The Proteome of the Isolated Chlamydia trachomatis Containing Vacuole Reveals a Complex Trafficking Platform Enriched for Retromer Components Article Swipe
 YOU?
·
· 2015
· Open Access
·
· DOI: https://doi.org/10.1371/journal.ppat.1004883
YOU?
·
· 2015
· Open Access
·
· DOI: https://doi.org/10.1371/journal.ppat.1004883
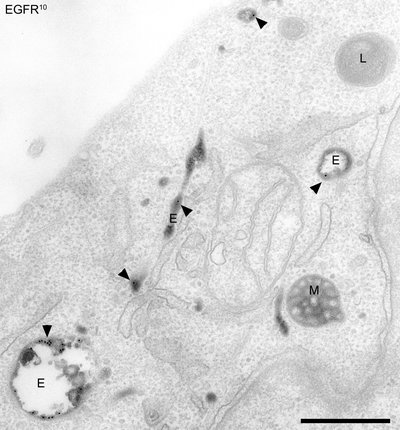
Chlamydia trachomatis is an important human pathogen that replicates inside the infected host cell in a unique vacuole, the inclusion. The formation of this intracellular bacterial niche is essential for productive Chlamydia infections. Despite its importance for Chlamydia biology, a holistic view on the protein composition of the inclusion, including its membrane, is currently missing. Here we describe the host cell-derived proteome of isolated C. trachomatis inclusions by quantitative proteomics. Computational analysis indicated that the inclusion is a complex intracellular trafficking platform that interacts with host cells' antero- and retrograde trafficking pathways. Furthermore, the inclusion is highly enriched for sorting nexins of the SNX-BAR retromer, a complex essential for retrograde trafficking. Functional studies showed that in particular, SNX5 controls the C. trachomatis infection and that retrograde trafficking is essential for infectious progeny formation. In summary, these findings suggest that C. trachomatis hijacks retrograde pathways for effective infection.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.1371/journal.ppat.1004883
- https://journals.plos.org/plospathogens/article/file?id=10.1371/journal.ppat.1004883&type=printable
- OA Status
- gold
- Cited By
- 76
- References
- 58
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W587240085
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W587240085Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1371/journal.ppat.1004883Digital Object Identifier
- Title
-
The Proteome of the Isolated Chlamydia trachomatis Containing Vacuole Reveals a Complex Trafficking Platform Enriched for Retromer ComponentsWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2015Year of publication
- Publication date
-
2015-06-04Full publication date if available
- Authors
-
Lukas Aeberhard, Sebastian Banhart, Martina Fischer, Nico Jehmlich, Laura J. Rose, Sophia Koch, Michael Laue, Bernhard Y. Renard, Frank Schmidt, Dagmar HeuerList of authors in order
- Landing page
-
https://doi.org/10.1371/journal.ppat.1004883Publisher landing page
- PDF URL
-
https://journals.plos.org/plospathogens/article/file?id=10.1371/journal.ppat.1004883&type=printableDirect link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
goldOpen access status per OpenAlex
- OA URL
-
https://journals.plos.org/plospathogens/article/file?id=10.1371/journal.ppat.1004883&type=printableDirect OA link when available
- Concepts
-
Retromer, Chlamydia trachomatis, Vacuole, Biology, Proteome, Cell biology, Endosome, Intracellular, Protein targeting, Inclusion bodies, Chlamydia, Proteomics, Transport protein, Host–pathogen interaction, Microbiology, Membrane protein, Virology, Genetics, Gene, Escherichia coli, Membrane, Cytoplasm, VirulenceTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
76Total citation count in OpenAlex
- Citations by year (recent)
-
2025: 6, 2024: 3, 2023: 4, 2022: 2, 2021: 6Per-year citation counts (last 5 years)
- References (count)
-
58Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W587240085 |
|---|---|
| doi | https://doi.org/10.1371/journal.ppat.1004883 |
| ids.doi | https://doi.org/10.1371/journal.ppat.1004883 |
| ids.mag | 587240085 |
| ids.pmid | https://pubmed.ncbi.nlm.nih.gov/26042774 |
| ids.openalex | https://openalex.org/W587240085 |
| fwci | 8.01033428 |
| mesh[0].qualifier_ui | |
| mesh[0].descriptor_ui | D002469 |
| mesh[0].is_major_topic | False |
| mesh[0].qualifier_name | |
| mesh[0].descriptor_name | Cell Separation |
| mesh[1].qualifier_ui | Q000378 |
| mesh[1].descriptor_ui | D002690 |
| mesh[1].is_major_topic | False |
| mesh[1].qualifier_name | metabolism |
| mesh[1].descriptor_name | Chlamydia Infections |
| mesh[2].qualifier_ui | Q000378 |
| mesh[2].descriptor_ui | D002692 |
| mesh[2].is_major_topic | False |
| mesh[2].qualifier_name | metabolism |
| mesh[2].descriptor_name | Chlamydia trachomatis |
| mesh[3].qualifier_ui | Q000472 |
| mesh[3].descriptor_ui | D002692 |
| mesh[3].is_major_topic | False |
| mesh[3].qualifier_name | pathogenicity |
| mesh[3].descriptor_name | Chlamydia trachomatis |
| mesh[4].qualifier_ui | |
| mesh[4].descriptor_ui | D005434 |
| mesh[4].is_major_topic | False |
| mesh[4].qualifier_name | |
| mesh[4].descriptor_name | Flow Cytometry |
| mesh[5].qualifier_ui | |
| mesh[5].descriptor_ui | D006367 |
| mesh[5].is_major_topic | False |
| mesh[5].qualifier_name | |
| mesh[5].descriptor_name | HeLa Cells |
| mesh[6].qualifier_ui | |
| mesh[6].descriptor_ui | D006801 |
| mesh[6].is_major_topic | False |
| mesh[6].qualifier_name | |
| mesh[6].descriptor_name | Humans |
| mesh[7].qualifier_ui | Q000378 |
| mesh[7].descriptor_ui | D002479 |
| mesh[7].is_major_topic | False |
| mesh[7].qualifier_name | metabolism |
| mesh[7].descriptor_name | Inclusion Bodies |
| mesh[8].qualifier_ui | |
| mesh[8].descriptor_ui | D021381 |
| mesh[8].is_major_topic | False |
| mesh[8].qualifier_name | |
| mesh[8].descriptor_name | Protein Transport |
| mesh[9].qualifier_ui | |
| mesh[9].descriptor_ui | D020543 |
| mesh[9].is_major_topic | False |
| mesh[9].qualifier_name | |
| mesh[9].descriptor_name | Proteome |
| mesh[10].qualifier_ui | |
| mesh[10].descriptor_ui | D040901 |
| mesh[10].is_major_topic | False |
| mesh[10].qualifier_name | |
| mesh[10].descriptor_name | Proteomics |
| mesh[11].qualifier_ui | |
| mesh[11].descriptor_ui | D034741 |
| mesh[11].is_major_topic | False |
| mesh[11].qualifier_name | |
