | education |
| authorships[5].institutions[0].lineage | https://openalex.org/I32971472 |
| authorships[5].institutions[0].country_code | US |
| authorships[5].institutions[0].display_name | Yale University |
| authorships[5].author_position | middle |
| authorships[5].raw_author_name | Yiming Hu |
| authorships[5].is_corresponding | False |
| authorships[5].raw_affiliation_strings | Department of Biostatistics, Yale School of Public Health, New Haven, CT, USA |
| authorships[6].author.id | https://openalex.org/A5085235353 |
| authorships[6].author.orcid | https://orcid.org/0000-0002-7960-5445 |
| authorships[6].author.display_name | Yisi Lu |
| authorships[6].countries | US |
| authorships[6].affiliations[0].institution_ids | https://openalex.org/I32971472 |
| authorships[6].affiliations[0].raw_affiliation_string | Department of Immunobiology, Yale University School of Medicine, New Haven, CT, USA |
| authorships[6].institutions[0].id | https://openalex.org/I32971472 |
| authorships[6].institutions[0].ror | https://ror.org/03v76x132 |
| authorships[6].institutions[0].type | education |
| authorships[6].institutions[0].lineage | https://openalex.org/I32971472 |
| authorships[6].institutions[0].country_code | US |
| authorships[6].institutions[0].display_name | Yale University |
| authorships[6].author_position | middle |
| authorships[6].raw_author_name | Yisi Lu |
| authorships[6].is_corresponding | False |
| authorships[6].raw_affiliation_strings | Department of Immunobiology, Yale University School of Medicine, New Haven, CT, USA |
| authorships[7].author.id | https://openalex.org/A5100432025 |
| authorships[7].author.orcid | https://orcid.org/0009-0007-4263-7383 |
| authorships[7].author.display_name | Wei Liu |
| authorships[7].countries | CN |
| authorships[7].affiliations[0].institution_ids | https://openalex.org/I20231570 |
| authorships[7].affiliations[0].raw_affiliation_string | School of Life Sciences, Peking University, Beijing, China |
| authorships[7].institutions[0].id | https://openalex.org/I20231570 |
| authorships[7].institutions[0].ror | https://ror.org/02v51f717 |
| authorships[7].institutions[0].type | education |
| authorships[7].institutions[0].lineage | https://openalex.org/I20231570 |
| authorships[7].institutions[0].country_code | CN |
| authorships[7].institutions[0].display_name | Peking University |
| authorships[7].author_position | middle |
| authorships[7].raw_author_name | Wei Liu |
| authorships[7].is_corresponding | False |
| authorships[7].raw_affiliation_strings | School of Life Sciences, Peking University, Beijing, China |
| authorships[8].author.id | https://openalex.org/A5100732751 |
| authorships[8].author.orcid | https://orcid.org/0000-0003-2808-9986 |
| authorships[8].author.display_name | Boyang Li |
| authorships[8].countries | US |
| authorships[8].affiliations[0].institution_ids | https://openalex.org/I32971472 |
| authorships[8].affiliations[0].raw_affiliation_string | Department of Biostatistics, Yale School of Public Health, New Haven, CT, USA |
| authorships[8].institutions[0].id | https://openalex.org/I32971472 |
| authorships[8].institutions[0].ror | https://ror.org/03v76x132 |
| authorships[8].institutions[0].type | education |
| authorships[8].institutions[0].lineage | https://openalex.org/I32971472 |
| authorships[8].institutions[0].country_code | US |
| authorships[8].institutions[0].display_name | Yale University |
| authorships[8].author_position | middle |
| authorships[8].raw_author_name | Boyang Li |
| authorships[8].is_corresponding | False |