| mesh[11].descriptor_name | RNA, Small Interfering |
| mesh[12].qualifier_ui | |
| mesh[12].descriptor_ui | D053719 |
| mesh[12].is_major_topic | False |
| mesh[12].qualifier_name | |
| mesh[12].descriptor_name | Tandem Mass Spectrometry |
| mesh[13].qualifier_ui | |
| mesh[13].descriptor_ui | D014162 |
| mesh[13].is_major_topic | False |
| mesh[13].qualifier_name | |
| mesh[13].descriptor_name | Transfection |
| mesh[14].qualifier_ui | Q000378 |
| mesh[14].descriptor_ui | D014617 |
| mesh[14].is_major_topic | False |
| mesh[14].qualifier_name | metabolism |
| mesh[14].descriptor_name | Vacuoles |
| mesh[15].qualifier_ui | |
| mesh[15].descriptor_ui | D002469 |
| mesh[15].is_major_topic | False |
| mesh[15].qualifier_name | |
| mesh[15].descriptor_name | Cell Separation |
| mesh[16].qualifier_ui | Q000378 |
| mesh[16].descriptor_ui | D002690 |
| mesh[16].is_major_topic | False |
| mesh[16].qualifier_name | metabolism |
| mesh[16].descriptor_name | Chlamydia Infections |
| mesh[17].qualifier_ui | Q000378 |
| mesh[17].descriptor_ui | D002692 |
| mesh[17].is_major_topic | False |
| mesh[17].qualifier_name | metabolism |
| mesh[17].descriptor_name | Chlamydia trachomatis |
| mesh[18].qualifier_ui | Q000472 |
| mesh[18].descriptor_ui | D002692 |
| mesh[18].is_major_topic | False |
| mesh[18].qualifier_name | pathogenicity |
| mesh[18].descriptor_name | Chlamydia trachomatis |
| mesh[19].qualifier_ui | |
| mesh[19].descriptor_ui | D005434 |
| mesh[19].is_major_topic | False |
| mesh[19].qualifier_name | |
| mesh[19].descriptor_name | Flow Cytometry |
| mesh[20].qualifier_ui | |
| mesh[20].descriptor_ui | D006367 |
| mesh[20].is_major_topic | False |
| mesh[20].qualifier_name | |
| mesh[20].descriptor_name | HeLa Cells |
| mesh[21].qualifier_ui | |
| mesh[21].descriptor_ui | D006801 |
| mesh[21].is_major_topic | False |
| mesh[21].qualifier_name | |
| mesh[21].descriptor_name | Humans |
| mesh[22].qualifier_ui | Q000378 |
| mesh[22].descriptor_ui | D002479 |
| mesh[22].is_major_topic | False |
| mesh[22].qualifier_name | metabolism |
| mesh[22].descriptor_name | Inclusion Bodies |
| mesh[23].qualifier_ui | |
| mesh[23].descriptor_ui | D021381 |
| mesh[23].is_major_topic | False |
| mesh[23].qualifier_name | |
| mesh[23].descriptor_name | Protein Transport |
| mesh[24].qualifier_ui | |
| mesh[24].descriptor_ui | D020543 |
| mesh[24].is_major_topic | False |
| mesh[24].qualifier_name | |
| mesh[24].descriptor_name | Proteome |
| mesh[25].qualifier_ui | |
| mesh[25].descriptor_ui | D040901 |
| mesh[25].is_major_topic | False |
| mesh[25].qualifier_name | |
| mesh[25].descriptor_name | Proteomics |
| mesh[26].qualifier_ui | |
| mesh[26].descriptor_ui | D034741 |
| mesh[26].is_major_topic | False |
| mesh[26].qualifier_name | |
| mesh[26].descriptor_name | RNA, Small Interfering |
| mesh[27].qualifier_ui | |
| mesh[27].descriptor_ui | D053719 |
| mesh[27].is_major_topic | False |
| mesh[27].qualifier_name | |
| mesh[27].descriptor_name | Tandem Mass Spectrometry |
| mesh[28].qualifier_ui | |
| mesh[28].descriptor_ui | D014162 |
| mesh[28].is_major_topic | False |
| mesh[28].qualifier_name | |
| mesh[28].descriptor_name | Transfection |
| mesh[29].qualifier_ui | Q000378 |
| mesh[29].descriptor_ui | D014617 |
| mesh[29].is_major_topic | False |
| mesh[29].qualifier_name | metabolism |
| mesh[29].descriptor_name | Vacuoles |
| type | article |
| title | The Proteome of the Isolated Chlamydia trachomatis Containing Vacuole Reveals a Complex Trafficking Platform Enriched for Retromer Components |
| biblio.issue | 6 |
| biblio.volume | 11 |
| biblio.last_page | e1004883 |
| biblio.first_page | e1004883 |
| topics[0].id | https://openalex.org/T10428 |
| topics[0].field.id | https://openalex.org/fields/24 |
| topics[0].field.display_name | Immunology and Microbiology |
| topics[0].score | 0.9997000098228455 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/2404 |
| topics[0].subfield.display_name | Microbiology |
| topics[0].display_name | Reproductive tract infections research |
| topics[1].id | https://openalex.org/T10066 |
| topics[1].field.id | https://openalex.org/fields/13 |
| topics[1].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[1].score | 0.9550999999046326 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1312 |
| topics[1].subfield.display_name | Molecular Biology |
| topics[1].display_name | Gut microbiota and health |
| topics[2].id | https://openalex.org/T11232 |
| topics[2].field.id | https://openalex.org/fields/27 |
| topics[2].field.display_name | Medicine |
| topics[2].score | 0.9222000241279602 |
| topics[2].domain.id | https://openalex.org/domains/4 |
| topics[2].domain.display_name | Health Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/2713 |
| topics[2].subfield.display_name | Epidemiology |
| topics[2].display_name | Urinary Tract Infections Management |
| is_xpac | False |
| apc_list.value | 2655 |
| apc_list.currency | USD |
| apc_list.value_usd | 2655 |
| apc_paid.value | 2655 |
| apc_paid.currency | USD |
| apc_paid.value_usd | 2655 |
| concepts[0].id | https://openalex.org/C145458877 |
| concepts[0].level | 4 |
| concepts[0].score | 0.9151167869567871 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q7317044 |
| concepts[0].display_name | Retromer |
| concepts[1].id | https://openalex.org/C2777391075 |
| concepts[1].level | 2 |
| concepts[1].score | 0.8632645010948181 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q131065 |
| concepts[1].display_name | Chlamydia trachomatis |
| concepts[2].id | https://openalex.org/C102568950 |
| concepts[2].level | 3 |
| concepts[2].score | 0.8042076230049133 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q127702 |
| concepts[2].display_name | Vacuole |
| concepts[3].id | https://openalex.org/C86803240 |
| concepts[3].level | 0 |
| concepts[3].score | 0.7202627658843994 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[3].display_name | Biology |
| concepts[4].id | https://openalex.org/C104397665 |
| concepts[4].level | 2 |
| concepts[4].score | 0.6935510039329529 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q860947 |
| concepts[4].display_name | Proteome |
| concepts[5].id | https://openalex.org/C95444343 |
| concepts[5].level | 1 |
| concepts[5].score | 0.587357759475708 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q7141 |
| concepts[5].display_name | Cell biology |
| concepts[6].id | https://openalex.org/C102747710 |
| concepts[6].level | 3 |
| concepts[6].score | 0.5636380910873413 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q824237 |
| concepts[6].display_name | Endosome |
| concepts[7].id | https://openalex.org/C79879829 |
| concepts[7].level | 2 |
| concepts[7].score | 0.5509365200996399 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q5571762 |
| concepts[7].display_name | Intracellular |
| concepts[8].id | https://openalex.org/C559089031 |
| concepts[8].level | 4 |
| concepts[8].score | 0.5207490921020508 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q658882 |
| concepts[8].display_name | Protein targeting |
| concepts[9].id | https://openalex.org/C203750385 |
| concepts[9].level | 4 |
| concepts[9].score | 0.5072476267814636 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q1308970 |
| concepts[9].display_name | Inclusion bodies |
| concepts[10].id | https://openalex.org/C2780122209 |
| concepts[10].level | 2 |
| concepts[10].score | 0.45755186676979065 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q153356 |
| concepts[10].display_name | Chlamydia |
| concepts[11].id | https://openalex.org/C46111723 |
| concepts[11].level | 3 |
| concepts[11].score | 0.44877469539642334 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q471857 |
| concepts[11].display_name | Proteomics |
| concepts[12].id | https://openalex.org/C57581600 |
| concepts[12].level | 2 |
| concepts[12].score | 0.4194521903991699 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q2449730 |
| concepts[12].display_name | Transport protein |
| concepts[13].id | https://openalex.org/C2778375072 |
| concepts[13].level | 4 |
| concepts[13].score | 0.41610926389694214 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q5909198 |
| concepts[13].display_name | Host–pathogen interaction |
| concepts[14].id | https://openalex.org/C89423630 |
| concepts[14].level | 1 |
| concepts[14].score | 0.32021045684814453 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q7193 |
| concepts[14].display_name | Microbiology |
| concepts[15].id | https://openalex.org/C144647389 |
| concepts[15].level | 3 |
| concepts[15].score | 0.2525373697280884 |
| concepts[15].wikidata | https://www.wikidata.org/wiki/Q423042 |
| concepts[15].display_name | Membrane protein |
| concepts[16].id | https://openalex.org/C159047783 |
| concepts[16].level | 1 |
| concepts[16].score | 0.24157431721687317 |
| concepts[16].wikidata | https://www.wikidata.org/wiki/Q7215 |
| concepts[16].display_name | Virology |
| concepts[17].id | https://openalex.org/C54355233 |
| concepts[17].level | 1 |
| concepts[17].score | 0.13036701083183289 |
| concepts[17].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[17].display_name | Genetics |
| concepts[18].id | https://openalex.org/C104317684 |
| concepts[18].level | 2 |
| concepts[18].score | 0.09542974829673767 |
| concepts[18].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[18].display_name | Gene |
| concepts[19].id | https://openalex.org/C547475151 |
| concepts[19].level | 3 |
| concepts[19].score | 0.0 |
| concepts[19].wikidata | https://www.wikidata.org/wiki/Q25419 |
| concepts[19].display_name | Escherichia coli |
| concepts[20].id | https://openalex.org/C41625074 |
| concepts[20].level | 2 |
| concepts[20].score | 0.0 |
| concepts[20].wikidata | https://www.wikidata.org/wiki/Q176088 |
| concepts[20].display_name | Membrane |
| concepts[21].id | https://openalex.org/C190062978 |
| concepts[21].level | 2 |
| concepts[21].score | 0.0 |
| concepts[21].wikidata | https://www.wikidata.org/wiki/Q79899 |
| concepts[21].display_name | Cytoplasm |
| concepts[22].id | https://openalex.org/C60987743 |
| concepts[22].level | 3 |
| concepts[22].score | 0.0 |
| concepts[22].wikidata | https://www.wikidata.org/wiki/Q1460232 |
| concepts[22].display_name | Virulence |
| keywords[0].id | https://openalex.org/keywords/retromer |
| keywords[0].score | 0.9151167869567871 |
| keywords[0].display_name | Retromer |
| keywords[1].id | https://openalex.org/keywords/chlamydia-trachomatis |
| keywords[1].score | 0.8632645010948181 |
| keywords[1].display_name | Chlamydia trachomatis |
| keywords[2].id | https://openalex.org/keywords/vacuole |
| keywords[2].score | 0.8042076230049133 |
| keywords[2].display_name | Vacuole |
| keywords[3].id | https://openalex.org/keywords/biology |
| keywords[3].score | 0.7202627658843994 |
| keywords[3].display_name | Biology |
| keywords[4].id | https://openalex.org/keywords/proteome |
| keywords[4].score | 0.6935510039329529 |
| keywords[4].display_name | Proteome |
| keywords[5].id | https://openalex.org/keywords/cell-biology |
| keywords[5].score | 0.587357759475708 |
| keywords[5].display_name | Cell biology |
| keywords[6].id | https://openalex.org/keywords/endosome |
| keywords[6].score | 0.5636380910873413 |
| keywords[6].display_name | Endosome |
| keywords[7].id | https://openalex.org/keywords/intracellular |
| keywords[7].score | 0.5509365200996399 |
| keywords[7].display_name | Intracellular |
| keywords[8].id | https://openalex.org/keywords/protein-targeting |
| keywords[8].score | 0.5207490921020508 |
| keywords[8].display_name | Protein targeting |
| keywords[9].id | https://openalex.org/keywords/inclusion-bodies |
| keywords[9].score | 0.5072476267814636 |
| keywords[9].display_name | Inclusion bodies |