| authorships[8].raw_affiliation_strings | Department of Biostatistics, Yale School of Public Health, New Haven, CT, USA |
| authorships[9].author.id | https://openalex.org/A5058279309 |
| authorships[9].author.orcid | https://orcid.org/0000-0003-2522-2884 |
| authorships[9].author.display_name | Shubhabrata Mukherjee |
| authorships[9].countries | US |
| authorships[9].affiliations[0].institution_ids | https://openalex.org/I201448701 |
| authorships[9].affiliations[0].raw_affiliation_string | Division of General Internal Medicine, Department of Medicine, University of Washington, Seattle, WA, USA |
| authorships[9].institutions[0].id | https://openalex.org/I201448701 |
| authorships[9].institutions[0].ror | https://ror.org/00cvxb145 |
| authorships[9].institutions[0].type | education |
| authorships[9].institutions[0].lineage | https://openalex.org/I201448701 |
| authorships[9].institutions[0].country_code | US |
| authorships[9].institutions[0].display_name | University of Washington |
| authorships[9].author_position | middle |
| authorships[9].raw_author_name | Shubhabrata Mukherjee |
| authorships[9].is_corresponding | False |
| authorships[9].raw_affiliation_strings | Division of General Internal Medicine, Department of Medicine, University of Washington, Seattle, WA, USA |
| authorships[10].author.id | https://openalex.org/A5050170858 |
| authorships[10].author.orcid | https://orcid.org/0000-0003-4278-7465 |
| authorships[10].author.display_name | Paul K. Crane |
| authorships[10].countries | US |
| authorships[10].affiliations[0].institution_ids | https://openalex.org/I201448701 |
| authorships[10].affiliations[0].raw_affiliation_string | Division of General Internal Medicine, Department of Medicine, University of Washington, Seattle, WA, USA |
| authorships[10].institutions[0].id | https://openalex.org/I201448701 |
| authorships[10].institutions[0].ror | https://ror.org/00cvxb145 |
| authorships[10].institutions[0].type | education |
| authorships[10].institutions[0].lineage | https://openalex.org/I201448701 |
| authorships[10].institutions[0].country_code | US |
| authorships[10].institutions[0].display_name | University of Washington |
| authorships[10].author_position | middle |
| authorships[10].raw_author_name | Paul K. Crane |
| authorships[10].is_corresponding | False |
| authorships[10].raw_affiliation_strings | Division of General Internal Medicine, Department of Medicine, University of Washington, Seattle, WA, USA |
| authorships[11].author.id | https://openalex.org/A5074828414 |
| authorships[11].author.orcid | https://orcid.org/0000-0003-1195-9607 |
| authorships[11].author.display_name | Hongyu Zhao |
| authorships[11].countries | US |
| authorships[11].affiliations[0].institution_ids | https://openalex.org/I4210120339 |
| authorships[11].affiliations[0].raw_affiliation_string | VA Cooperative Studies Program Coordinating Center, West Haven, CT, USA |
| authorships[11].affiliations[1].institution_ids | https://openalex.org/I32971472 |
| authorships[11].affiliations[1].raw_affiliation_string | Department of Biostatistics, Yale School of Public Health, New Haven, CT, USA |
| authorships[11].affiliations[2].institution_ids | https://openalex.org/I32971472 |
| authorships[11].affiliations[2].raw_affiliation_string | Program of Computational Biology and Bioinformatics, Yale University, New Haven, CT, USA |
| authorships[11].institutions[0].id | https://openalex.org/I4210120339 |
| authorships[11].institutions[0].ror | https://ror.org/02r0j1w74 |
| authorships[11].institutions[0].type | government |
| authorships[11].institutions[0].lineage | https://openalex.org/I1299022934, https://openalex.org/I4210120339 |