| keywords[10].id | https://openalex.org/keywords/chlamydia |
| keywords[10].score | 0.45755186676979065 |
| keywords[10].display_name | Chlamydia |
| keywords[11].id | https://openalex.org/keywords/proteomics |
| keywords[11].score | 0.44877469539642334 |
| keywords[11].display_name | Proteomics |
| keywords[12].id | https://openalex.org/keywords/transport-protein |
| keywords[12].score | 0.4194521903991699 |
| keywords[12].display_name | Transport protein |
| keywords[13].id | https://openalex.org/keywords/host–pathogen-interaction |
| keywords[13].score | 0.41610926389694214 |
| keywords[13].display_name | Host–pathogen interaction |
| keywords[14].id | https://openalex.org/keywords/microbiology |
| keywords[14].score | 0.32021045684814453 |
| keywords[14].display_name | Microbiology |
| keywords[15].id | https://openalex.org/keywords/membrane-protein |
| keywords[15].score | 0.2525373697280884 |
| keywords[15].display_name | Membrane protein |
| keywords[16].id | https://openalex.org/keywords/virology |
| keywords[16].score | 0.24157431721687317 |
| keywords[16].display_name | Virology |
| keywords[17].id | https://openalex.org/keywords/genetics |
| keywords[17].score | 0.13036701083183289 |
| keywords[17].display_name | Genetics |
| keywords[18].id | https://openalex.org/keywords/gene |
| keywords[18].score | 0.09542974829673767 |
| keywords[18].display_name | Gene |
| language | en |
| locations[0].id | doi:10.1371/journal.ppat.1004883 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S2004986 |
| locations[0].source.issn | 1553-7366, 1553-7374 |
| locations[0].source.type | journal |
| locations[0].source.is_oa | True |
| locations[0].source.issn_l | 1553-7366 |
| locations[0].source.is_core | True |
| locations[0].source.is_in_doaj | True |
| locations[0].source.display_name | PLoS Pathogens |
| locations[0].source.host_organization | https://openalex.org/P4310315706 |
| locations[0].source.host_organization_name | Public Library of Science |
| locations[0].source.host_organization_lineage | https://openalex.org/P4310315706 |
| locations[0].license | cc-by |
| locations[0].pdf_url | https://journals.plos.org/plospathogens/article/file?id=10.1371/journal.ppat.1004883&type=printable |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | PLOS Pathogens |
| locations[0].landing_page_url | https://doi.org/10.1371/journal.ppat.1004883 |
| locations[1].id | pmid:26042774 |
| locations[1].is_oa | False |
| locations[1].source.id | https://openalex.org/S4306525036 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | PubMed |
| locations[1].source.host_organization | https://openalex.org/I1299303238 |
| locations[1].source.host_organization_name | National Institutes of Health |
| locations[1].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[1].license | |
| locations[1].pdf_url | |
| locations[1].version | publishedVersion |
| locations[1].raw_type | |
| locations[1].license_id | |
| locations[1].is_accepted | True |
| locations[1].is_published | True |
| locations[1].raw_source_name | PLoS pathogens |
| locations[1].landing_page_url | https://pubmed.ncbi.nlm.nih.gov/26042774 |
| locations[2].id | pmh:oai:doaj.org/article:17721e1ba89a4290b10a63c1b4d69283 |
| locations[2].is_oa | True |
| locations[2].source.id | https://openalex.org/S4306401280 |
| locations[2].source.issn | |
| locations[2].source.type | repository |
| locations[2].source.is_oa | False |
| locations[2].source.issn_l | |
| locations[2].source.is_core | False |
| locations[2].source.is_in_doaj | False |
| locations[2].source.display_name | DOAJ (DOAJ: Directory of Open Access Journals) |
| locations[2].source.host_organization | |
| locations[2].source.host_organization_name | |
| locations[2].source.host_organization_lineage | |
| locations[2].license | cc-by-sa |
| locations[2].pdf_url | |
| locations[2].version | submittedVersion |
| locations[2].raw_type | article |
| locations[2].license_id | https://openalex.org/licenses/cc-by-sa |
| locations[2].is_accepted | False |
| locations[2].is_published | False |
| locations[2].raw_source_name | PLoS Pathogens, Vol 11, Iss 6, p e1004883 (2015) |
| locations[2].landing_page_url | https://doaj.org/article/17721e1ba89a4290b10a63c1b4d69283 |
| locations[3].id | pmh:oai:pubmedcentral.nih.gov:4456400 |
| locations[3].is_oa | True |
| locations[3].source.id | https://openalex.org/S2764455111 |
| locations[3].source.issn | |
| locations[3].source.type | repository |
| locations[3].source.is_oa | False |
| locations[3].source.issn_l | |
| locations[3].source.is_core | False |
| locations[3].source.is_in_doaj | False |
| locations[3].source.display_name | PubMed Central |
| locations[3].source.host_organization | https://openalex.org/I1299303238 |
| locations[3].source.host_organization_name | National Institutes of Health |
| locations[3].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[3].license | cc-by |
| locations[3].pdf_url | |
| locations[3].version | submittedVersion |
| locations[3].raw_type | Text |
| locations[3].license_id | https://openalex.org/licenses/cc-by |
| locations[3].is_accepted | False |
| locations[3].is_published | False |
| locations[3].raw_source_name | PLoS Pathog |
| locations[3].landing_page_url | https://www.ncbi.nlm.nih.gov/pmc/articles/4456400 |
| locations[4].id | pmh:oai:figshare.com:article/1437231 |
| locations[4].is_oa | True |
| locations[4].source.id | https://openalex.org/S4306400572 |
| locations[4].source.issn | |
| locations[4].source.type | repository |
| locations[4].source.is_oa | False |
| locations[4].source.issn_l | |
| locations[4].source.is_core | False |
| locations[4].source.is_in_doaj | False |
| locations[4].source.display_name | OPAL (Open@LaTrobe) (La Trobe University) |
| locations[4].source.host_organization | https://openalex.org/I196829312 |
| locations[4].source.host_organization_name | La Trobe University |
| locations[4].source.host_organization_lineage | https://openalex.org/I196829312 |
| locations[4].license | cc-by |
| locations[4].pdf_url | |
| locations[4].version | submittedVersion |
| locations[4].raw_type | Dataset |
| locations[4].license_id | https://openalex.org/licenses/cc-by |
| locations[4].is_accepted | False |
| locations[4].is_published | False |
| locations[4].raw_source_name | |
| locations[4].landing_page_url | https://figshare.com/articles/dataset/The_Proteome_of_the_Isolated_Chlamydia_trachomatis_Containing_Vacuole_Reveals_a_Complex_Trafficking_Platform_Enriched_for_Retromer_Components/1437231 |
| indexed_in | crossref, doaj, pubmed |
| authorships[0].author.id | https://openalex.org/A5012124891 |
| authorships[0].author.orcid | |
| authorships[0].author.display_name | Lukas Aeberhard |
| authorships[0].countries | DE |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I24359323 |
| authorships[0].affiliations[0].raw_affiliation_string | Junior Research Group `Sexually Transmitted Bacterial Pathogens´, Robert Koch Institute, Berlin, Germany |
| authorships[0].institutions[0].id | https://openalex.org/I24359323 |
| authorships[0].institutions[0].ror | https://ror.org/01k5qnb77 |
| authorships[0].institutions[0].type | government |
| authorships[0].institutions[0].lineage | https://openalex.org/I24359323 |
| authorships[0].institutions[0].country_code | DE |
| authorships[0].institutions[0].display_name | Robert Koch Institute |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Lukas Aeberhard |
| authorships[0].is_corresponding | True |
| authorships[0].raw_affiliation_strings | Junior Research Group `Sexually Transmitted Bacterial Pathogens´, Robert Koch Institute, Berlin, Germany |
| authorships[1].author.id | https://openalex.org/A5049628915 |
| authorships[1].author.orcid | https://orcid.org/0000-0003-4720-9330 |
| authorships[1].author.display_name | Sebastian Banhart |
| authorships[1].countries | DE |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I24359323 |
| authorships[1].affiliations[0].raw_affiliation_string | Junior Research Group `Sexually Transmitted Bacterial Pathogens´, Robert Koch Institute, Berlin, Germany |
| authorships[1].institutions[0].id | https://openalex.org/I24359323 |
| authorships[1].institutions[0].ror | https://ror.org/01k5qnb77 |
| authorships[1].institutions[0].type | government |
| authorships[1].institutions[0].lineage | https://openalex.org/I24359323 |
| authorships[1].institutions[0].country_code | DE |
| authorships[1].institutions[0].display_name | Robert Koch Institute |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Sebastian Banhart |
| authorships[1].is_corresponding | True |
| authorships[1].raw_affiliation_strings | Junior Research Group `Sexually Transmitted Bacterial Pathogens´, Robert Koch Institute, Berlin, Germany |
| authorships[2].author.id | https://openalex.org/A5110966455 |
| authorships[2].author.orcid | |
| authorships[2].author.display_name | Martina Fischer |
| authorships[2].countries | DE |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I24359323 |
| authorships[2].affiliations[0].raw_affiliation_string | Junior Research Group `Bioinformatics´, Robert Koch Institute, Berlin, Germany |
| authorships[2].institutions[0].id | https://openalex.org/I24359323 |
| authorships[2].institutions[0].ror | https://ror.org/01k5qnb77 |
| authorships[2].institutions[0].type | government |
| authorships[2].institutions[0].lineage | https://openalex.org/I24359323 |
| authorships[2].institutions[0].country_code | DE |
| authorships[2].institutions[0].display_name | Robert Koch Institute |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Martina Fischer |
| authorships[2].is_corresponding | True |
| authorships[2].raw_affiliation_strings | Junior Research Group `Bioinformatics´, Robert Koch Institute, Berlin, Germany |
| authorships[3].author.id | https://openalex.org/A5018412626 |
| authorships[3].author.orcid | https://orcid.org/0000-0002-5638-6868 |
| authorships[3].author.display_name | Nico Jehmlich |
| authorships[3].countries | DE |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I36522303 |
| authorships[3].affiliations[0].raw_affiliation_string | Interfaculty Institute for Genetics and Functional Genomics, Ernst-Moritz-Arndt-University, Greifswald, Germany |
| authorships[3].institutions[0].id | https://openalex.org/I36522303 |
| authorships[3].institutions[0].ror | https://ror.org/00r1edq15 |
| authorships[3].institutions[0].type | education |
| authorships[3].institutions[0].lineage | https://openalex.org/I36522303 |
| authorships[3].institutions[0].country_code | DE |
| authorships[3].institutions[0].display_name | Universität Greifswald |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Nico Jehmlich |
| authorships[3].is_corresponding | True |
| authorships[3].raw_affiliation_strings | Interfaculty Institute for Genetics and Functional Genomics, Ernst-Moritz-Arndt-University, Greifswald, Germany |
| authorships[4].author.id | https://openalex.org/A5066787801 |
| authorships[4].author.orcid | https://orcid.org/0000-0002-1428-6594 |
| authorships[4].author.display_name | Laura J. Rose |
| authorships[4].countries | DE |
| authorships[4].affiliations[0].institution_ids | https://openalex.org/I24359323 |
| authorships[4].affiliations[0].raw_affiliation_string | Junior Research Group `Sexually Transmitted Bacterial Pathogens´, Robert Koch Institute, Berlin, Germany |
| authorships[4].institutions[0].id | https://openalex.org/I24359323 |
| authorships[4].institutions[0].ror | https://ror.org/01k5qnb77 |
| authorships[4].institutions[0].type | government |
| authorships[4].institutions[0].lineage | https://openalex.org/I24359323 |
| authorships[4].institutions[0].country_code | DE |
| authorships[4].institutions[0].display_name | Robert Koch Institute |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Laura Rose |