| authorships[11].institutions[0].country_code | US |
| authorships[11].institutions[0].display_name | Program Support Center |
| authorships[11].institutions[1].id | https://openalex.org/I32971472 |
| authorships[11].institutions[1].ror | https://ror.org/03v76x132 |
| authorships[11].institutions[1].type | education |
| authorships[11].institutions[1].lineage | https://openalex.org/I32971472 |
| authorships[11].institutions[1].country_code | US |
| authorships[11].institutions[1].display_name | Yale University |
| authorships[11].author_position | last |
| authorships[11].raw_author_name | Hongyu Zhao |
| authorships[11].is_corresponding | True |
| authorships[11].raw_affiliation_strings | Department of Biostatistics, Yale School of Public Health, New Haven, CT, USA, Program of Computational Biology and Bioinformatics, Yale University, New Haven, CT, USA, VA Cooperative Studies Program Coordinating Center, West Haven, CT, USA |
| has_content.pdf | True |
| has_content.grobid_xml | True |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://www.biorxiv.org/content/biorxiv/early/2017/05/28/078865.full.pdf |
| open_access.oa_status | green |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Systematic tissue-specific functional annotation of the human genome highlights immune-related DNA elements for late-onset Alzheimer’s disease |
| has_fulltext | True |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T10269 |
| primary_topic.field.id | https://openalex.org/fields/13 |
| primary_topic.field.display_name | Biochemistry, Genetics and Molecular Biology |
| primary_topic.score | 0.9987000226974487 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1312 |
| primary_topic.subfield.display_name | Molecular Biology |
| primary_topic.display_name | Epigenetics and DNA Methylation |
| related_works | https://openalex.org/W2424092526, https://openalex.org/W2507790420, https://openalex.org/W2212710981, https://openalex.org/W2795223073, https://openalex.org/W4200376839, https://openalex.org/W2151543286, https://openalex.org/W2418239907, https://openalex.org/W4280600268, https://openalex.org/W2474086683, https://openalex.org/W3217179662 |
| cited_by_count | 8 |
| counts_by_year[0].year | 2025 |
| counts_by_year[0].cited_by_count | 1 |
| counts_by_year[1].year | 2023 |
| counts_by_year[1].cited_by_count | 2 |
| counts_by_year[2].year | 2021 |
| counts_by_year[2].cited_by_count | 1 |
| counts_by_year[3].year | 2020 |
| counts_by_year[3].cited_by_count | 3 |
| counts_by_year[4].year | 2017 |
| counts_by_year[4].cited_by_count | 1 |
| locations_count | 1 |
| best_oa_location.id | doi:10.1101/078865 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4306402567 |
| best_oa_location.source.issn | |
| best_oa_location.source.type | repository |
| best_oa_location.source.is_oa | False |
| best_oa_location.source.issn_l | |
| best_oa_location.source.is_core | False |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| best_oa_location.source.host_organization | https://openalex.org/I2750212522 |
| best_oa_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| best_oa_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| best_oa_location.license | cc-by-nc-nd |
| best_oa_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2017/05/28/078865.full.pdf |
| best_oa_location.version | acceptedVersion |
| best_oa_location.raw_type | posted-content |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by-nc-nd |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | False |
| best_oa_location.raw_source_name | |
| best_oa_location.landing_page_url | https://doi.org/10.1101/078865 |
| primary_location.id | doi:10.1101/078865 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4306402567 |
| primary_location.source.issn | |
| primary_location.source.type | repository |
| primary_location.source.is_oa | False |
| primary_location.source.issn_l | |
| primary_location.source.is_core | False |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| primary_location.source.host_organization | https://openalex.org/I2750212522 |
| primary_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| primary_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| primary_location.license | cc-by-nc-nd |
| primary_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2017/05/28/078865.full.pdf |
| primary_location.version | acceptedVersion |
| primary_location.raw_type | posted-content |
| primary_location.license_id | https://openalex.org/licenses/cc-by-nc-nd |
| primary_location.is_accepted | True |
| primary_location.is_published | False |
| primary_location.raw_source_name | |
| primary_location.landing_page_url | https://doi.org/10.1101/078865 |
| publication_date | 2016-10-02 |
| publication_year | 2016 |
| referenced_works | https://openalex.org/W2259938310, https://openalex.org/W2076154138, https://openalex.org/W4213327946, https://openalex.org/W2078059415, https://openalex.org/W6652004108, https://openalex.org/W2951962793, https://openalex.org/W2330422707, https://openalex.org/W2115779804, https://openalex.org/W2101280135, https://openalex.org/W2064874359, https://openalex.org/W1841850981, https://openalex.org/W2257370818, https://openalex.org/W2049118388, https://openalex.org/W2133009026, https://openalex.org/W2113209482, https://openalex.org/W2130925862, https://openalex.org/W2288482887, https://openalex.org/W2030047242, https://openalex.org/W2141831115, https://openalex.org/W2022993325, https://openalex.org/W2015845481, https://openalex.org/W2024490058, https://openalex.org/W2054344840, https://openalex.org/W2109398701, https://openalex.org/W2095148075, https://openalex.org/W1844405070, https://openalex.org/W2139852278, https://openalex.org/W2152789823, https://openalex.org/W2148641246, https://openalex.org/W2065678033, https://openalex.org/W2166511841, https://openalex.org/W2160995259, https://openalex.org/W2018189081, https://openalex.org/W2225726427, https://openalex.org/W2010192073, https://openalex.org/W2070181801, https://openalex.org/W1901319035, https://openalex.org/W2141922205, https://openalex.org/W2013478372, https://openalex.org/W1983379765, https://openalex.org/W1965512299, https://openalex.org/W1993719373, https://openalex.org/W2168458391, https://openalex.org/W1989590412, https://openalex.org/W2102690320, https://openalex.org/W1014257459, https://openalex.org/W2112714218, https://openalex.org/W2162988145, https://openalex.org/W2028546773, https://openalex.org/W2479085189, https://openalex.org/W1775067635, https://openalex.org/W1578127299, https://openalex.org/W2141459724, https://openalex.org/W2098597355, https://openalex.org/W2014862380, https://openalex.org/W2169503014, https://openalex.org/W1963919142, https://openalex.org/W1865111710, https://openalex.org/W2138329729, https://openalex.org/W1997401532, https://openalex.org/W2035746095, https://openalex.org/W2112658198, https://openalex.org/W1989568128, https://openalex.org/W2120402847, https://openalex.org/W2100731712, https://openalex.org/W2128613449, https://openalex.org/W2124490243, https://openalex.org/W2531792466, https://openalex.org/W2340912524, https://openalex.org/W2587151667, https://openalex.org/W2469213152, https://openalex.org/W2209106767, https://openalex.org