| authorships[4].is_corresponding | True |
| authorships[4].raw_affiliation_strings | Junior Research Group `Sexually Transmitted Bacterial Pathogens´, Robert Koch Institute, Berlin, Germany |
| authorships[5].author.id | https://openalex.org/A5109803468 |
| authorships[5].author.orcid | |
| authorships[5].author.display_name | Sophia Koch |
| authorships[5].countries | DE |
| authorships[5].affiliations[0].institution_ids | https://openalex.org/I24359323 |
| authorships[5].affiliations[0].raw_affiliation_string | Junior Research Group `Sexually Transmitted Bacterial Pathogens´, Robert Koch Institute, Berlin, Germany |
| authorships[5].institutions[0].id | https://openalex.org/I24359323 |
| authorships[5].institutions[0].ror | https://ror.org/01k5qnb77 |
| authorships[5].institutions[0].type | government |
| authorships[5].institutions[0].lineage | https://openalex.org/I24359323 |
| authorships[5].institutions[0].country_code | DE |
| authorships[5].institutions[0].display_name | Robert Koch Institute |
| authorships[5].author_position | middle |
| authorships[5].raw_author_name | Sophia Koch |
| authorships[5].is_corresponding | True |
| authorships[5].raw_affiliation_strings | Junior Research Group `Sexually Transmitted Bacterial Pathogens´, Robert Koch Institute, Berlin, Germany |
| authorships[6].author.id | https://openalex.org/A5031672904 |
| authorships[6].author.orcid | https://orcid.org/0000-0002-6474-9139 |
| authorships[6].author.display_name | Michael Laue |
| authorships[6].countries | DE |
| authorships[6].affiliations[0].institution_ids | https://openalex.org/I24359323 |
| authorships[6].affiliations[0].raw_affiliation_string | Advanced Light and Electron Microscopy (ZBS 4), Robert Koch Institute, Berlin, Germany |
| authorships[6].institutions[0].id | https://openalex.org/I24359323 |
| authorships[6].institutions[0].ror | https://ror.org/01k5qnb77 |
| authorships[6].institutions[0].type | government |
| authorships[6].institutions[0].lineage | https://openalex.org/I24359323 |
| authorships[6].institutions[0].country_code | DE |
| authorships[6].institutions[0].display_name | Robert Koch Institute |
| authorships[6].author_position | middle |
| authorships[6].raw_author_name | Michael Laue |
| authorships[6].is_corresponding | True |
| authorships[6].raw_affiliation_strings | Advanced Light and Electron Microscopy (ZBS 4), Robert Koch Institute, Berlin, Germany |
| authorships[7].author.id | https://openalex.org/A5005996110 |
| authorships[7].author.orcid | https://orcid.org/0000-0003-4589-9809 |
| authorships[7].author.display_name | Bernhard Y. Renard |
| authorships[7].countries | DE |
| authorships[7].affiliations[0].institution_ids | https://openalex.org/I24359323 |
| authorships[7].affiliations[0].raw_affiliation_string | Junior Research Group `Bioinformatics´, Robert Koch Institute, Berlin, Germany |
| authorships[7].institutions[0].id | https://openalex.org/I24359323 |
| authorships[7].institutions[0].ror | https://ror.org/01k5qnb77 |
| authorships[7].institutions[0].type | government |
| authorships[7].institutions[0].lineage | https://openalex.org/I24359323 |
| authorships[7].institutions[0].country_code | DE |
| authorships[7].institutions[0].display_name | Robert Koch Institute |
| authorships[7].author_position | middle |
| authorships[7].raw_author_name | Bernhard Y. Renard |
| authorships[7].is_corresponding | True |
| authorships[7].raw_affiliation_strings | Junior Research Group `Bioinformatics´, Robert Koch Institute, Berlin, Germany |
| authorships[8].author.id | https://openalex.org/A5045874625 |
| authorships[8].author.orcid | https://orcid.org/0000-0001-5406-3988 |
| authorships[8].author.display_name | Frank Schmidt |
| authorships[8].countries | DE |
| authorships[8].affiliations[0].institution_ids | https://openalex.org/I36522303 |
| authorships[8].affiliations[0].raw_affiliation_string | Interfaculty Institute for Genetics and Functional Genomics, Ernst-Moritz-Arndt-University, Greifswald, Germany |
| authorships[8].affiliations[1].institution_ids | https://openalex.org/I36522303 |
| authorships[8].affiliations[1].raw_affiliation_string | ZIK-FunGene Junior Research Group `Applied Proteomics´, Interfaculty Institute for Genetics and Functional Genomics, Ernst-Moritz-Arndt-University, Greifswald, Germany |
| authorships[8].institutions[0].id | https://openalex.org/I36522303 |
| authorships[8].institutions[0].ror | https://ror.org/00r1edq15 |
| authorships[8].institutions[0].type | education |
| authorships[8].institutions[0].lineage | https://openalex.org/I36522303 |
| authorships[8].institutions[0].country_code | DE |
| authorships[8].institutions[0].display_name | Universität Greifswald |
| authorships[8].author_position | middle |
| authorships[8].raw_author_name | Frank Schmidt |
| authorships[8].is_corresponding | True |
| authorships[8].raw_affiliation_strings | Interfaculty Institute for Genetics and Functional Genomics, Ernst-Moritz-Arndt-University, Greifswald, Germany, ZIK-FunGene Junior Research Group `Applied Proteomics´, Interfaculty Institute for Genetics and Functional Genomics, Ernst-Moritz-Arndt-University, Greifswald, Germany |
| authorships[9].author.id | https://openalex.org/A5063838858 |
| authorships[9].author.orcid | https://orcid.org/0000-0003-2551-1299 |
| authorships[9].author.display_name | Dagmar Heuer |
| authorships[9].countries | DE |
| authorships[9].affiliations[0].institution_ids | https://openalex.org/I24359323 |
| authorships[9].affiliations[0].raw_affiliation_string | Junior Research Group `Sexually Transmitted Bacterial Pathogens´, Robert Koch Institute, Berlin, Germany |
| authorships[9].institutions[0].id | https://openalex.org/I24359323 |
| authorships[9].institutions[0].ror | https://ror.org/01k5qnb77 |