/W2264585211, https://openalex.org/W2591470648, https://openalex.org/W2019251848, https://openalex.org/W1986223165, https://openalex.org/W2096791516, https://openalex.org/W2033151532, https://openalex.org/W2133520037, https://openalex.org/W2918086501, https://openalex.org/W2163013660, https://openalex.org/W2306744213, https://openalex.org/W2122164926, https://openalex.org/W2949356259, https://openalex.org/W2586698568, https://openalex.org/W2486785549, https://openalex.org/W2007203150, https://openalex.org/W2519244818, https://openalex.org/W2064257734 |
| referenced_works_count | 89 |
| abstract_inverted_index.. | 235 |
| abstract_inverted_index.a | 18, 41, 75, 168, 192, 216, 302 |
| abstract_inverted_index.45 | 107, 338 |
| abstract_inverted_index.As | 270 |
| abstract_inverted_index.In | 296 |
| abstract_inverted_index.We | 111 |
| abstract_inverted_index.an | 47, 56, 101, 156, 342 |
| abstract_inverted_index.at | 210, 233 |
| abstract_inverted_index.be | 285 |
| abstract_inverted_index.in | 83, 124, 155, 178, 184, 267, 292, 314, 360 |
| abstract_inverted_index.of | 20, 27, 49, 55, 59, 77, 103, 135, 160, 180, 195, 223, 240, 256, 320, 329, 346, 353, 373 |
| abstract_inverted_index.on | 337 |
| abstract_inverted_index.or | 141 |
| abstract_inverted_index.to | 34, 64, 99, 197, 306, 355 |
| abstract_inverted_index.we | 44, 86, 150, 187, 204, 299, 349 |
| abstract_inverted_index.127 | 93 |
| abstract_inverted_index.DNA | 282 |
| abstract_inverted_index.Our | 165, 227 |
| abstract_inverted_index.and | 11, 22, 61, 96, 174, 245, 251, 309, 324, 341, 364 |
| abstract_inverted_index.are | 230 |
| abstract_inverted_index.for | 17, 130, 219, 253 |
| abstract_inverted_index.may | 284 |
| abstract_inverted_index.our | 50, 72, 88, 152 |
| abstract_inverted_index.set | 58 |
| abstract_inverted_index.the | 36, 84, 181, 211, 221, 257, 286, 315, 351, 361, 370 |
| abstract_inverted_index.use | 151 |
| abstract_inverted_index.yet | 260 |
| abstract_inverted_index.GWAS | 109 |
| abstract_inverted_index.LOAD | 172, 190 |
| abstract_inverted_index.SNPs | 196 |
| abstract_inverted_index.been | 249 |
| abstract_inverted_index.case | 158, 344 |
| abstract_inverted_index.cell | 21, 95, 137, 310 |
| abstract_inverted_index.data | 14, 91 |
| abstract_inverted_index.from | 3, 92 |
| abstract_inverted_index.have | 7, 248 |
| abstract_inverted_index.into | 30, 146, 369 |
| abstract_inverted_index.made | 8 |
| abstract_inverted_index.many | 243 |
| abstract_inverted_index.risk | 290 |
| abstract_inverted_index.show | 112, 188 |
| abstract_inverted_index.that | 113, 189, 206, 279 |
| abstract_inverted_index.this | 297 |
| abstract_inverted_index.tool | 218 |
| abstract_inverted_index.with | 74, 200 |
| abstract_inverted_index.work | 52 |
| abstract_inverted_index.(e.g. | 118, 139 |
| abstract_inverted_index.After | 70, 238 |
| abstract_inverted_index.Here, | 43 |
| abstract_inverted_index.apply | 87 |
| abstract_inverted_index.atlas | 102 |
| abstract_inverted_index.basis | 372 |
| abstract_inverted_index.data. | 326 |
| abstract_inverted_index.human | 225, 258, 293, 316, 362, 374 |
| abstract_inverted_index.known | 330 |
| abstract_inverted_index.large | 4 |
| abstract_inverted_index.level | 214 |
| abstract_inverted_index.organ | 115 |
| abstract_inverted_index.power | 123 |
| abstract_inverted_index.still | 263 |
| abstract_inverted_index.study | 159, 345 |
| abstract_inverted_index.these | 28 |
| abstract_inverted_index.types | 98, 129, 138 |
| abstract_inverted_index.using | 90 |
| abstract_inverted_index.while | 133 |
| abstract_inverted_index.years | 239 |
| abstract_inverted_index.Author | 236 |