| authorships[9].institutions[0].type | government |
| authorships[9].institutions[0].lineage | https://openalex.org/I24359323 |
| authorships[9].institutions[0].country_code | DE |
| authorships[9].institutions[0].display_name | Robert Koch Institute |
| authorships[9].author_position | last |
| authorships[9].raw_author_name | Dagmar Heuer |
| authorships[9].is_corresponding | True |
| authorships[9].raw_affiliation_strings | Junior Research Group `Sexually Transmitted Bacterial Pathogens´, Robert Koch Institute, Berlin, Germany |
| has_content.pdf | True |
| has_content.grobid_xml | True |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://journals.plos.org/plospathogens/article/file?id=10.1371/journal.ppat.1004883&type=printable |
| open_access.oa_status | gold |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | The Proteome of the Isolated Chlamydia trachomatis Containing Vacuole Reveals a Complex Trafficking Platform Enriched for Retromer Components |
| has_fulltext | True |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T10428 |
| primary_topic.field.id | https://openalex.org/fields/24 |
| primary_topic.field.display_name | Immunology and Microbiology |
| primary_topic.score | 0.9997000098228455 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/2404 |
| primary_topic.subfield.display_name | Microbiology |
| primary_topic.display_name | Reproductive tract infections research |
| related_works | https://openalex.org/W2550012727, https://openalex.org/W4224115480, https://openalex.org/W2121939655, https://openalex.org/W4223926836, https://openalex.org/W1975585204, https://openalex.org/W1992461042, https://openalex.org/W2765884106, https://openalex.org/W2061111237, https://openalex.org/W2994871160, https://openalex.org/W2000999497 |
| cited_by_count | 76 |
| counts_by_year[0].year | 2025 |
| counts_by_year[0].cited_by_count | 6 |
| counts_by_year[1].year | 2024 |
| counts_by_year[1].cited_by_count | 3 |
| counts_by_year[2].year | 2023 |
| counts_by_year[2].cited_by_count | 4 |
| counts_by_year[3].year | 2022 |
| counts_by_year[3].cited_by_count | 2 |
| counts_by_year[4].year | 2021 |
| counts_by_year[4].cited_by_count | 6 |
| counts_by_year[5].year | 2020 |
| counts_by_year[5].cited_by_count | 7 |
| counts_by_year[6].year | 2019 |
| counts_by_year[6].cited_by_count | 13 |
| counts_by_year[7].year | 2018 |
| counts_by_year[7].cited_by_count | 12 |
| counts_by_year[8].year | 2017 |
| counts_by_year[8].cited_by_count | 16 |
| counts_by_year[9].year | 2016 |
| counts_by_year[9].cited_by_count | 5 |
| counts_by_year[10].year | 2015 |
| counts_by_year[10].cited_by_count | 2 |
| locations_count | 5 |
| best_oa_location.id | doi:10.1371/journal.ppat.1004883 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S2004986 |
| best_oa_location.source.issn | 1553-7366, 1553-7374 |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | True |
| best_oa_location.source.issn_l | 1553-7366 |
| best_oa_location.source.is_core | True |
| best_oa_location.source.is_in_doaj | True |
| best_oa_location.source.display_name | PLoS Pathogens |
| best_oa_location.source.host_organization | https://openalex.org/P4310315706 |
| best_oa_location.source.host_organization_name | Public Library of Science |
| best_oa_location.source.host_organization_lineage | https://openalex.org/P4310315706 |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | https://journals.plos.org/plospathogens/article/file?id=10.1371/journal.ppat.1004883&type=printable |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | journal-article |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | PLOS Pathogens |
| best_oa_location.landing_page_url | https://doi.org/10.1371/journal.ppat.1004883 |
| primary_location.id | doi:10.1371/journal.ppat.1004883 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S2004986 |
| primary_location.source.issn | 1553-7366, 1553-7374 |
| primary_location.source.type | journal |
| primary_location.source.is_oa | True |
| primary_location.source.issn_l | 1553-7366 |
| primary_location.source.is_core | True |
| primary_location.source.is_in_doaj | True |
| primary_location.source.display_name | PLoS Pathogens |
| primary_location.source.host_organization | https://openalex.org/P4310315706 |
| primary_location.source.host_organization_name | Public Library of Science |
| primary_location.source.host_organization_lineage | https://openalex.org/P4310315706 |
| primary_location.license | cc-by |
| primary_location.pdf_url | https://journals.plos.org/plospathogens/article/file?id=10.1371/journal.ppat.1004883&type=printable |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | PLOS Pathogens |
| primary_location.landing_page_url | https://doi.org/10.1371/journal.ppat.1004883 |
| publication_date | 2015-06-04 |
| publication_year | 2015 |
| referenced_works | https://openalex.org/W2027095867, https://openalex.org/W4248470101, https://openalex.org/W2110331556, https://openalex.org/W1534004966, https://openalex.org/W2116002414, https://openalex.org/W2011520932, https://openalex.org/W1999396012, https://openalex.org/W2034806371, https://openalex.org/W2019698535, https://openalex.org/W2029987169, https://openalex.org/W2123119856, https://openalex.org/W2171251887, https://openalex.org/W2098676854, https://openalex.org/W1966811207, https://openalex.org/W1511309704, https://openalex.org/W2034969129, https://openalex.org/W2041188644, https://openalex.org/W2129551826, https://openalex.org/W2161973037, https://openalex.org/W2140946052, https://openalex.org/W2128551987, https://openalex.org/W2107578201, https://openalex.org/W2605068739, https://openalex.org/W2073352515, https://openalex.org/W2061771331, https://openalex.org/W1990123554, https://openalex.org/W2037617099, https://openalex.org/W2105924489, https://openalex.org/W2079384116, https://openalex.org/W1542673149, https://openalex.org/W2108579458, https://