| abstract_inverted_index.across | 106 |
| abstract_inverted_index.deeper | 144 |
| abstract_inverted_index.freely | 231 |
| abstract_inverted_index.genome | 39, 182, 208, 317, 363 |
| abstract_inverted_index.immune | 119 |
| abstract_inverted_index.method | 89 |
| abstract_inverted_index.paper, | 298 |
| abstract_inverted_index.proper | 261 |
| abstract_inverted_index.shares | 191 |
| abstract_inverted_index.single | 67, 212 |
| abstract_inverted_index.strong | 169 |
| abstract_inverted_index.system | 116 |
| abstract_inverted_index.tissue | 23, 68, 97, 128, 213, 308 |
| abstract_inverted_index.types. | 24 |
| abstract_inverted_index.(LOAD). | 164 |
| abstract_inverted_index.Summary | 237 |
| abstract_inverted_index.Through | 327 |
| abstract_inverted_index.ability | 352 |
| abstract_inverted_index.applied | 252 |
| abstract_inverted_index.between | 171 |
| abstract_inverted_index.broader | 114 |
| abstract_inverted_index.catalog | 76 |
| abstract_inverted_index.complex | 131, 224, 271, 294, 339, 375 |
| abstract_inverted_index.disease | 147, 163, 272 |
| abstract_inverted_index.diverse | 321 |
| abstract_inverted_index.efforts | 2 |
| abstract_inverted_index.genetic | 175, 371 |
| abstract_inverted_index.genome, | 259 |
| abstract_inverted_index.metrics | 33 |
| abstract_inverted_index.present | 45, 100 |
| abstract_inverted_index.primary | 287 |
| abstract_inverted_index.produce | 65 |
| abstract_inverted_index.provide | 143, 215, 365 |
| abstract_inverted_index.rapidly | 274 |
| abstract_inverted_index.regions | 179, 199, 288, 313 |
| abstract_inverted_index.remains | 40, 264 |
| abstract_inverted_index.similar | 193 |
| abstract_inverted_index.suggest | 167 |
| abstract_inverted_index.summary | 32 |
| abstract_inverted_index.system) | 120 |
| abstract_inverted_index.through | 53, 318 |
| abstract_inverted_index.traits, | 340 |
| abstract_inverted_index.traits. | 110 |
| abstract_inverted_index.variety | 19 |
| abstract_inverted_index.Abstract | 0 |
| abstract_inverted_index.B-cells) | 142 |
| abstract_inverted_index.However, | 25 |
| abstract_inverted_index.Overall, | 203 |
| abstract_inverted_index.analyses | 166, 336 |
| abstract_inverted_index.datasets | 29 |
| abstract_inverted_index.disease. | 202 |
| abstract_inverted_index.diseases | 132 |
| abstract_inverted_index.efforts, | 242 |
| abstract_inverted_index.elements | 80, 283 |
| abstract_inverted_index.etiology | 222 |
| abstract_inverted_index.evidence | 277 |
| abstract_inverted_index.expanded | 57 |
| abstract_inverted_index.identify | 307, 357 |
| abstract_inverted_index.in-depth | 157, 343 |
| abstract_inverted_index.increase | 121 |
| abstract_inverted_index.insights | 145, 368 |
| abstract_inverted_index.previous | 51 |
| abstract_inverted_index.publicly | 15 |
| abstract_inverted_index.regions, | 334 |
| abstract_inverted_index.regions. | 269 |
| abstract_inverted_index.relevant | 127 |
| abstract_inverted_index.research | 273 |
| abstract_inverted_index.suggests | 278 |
| abstract_inverted_index.valuable | 217 |
| abstract_inverted_index.variants | 176, 291 |
| abstract_inverted_index.advances, | 275 |
| abstract_inverted_index.available | 16, 232 |
| abstract_inverted_index.community | 241 |
| abstract_inverted_index.consortia | 6 |
| abstract_inverted_index.contained | 177 |
| abstract_inverted_index.developed | 250 |
| abstract_inverted_index.different | 94, 108 |
| abstract_inverted_index.diseases, | 348 |
| abstract_inverted_index.diseases. | 226, 295, 376 |
| abstract_inverted_index.effective | 31 |
| abstract_inverted_index.etiology. | 148 |
| abstract_inverted_index.extension | 48 |