openalex.org/W2064380139, https://openalex.org/W2150152371, https://openalex.org/W2160885923, https://openalex.org/W2115780677, https://openalex.org/W2106804776, https://openalex.org/W1965015341, https://openalex.org/W1969987939, https://openalex.org/W2142778943, https://openalex.org/W2108817086, https://openalex.org/W2108586733, https://openalex.org/W2114912316, https://openalex.org/W2062348048, https://openalex.org/W2092250470, https://openalex.org/W1987990206, https://openalex.org/W1996038248, https://openalex.org/W2115490832, https://openalex.org/W2138582859, https://openalex.org/W1999056400, https://openalex.org/W2031406661, https://openalex.org/W1903000184, https://openalex.org/W2082825744, https://openalex.org/W2050471757, https://openalex.org/W2127969245, https://openalex.org/W2114150309, https://openalex.org/W2080752012, https://openalex.org/W2038963685, https://openalex.org/W1890701869 |
| referenced_works_count | 58 |
| abstract_inverted_index.a | 15, 39, 77, 105 |
| abstract_inverted_index.C. | 64, 120, 139 |
| abstract_inverted_index.In | 133 |
| abstract_inverted_index.an | 3 |
| abstract_inverted_index.by | 67 |
| abstract_inverted_index.in | 14, 115 |
| abstract_inverted_index.is | 2, 27, 52, 76, 95, 127 |
| abstract_inverted_index.of | 22, 46, 62, 101 |
| abstract_inverted_index.on | 42 |
| abstract_inverted_index.we | 56 |
| abstract_inverted_index.The | 20 |
| abstract_inverted_index.and | 88, 123 |
| abstract_inverted_index.for | 29, 36, 98, 108, 129, 144 |
| abstract_inverted_index.its | 34, 50 |
| abstract_inverted_index.the | 10, 18, 43, 47, 58, 74, 93, 102, 119 |
| abstract_inverted_index.Here | 55 |
| abstract_inverted_index.SNX5 | 117 |
| abstract_inverted_index.cell | 13 |
| abstract_inverted_index.host | 12, 59, 85 |
| abstract_inverted_index.that | 7, 73, 82, 114, 124, 138 |
| abstract_inverted_index.this | 23 |
| abstract_inverted_index.view | 41 |
| abstract_inverted_index.with | 84 |
| abstract_inverted_index.human | 5 |
| abstract_inverted_index.niche | 26 |
| abstract_inverted_index.these | 135 |
| abstract_inverted_index.cells' | 86 |
| abstract_inverted_index.highly | 96 |
| abstract_inverted_index.inside | 9 |
| abstract_inverted_index.nexins | 100 |
| abstract_inverted_index.showed | 113 |
| abstract_inverted_index.unique | 16 |
| abstract_inverted_index.Despite | 33 |
| abstract_inverted_index.SNX-BAR | 103 |
| abstract_inverted_index.antero- | 87 |
| abstract_inverted_index.complex | 78, 106 |
| abstract_inverted_index.hijacks | 141 |
| abstract_inverted_index.progeny | 131 |
| abstract_inverted_index.protein | 44 |
| abstract_inverted_index.sorting | 99 |
| abstract_inverted_index.studies | 112 |
| abstract_inverted_index.suggest | 137 |
| abstract_inverted_index.analysis | 71 |
| abstract_inverted_index.biology, | 38 |
| abstract_inverted_index.controls | 118 |
| abstract_inverted_index.describe | 57 |
| abstract_inverted_index.enriched | 97 |
| abstract_inverted_index.findings | 136 |
| abstract_inverted_index.holistic | 40 |
| abstract_inverted_index.infected | 11 |
| abstract_inverted_index.isolated | 63 |
| abstract_inverted_index.missing. | 54 |
| abstract_inverted_index.pathogen | 6 |
| abstract_inverted_index.pathways | 143 |
| abstract_inverted_index.platform | 81 |
| abstract_inverted_index.proteome | 61 |
| abstract_inverted_index.summary, | 134 |
| abstract_inverted_index.vacuole, | 17 |
| abstract_inverted_index.Chlamydia | 0, 31, 37 |
| abstract_inverted_index.bacterial | 25 |
| abstract_inverted_index.currently | 53 |
| abstract_inverted_index.effective | 145 |
| abstract_inverted_index.essential | 28, 107, 128 |
| abstract_inverted_index.formation | 21 |
| abstract_inverted_index.important | 4 |
| abstract_inverted_index.including | 49 |
| abstract_inverted_index.inclusion | 75, 94 |
| abstract_inverted_index.indicated | 72 |
| abstract_inverted_index.infection | 122 |
| abstract_inverted_index.interacts | 83 |
| abstract_inverted_index.membrane, | 51 |
| abstract_inverted_index.pathways. | 91 |
| abstract_inverted_index.retromer, | 104 |
| abstract_inverted_index.Functional | 111 |
| abstract_inverted_index.formation. | 132 |
| abstract_inverted_index.importance | 35 |
| abstract_inverted_index.inclusion, | 48 |
| abstract_inverted_index.inclusion. | 19 |
| abstract_inverted_index.inclusions | 66 |
| abstract_inverted_index.infection. | 146 |
| abstract_inverted_index.infectious | 130 |
| abstract_inverted_index.productive | 30 |
| abstract_inverted_index.replicates | 8 |
| abstract_inverted_index.retrograde | 89, 109, 125, 142 |
| abstract_inverted_index.composition | 45 |
| abstract_inverted_index.infections. | 32 |
| abstract_inverted_index.particular, | 116 |
| abstract_inverted_index.proteomics. | 69 |
| abstract_inverted_index.trachomatis | 1, 65, 121, 140 |
| abstract_inverted_index.trafficking | 80, 90, 126 |
| abstract_inverted_index.Furthermore, | 92 |
| abstract_inverted_index.cell-derived | 60 |
| abstract_inverted_index.quantitative | 68 |
| abstract_inverted_index.trafficking. | 110 |
| abstract_inverted_index.Computational | 70 |
| abstract_inverted_index.intracellular | 24, 79 |
| cited_by_percentile_year.max | 100 |
| cited_by_percentile_year.min | 94 |
| corresponding_author_ids | https://openalex.org/A5110966455, https://openalex.org/A5031672904, https://openalex.org/A5109803468, https://openalex.org/A5049628915, https://openalex.org/A5018412626, https://openalex.org/A5005996110, https://openalex.org/A5012124891, https://openalex.org/A5063838858, https://openalex.org/A5045874625, https://openalex.org/A5066787801 |
| countries_distinct_count | 1 |
| institutions_distinct_count | 10 |
| corresponding_institution_ids | https://openalex.org/I24359323, https://openalex.org/I36522303 |
| citation_normalized_percentile.value | 0.98377657 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | True |