| abstract_inverted_index.framework | 305 |
| abstract_inverted_index.harboring | 289 |
| abstract_inverted_index.introduce | 300 |
| abstract_inverted_index.monocytes | 140 |
| abstract_inverted_index.synthesis | 26 |
| abstract_inverted_index.unbiased, | 366 |
| abstract_inverted_index.Continuing | 1 |
| abstract_inverted_index.accurately | 356 |
| abstract_inverted_index.annotation | 13, 255, 262, 304 |
| abstract_inverted_index.approaches | 247 |
| abstract_inverted_index.categories | 117 |
| abstract_inverted_index.challenge. | 42 |
| abstract_inverted_index.connection | 170 |
| abstract_inverted_index.enrichment | 105, 335 |
| abstract_inverted_index.epigenomic | 10, 60, 323 |
| abstract_inverted_index.especially | 266 |
| abstract_inverted_index.functional | 37, 183, 254, 312 |
| abstract_inverted_index.identified | 82 |
| abstract_inverted_index.increasing | 276 |
| abstract_inverted_index.individual | 136 |
| abstract_inverted_index.integrated | 207 |
| abstract_inverted_index.late-onset | 161 |
| abstract_inverted_index.monocytes. | 185 |
| abstract_inverted_index.non-coding | 38, 79, 268, 280, 331 |
| abstract_inverted_index.previously | 81 |
| abstract_inverted_index.principled | 303 |
| abstract_inverted_index.regulatory | 281, 333 |
| abstract_inverted_index.validating | 71 |
| abstract_inverted_index.validation | 328 |
| abstract_inverted_index.annotations | 63, 73, 134, 154, 209, 229 |
| abstract_inverted_index.demonstrate | 205, 350 |
| abstract_inverted_index.genome-wide | 9, 367 |
| abstract_inverted_index.identifying | 125 |
| abstract_inverted_index.integration | 54, 319 |
| abstract_inverted_index.literature, | 85 |
| abstract_inverted_index.statistical | 122 |
| abstract_inverted_index.Furthermore, | 186 |
| abstract_inverted_index.annotations. | 69 |
| abstract_inverted_index.biologically | 126 |
| abstract_inverted_index.challenging, | 265 |
| abstract_inverted_index.characterize | 35 |
| abstract_inverted_index.experimental | 244 |
| abstract_inverted_index.heritability | 104, 173 |
| abstract_inverted_index.localization | 194 |
| abstract_inverted_index.Additionally, | 149 |
| abstract_inverted_index.Alzheimer’s | 162 |
| abstract_inverted_index.Parkinson’s | 201 |
| abstract_inverted_index.computational | 246 |
| abstract_inverted_index.functionality | 359 |
| abstract_inverted_index.international | 5 |
| abstract_inverted_index.type-specific | 311 |
| abstract_inverted_index.understanding | 220 |
| abstract_inverted_index.transcriptomic | 12, 62, 325 |
| abstract_inverted_index.high-throughput | 322 |
| abstract_inverted_index.tissue-specific | 78, 332, 358 |
| abstract_inverted_index.GenoSkyline-Plus | 153, 228, 354 |
| abstract_inverted_index.high-resolution, | 66 |
| abstract_inverted_index.GenoSkyline-Plus, | 46, 301 |
| abstract_inverted_index.neurodegenerative | 347 |
| abstract_inverted_index.monocyte-functional | 198 |
| abstract_inverted_index.http://genocanyon.med.yale.edu/GenoSkyline | 234 |
| cited_by_percentile_year.max | 97 |
| cited_by_percentile_year.min | 89 |
| corresponding_author_ids | https://openalex.org/A5074828414 |
| countries_distinct_count | 2 |
| institutions_distinct_count | 12 |
| corresponding_institution_ids | https://openalex.org/I32971472, https://openalex.org/I4210120339 |
| sustainable_development_goals[0].id | https://metadata.un.org/sdg/17 |
| sustainable_development_goals[0].score | 0.5699999928474426 |
| sustainable_development_goals[0].display_name | Partnerships for the goals |
| citation_normalized_percentile.value | 0.7135641 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |