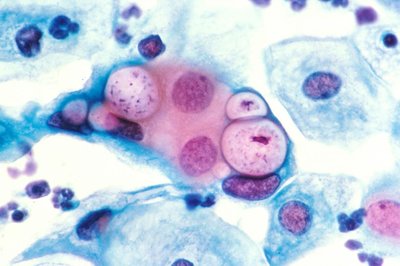
The second messenger signaling molecule cyclic di-AMP drives developmental cycle progression in Chlamydia trachomatis Article Swipe
 YOU?
·
· 2025
· Open Access
·
· DOI: https://doi.org/10.7554/elife.104240
YOU?
·
· 2025
· Open Access
·
· DOI: https://doi.org/10.7554/elife.104240
The obligate intracellular bacterium Chlamydia alternates between two functional forms during its developmental cycle: elementary body (EB) and reticulate body (RB). However, the molecular mechanisms governing the transitions between these forms are unknown. Here, we present evidence that cyclic di-AMP (c-di-AMP) is a key factor in triggering the transition from RB to EB (i.e., secondary differentiation) in the chlamydial developmental cycle. By overexpressing or knocking down expression of c-di-AMP synthase genes, we made strains producing different levels of c-di-AMP, which we linked to changes in secondary differentiation status. Increases in c-di-AMP resulted in an earlier increase in transcription of EB-associated genes, and this was further manifested in earlier production of EBs. In contrast, when c-di-AMP levels were decreased, developmental cycle progression was delayed. Based on these data, we conclude there is a threshold level of c-di-AMP needed to trigger secondary differentiation in Chlamydia . This study identifies a mechanism by which secondary differentiation is initiated in Chlamydia and reveals a critical role for the second messenger signaling molecule c-di-AMP in this process.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.7554/elife.104240
- OA Status
- gold
- References
- 64
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4406895662
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4406895662Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.7554/elife.104240Digital Object Identifier
- Title
-
The second messenger signaling molecule cyclic di-AMP drives developmental cycle progression in Chlamydia trachomatisWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2025Year of publication
- Publication date
-
2025-01-28Full publication date if available
- Authors
-
Junghoon Lee, Scot P. OuelletteList of authors in order
- Landing page
-
https://doi.org/10.7554/elife.104240Publisher landing page
- Open access
-
YesWhether a free full text is available
- OA status
-
goldOpen access status per OpenAlex
- OA URL
-
https://doi.org/10.7554/elife.104240Direct OA link when available
- Concepts
-
Chlamydia trachomatis, Biology, Chlamydia, Cell biology, Physics, Virology, GeneticsTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
0Total citation count in OpenAlex
- References (count)
-
64Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4406895662 |
|---|---|
| doi | https://doi.org/10.7554/elife.104240 |
| ids.doi | https://doi.org/10.7554/elife.104240 |
| ids.pmid | https://pubmed.ncbi.nlm.nih.gov/40928359 |
| ids.openalex | https://openalex.org/W4406895662 |
| fwci | 0.0 |
| mesh[0].qualifier_ui | Q000254 |
| mesh[0].descriptor_ui | D002692 |
| mesh[0].is_major_topic | True |
| mesh[0].qualifier_name | growth & development |
| mesh[0].descriptor_name | Chlamydia trachomatis |
| mesh[1].qualifier_ui | Q000235 |
| mesh[1].descriptor_ui | D002692 |
| mesh[1].is_major_topic | True |
| mesh[1].qualifier_name | genetics |
| mesh[1].descriptor_name | Chlamydia trachomatis |
| mesh[2].qualifier_ui | Q000378 |
| mesh[2].descriptor_ui | D002692 |
| mesh[2].is_major_topic | True |
| mesh[2].qualifier_name | metabolism |
| mesh[2].descriptor_name | Chlamydia trachomatis |
| mesh[3].qualifier_ui | |
| mesh[3].descriptor_ui | D015290 |
| mesh[3].is_major_topic | True |
| mesh[3].qualifier_name | |
| mesh[3].descriptor_name | Second Messenger Systems |
| mesh[4].qualifier_ui | |
| mesh[4].descriptor_ui | D015964 |
| mesh[4].is_major_topic | False |
| mesh[4].qualifier_name | |
| mesh[4].descriptor_name | Gene Expression Regulation, Bacterial |
| mesh[5].qualifier_ui | Q000378 |
| mesh[5].descriptor_ui | D015226 |
| mesh[5].is_major_topic | True |
| mesh[5].qualifier_name | metabolism |
| mesh[5].descriptor_name | Dinucleoside Phosphates |
| mesh[6].qualifier_ui | |
| mesh[6].descriptor_ui | D006801 |
| mesh[6].is_major_topic | False |
| mesh[6].qualifier_name | |
| mesh[6].descriptor_name | Humans |
| mesh[7].qualifier_ui | Q000254 |
| mesh[7].descriptor_ui | D002692 |
| mesh[7].is_major_topic | True |
| mesh[7].qualifier_name | growth & development |
| mesh[7].descriptor_name | Chlamydia trachomatis |
| mesh[8].qualifier_ui | Q000235 |
| mesh[8].descriptor_ui | D002692 |
| mesh[8].is_major_topic | True |
| mesh[8].qualifier_name | genetics |
| mesh[8].descriptor_name | Chlamydia trachomatis |
| mesh[9].qualifier_ui | Q000378 |
| mesh[9].descriptor_ui | D002692 |
| mesh[9].is_major_topic | True |
| mesh[9].qualifier_name | metabolism |
| mesh[9].descriptor_name | Chlamydia trachomatis |
| mesh[10].qualifier_ui | |
| mesh[10].descriptor_ui | D015290 |
| mesh[10].is_major_topic | True |
| mesh[10].qualifier_name | |
| mesh[10].descriptor_name | Second Messenger Systems |
| mesh[11].qualifier_ui | |
| mesh[11].descriptor_ui | D015964 |
| mesh[11].is_major_topic | False |
| mesh[11].qualifier_name | |
| mesh[11].descriptor_name | Gene Expression Regulation, Bacterial |
| mesh[12].qualifier_ui | Q000378 |
| mesh[12].descriptor_ui | D015226 |
| mesh[12].is_major_topic | True |
| mesh[12].qualifier_name | metabolism |
| mesh[12].descriptor_name | Dinucleoside Phosphates |
| mesh[13].qualifier_ui | |
| mesh[13].descriptor_ui | D006801 |
| mesh[13].is_major_topic | False |
| mesh[13].qualifier_name | |
| mesh[13].descriptor_name | Humans |
| mesh[14].qualifier_ui | Q000254 |
| mesh[14].descriptor_ui | D002692 |
| mesh[14].is_major_topic | True |
| mesh[14].qualifier_name | growth & development |
| mesh[14].descriptor_name | Chlamydia trachomatis |
| mesh[15].qualifier_ui | Q000235 |
| mesh[15].descriptor_ui | D002692 |
| mesh[15].is_major_topic | True |
| mesh[15].qualifier_name | genetics |
| mesh[15].descriptor_name | Chlamydia trachomatis |
| mesh[16].qualifier_ui | Q000378 |
| mesh[16].descriptor_ui | D002692 |
| mesh[16].is_major_topic | True |
| mesh[16].qualifier_name | metabolism |
| mesh[16].descriptor_name | Chlamydia trachomatis |
| mesh[17].qualifier_ui | |
| mesh[17].descriptor_ui | D015290 |
| mesh[17].is_major_topic | True |
| mesh[17].qualifier_name | |
| mesh[17].descriptor_name | Second Messenger Systems |
| mesh[18].qualifier_ui | |
| mesh[18].descriptor_ui | D015964 |
| mesh[18].is_major_topic | False |
| mesh[18].qualifier_name | |
| mesh[18].descriptor_name | Gene Expression Regulation, Bacterial |
| mesh[19].qualifier_ui | Q000378 |
| mesh[19].descriptor_ui | D015226 |
| mesh[19].is_major_topic | True |
| mesh[19].qualifier_name | metabolism |
| mesh[19].descriptor_name | Dinucleoside Phosphates |
| mesh[20].qualifier_ui | |
| mesh[20].descriptor_ui | D006801 |
| mesh[20].is_major_topic | False |
| mesh[20].qualifier_name | |
| mesh[20].descriptor_name | Humans |
| type | article |
| title | The second messenger signaling molecule cyclic di-AMP drives developmental cycle progression in Chlamydia trachomatis |
| biblio.issue | |
| biblio.volume | 14 |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T10428 |
| topics[0].field.id | https://openalex.org/fields/24 |
| topics[0].field.display_name | Immunology and Microbiology |
| topics[0].score | 0.9998000264167786 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/2404 |
| topics[0].subfield.display_name | Microbiology |
| topics[0].display_name | Reproductive tract infections research |
| topics[1].id | https://openalex.org/T11232 |
| topics[1].field.id | https://openalex.org/fields/27 |
| topics[1].field.display_name | Medicine |
| topics[1].score | 0.9901999831199646 |
| topics[1].domain.id | https://openalex.org/domains/4 |
| topics[1].domain.display_name | Health Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/2713 |
| topics[1].subfield.display_name | Epidemiology |
| topics[1].display_name | Urinary Tract Infections Management |
| topics[2].id | https://openalex.org/T10066 |
| topics[2].field.id | https://openalex.org/fields/13 |
| topics[2].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[2].score | 0.9506000280380249 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1312 |
| topics[2].subfield.display_name | Molecular Biology |
| topics[2].display_name | Gut microbiota and health |
| is_xpac | False |
| apc_list.value | 3000 |
| apc_list.currency | USD |
| apc_list.value_usd | 2500 |
| apc_paid.value | 3000 |
| apc_paid.currency | USD |
| apc_paid.value_usd | 2500 |
| concepts[0].id | https://openalex.org/C2777391075 |
| concepts[0].level | 2 |
| concepts[0].score | 0.8562831878662109 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q131065 |
| concepts[0].display_name | Chlamydia trachomatis |
| concepts[1].id | https://openalex.org/C86803240 |
| concepts[1].level | 0 |
| concepts[1].score | 0.4166698455810547 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[1].display_name | Biology |
| concepts[2].id | https://openalex.org/C2780122209 |
| concepts[2].level | 2 |
| concepts[2].score | 0.4110642373561859 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q153356 |
| concepts[2].display_name | Chlamydia |
| concepts[3].id | https://openalex.org/C95444343 |
| concepts[3].level | 1 |
| concepts[3].score | 0.33628207445144653 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q7141 |
| concepts[3].display_name | Cell biology |
| concepts[4].id | https://openalex.org/C121332964 |
| concepts[4].level | 0 |
| concepts[4].score | 0.3241056799888611 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q413 |
| concepts[4].display_name | Physics |
| concepts[5].id | https://openalex.org/C159047783 |
| concepts[5].level | 1 |
| concepts[5].score | 0.24346357583999634 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q7215 |
| concepts[5].display_name | Virology |
| concepts[6].id | https://openalex.org/C54355233 |
| concepts[6].level | 1 |
| concepts[6].score | 0.20078012347221375 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[6].display_name | Genetics |
| keywords[0].id | https://openalex.org/keywords/chlamydia-trachomatis |
| keywords[0].score | 0.8562831878662109 |
| keywords[0].display_name | Chlamydia trachomatis |
| keywords[1].id | https://openalex.org/keywords/biology |
| keywords[1].score | 0.4166698455810547 |
| keywords[1].display_name | Biology |
| keywords[2].id | https://openalex.org/keywords/chlamydia |
| keywords[2].score | 0.4110642373561859 |
| keywords[2].display_name | Chlamydia |
| keywords[3].id | https://openalex.org/keywords/cell-biology |
| keywords[3].score | 0.33628207445144653 |
| keywords[3].display_name | Cell biology |
| keywords[4].id | https://openalex.org/keywords/physics |
| keywords[4].score | 0.3241056799888611 |
| keywords[4].display_name | Physics |
| keywords[5].id | https://openalex.org/keywords/virology |
| keywords[5].score | 0.24346357583999634 |
| keywords[5].display_name | Virology |
| keywords[6].id | https://openalex.org/keywords/genetics |
| keywords[6].score | 0.20078012347221375 |
| keywords[6].display_name | Genetics |
| language | en |
| locations[0].id | doi:10.7554/elife.104240 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S1336409049 |
| locations[0].source.issn | 2050-084X |
| locations[0].source.type | journal |
| locations[0].source.is_oa | True |
| locations[0].source.issn_l | 2050-084X |
| locations[0].source.is_core | True |
| locations[0].source.is_in_doaj | True |
| locations[0].source.display_name | eLife |
| locations[0].source.host_organization | https://openalex.org/P4310311710 |
| locations[0].source.host_organization_name | eLife Sciences Publications Ltd |
| locations[0].source.host_organization_lineage | https://openalex.org/P4310311710 |
| locations[0].license | cc-by |
| locations[0].pdf_url | |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | eLife |
| locations[0].landing_page_url | https://doi.org/10.7554/elife.104240 |
| locations[1].id | pmid:40928359 |
| locations[1].is_oa | False |
| locations[1].source.id | https://openalex.org/S4306525036 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | PubMed |
| locations[1].source.host_organization | https://openalex.org/I1299303238 |
| locations[1].source.host_organization_name | National Institutes of Health |
| locations[1].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[1].license | |
| locations[1].pdf_url | |
| locations[1].version | publishedVersion |
| locations[1].raw_type | |
| locations[1].license_id | |
| locations[1].is_accepted | True |
| locations[1].is_published | True |
| locations[1].raw_source_name | eLife |
| locations[1].landing_page_url | https://pubmed.ncbi.nlm.nih.gov/40928359 |
| locations[2].id | pmh:oai:doaj.org/article:1078e4dcdb3b4a9fbad1e61f322d62bf |
| locations[2].is_oa | False |
| locations[2].source.id | https://openalex.org/S4306401280 |
| locations[2].source.issn | |
| locations[2].source.type | repository |
| locations[2].source.is_oa | False |
| locations[2].source.issn_l | |
| locations[2].source.is_core | False |
| locations[2].source.is_in_doaj | False |
| locations[2].source.display_name | DOAJ (DOAJ: Directory of Open Access Journals) |
| locations[2].source.host_organization | |
| locations[2].source.host_organization_name | |
| locations[2].source.host_organization_lineage | |
| locations[2].license | |
| locations[2].pdf_url | |
| locations[2].version | submittedVersion |
| locations[2].raw_type | article |
| locations[2].license_id | |
| locations[2].is_accepted | False |
| locations[2].is_published | False |
| locations[2].raw_source_name | eLife, Vol 14 (2025) |
| locations[2].landing_page_url | https://doaj.org/article/1078e4dcdb3b4a9fbad1e61f322d62bf |
| indexed_in | crossref, doaj, pubmed |
| authorships[0].author.id | https://openalex.org/A5100626272 |
| authorships[0].author.orcid | https://orcid.org/0000-0002-6416-8975 |
| authorships[0].author.display_name | Junghoon Lee |
| authorships[0].countries | US |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I59130452 |
| authorships[0].affiliations[0].raw_affiliation_string | Department of Pathology, Microbiology, and Immunology, College of Medicine, University of Nebraska Medical Center |
| authorships[0].institutions[0].id | https://openalex.org/I59130452 |
| authorships[0].institutions[0].ror | https://ror.org/00thqtb16 |
| authorships[0].institutions[0].type | healthcare |
| authorships[0].institutions[0].lineage | https://openalex.org/I59130452 |
| authorships[0].institutions[0].country_code | US |
| authorships[0].institutions[0].display_name | University of Nebraska Medical Center |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Junghoon Lee |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Department of Pathology, Microbiology, and Immunology, College of Medicine, University of Nebraska Medical Center |
| authorships[1].author.id | https://openalex.org/A5026285532 |
| authorships[1].author.orcid | https://orcid.org/0000-0002-3721-6839 |
| authorships[1].author.display_name | Scot P. Ouellette |
| authorships[1].countries | US |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I59130452 |
| authorships[1].affiliations[0].raw_affiliation_string | Department of Pathology, Microbiology, and Immunology, College of Medicine, University of Nebraska Medical Center |
| authorships[1].institutions[0].id | https://openalex.org/I59130452 |
| authorships[1].institutions[0].ror | https://ror.org/00thqtb16 |
| authorships[1].institutions[0].type | healthcare |
| authorships[1].institutions[0].lineage | https://openalex.org/I59130452 |
| authorships[1].institutions[0].country_code | US |
| authorships[1].institutions[0].display_name | University of Nebraska Medical Center |
| authorships[1].author_position | last |
| authorships[1].raw_author_name | Scot P Ouellette |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | Department of Pathology, Microbiology, and Immunology, College of Medicine, University of Nebraska Medical Center |
| has_content.pdf | False |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://doi.org/10.7554/elife.104240 |
| open_access.oa_status | gold |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-01-29T00:00:00 |
| display_name | The second messenger signaling molecule cyclic di-AMP drives developmental cycle progression in Chlamydia trachomatis |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T10428 |
| primary_topic.field.id | https://openalex.org/fields/24 |
| primary_topic.field.display_name | Immunology and Microbiology |
| primary_topic.score | 0.9998000264167786 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/2404 |
| primary_topic.subfield.display_name | Microbiology |
| primary_topic.display_name | Reproductive tract infections research |
| related_works | https://openalex.org/W4391068763, https://openalex.org/W4231476175, https://openalex.org/W2110420597, https://openalex.org/W2130554869, https://openalex.org/W2106767833, https://openalex.org/W2969526378, https://openalex.org/W2472116282, https://openalex.org/W1983396513, https://openalex.org/W1949477813, https://openalex.org/W2028096858 |
| cited_by_count | 0 |
| locations_count | 3 |
| best_oa_location.id | doi:10.7554/elife.104240 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S1336409049 |
| best_oa_location.source.issn | 2050-084X |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | True |
| best_oa_location.source.issn_l | 2050-084X |
| best_oa_location.source.is_core | True |
| best_oa_location.source.is_in_doaj | True |
| best_oa_location.source.display_name | eLife |
| best_oa_location.source.host_organization | https://openalex.org/P4310311710 |
| best_oa_location.source.host_organization_name | eLife Sciences Publications Ltd |
| best_oa_location.source.host_organization_lineage | https://openalex.org/P4310311710 |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | journal-article |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | eLife |
| best_oa_location.landing_page_url | https://doi.org/10.7554/elife.104240 |
| primary_location.id | doi:10.7554/elife.104240 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S1336409049 |
| primary_location.source.issn | 2050-084X |
| primary_location.source.type | journal |
| primary_location.source.is_oa | True |
| primary_location.source.issn_l | 2050-084X |
| primary_location.source.is_core | True |
| primary_location.source.is_in_doaj | True |
| primary_location.source.display_name | eLife |
| primary_location.source.host_organization | https://openalex.org/P4310311710 |
| primary_location.source.host_organization_name | eLife Sciences Publications Ltd |
| primary_location.source.host_organization_lineage | https://openalex.org/P4310311710 |
| primary_location.license | cc-by |
| primary_location.pdf_url | |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | eLife |
| primary_location.landing_page_url | https://doi.org/10.7554/elife.104240 |
| publication_date | 2025-01-28 |
| publication_year | 2025 |
| referenced_works | https://openalex.org/W2119656536, https://openalex.org/W1719723029, https://openalex.org/W2011930613, https://openalex.org/W6634771800, https://openalex.org/W1869726924, https://openalex.org/W1783285978, https://openalex.org/W2510789424, https://openalex.org/W2346633051, https://openalex.org/W2139972143, https://openalex.org/W3006240911, https://openalex.org/W2017111499, https://openalex.org/W2121701946, https://openalex.org/W2135783899, https://openalex.org/W2075583035, https://openalex.org/W1843541362, https://openalex.org/W2142105619, https://openalex.org/W1780853035, https://openalex.org/W2972929810, https://openalex.org/W2914875038, https://openalex.org/W2196923764, https://openalex.org/W2157046025, https://openalex.org/W2942856089, https://openalex.org/W2165079807, https://openalex.org/W2152861858, https://openalex.org/W2137288582, https://openalex.org/W2611425968, https://openalex.org/W2054871938, https://openalex.org/W2115780677, https://openalex.org/W3154164226, https://openalex.org/W3092217152, https://openalex.org/W2124733101, https://openalex.org/W2150737519, https://openalex.org/W4387003247, https://openalex.org/W3084277729, https://openalex.org/W4404637955, https://openalex.org/W4297218728, https://openalex.org/W4406529894, https://openalex.org/W1670617703, https://openalex.org/W4309245464, https://openalex.org/W2981679577, https://openalex.org/W2093259951, https://openalex.org/W2118805096, https://openalex.org/W1847018147, https://openalex.org/W1929658003, https://openalex.org/W2153833219, https://openalex.org/W4366821613, https://openalex.org/W4220731037, https://openalex.org/W3121064739, https://openalex.org/W2912371761, https://openalex.org/W2059833435, https://openalex.org/W2126922170, https://openalex.org/W4327583115, https://openalex.org/W2046684285, https://openalex.org/W1938173378, https://openalex.org/W1560410275, https://openalex.org/W2158573520, https://openalex.org/W2170331865, https://openalex.org/W2602011001, https://openalex.org/W2152434206, https://openalex.org/W4395694956, https://openalex.org/W1581158476, https://openalex.org/W2793996708, https://openalex.org/W1923725911, https://openalex.org/W2398545537 |
| referenced_works_count | 64 |
| abstract_inverted_index.. | 143 |
| abstract_inverted_index.a | 42, 131, 147, 159 |
| abstract_inverted_index.By | 61 |
| abstract_inverted_index.EB | 52 |
| abstract_inverted_index.In | 111 |
| abstract_inverted_index.RB | 50 |
| abstract_inverted_index.an | 93 |
| abstract_inverted_index.by | 149 |
| abstract_inverted_index.in | 45, 56, 84, 89, 92, 96, 106, 141, 155, 169 |
| abstract_inverted_index.is | 41, 130, 153 |
| abstract_inverted_index.of | 67, 77, 98, 109, 134 |
| abstract_inverted_index.on | 124 |
| abstract_inverted_index.or | 63 |
| abstract_inverted_index.to | 51, 82, 137 |
| abstract_inverted_index.we | 34, 71, 80, 127 |
| abstract_inverted_index.The | 0 |
| abstract_inverted_index.and | 17, 101, 157 |
| abstract_inverted_index.are | 31 |
| abstract_inverted_index.for | 162 |
| abstract_inverted_index.its | 11 |
| abstract_inverted_index.key | 43 |
| abstract_inverted_index.the | 22, 26, 47, 57, 163 |
| abstract_inverted_index.two | 7 |
| abstract_inverted_index.was | 103, 121 |
| abstract_inverted_index.(EB) | 16 |
| abstract_inverted_index.EBs. | 110 |
| abstract_inverted_index.This | 144 |
| abstract_inverted_index.body | 15, 19 |
| abstract_inverted_index.down | 65 |
| abstract_inverted_index.from | 49 |
| abstract_inverted_index.made | 72 |
| abstract_inverted_index.role | 161 |
| abstract_inverted_index.that | 37 |
| abstract_inverted_index.this | 102, 170 |
| abstract_inverted_index.were | 116 |
| abstract_inverted_index.when | 113 |
| abstract_inverted_index.(RB). | 20 |
| abstract_inverted_index.Based | 123 |
| abstract_inverted_index.Here, | 33 |
| abstract_inverted_index.cycle | 119 |
| abstract_inverted_index.data, | 126 |
| abstract_inverted_index.forms | 9, 30 |
| abstract_inverted_index.level | 133 |
| abstract_inverted_index.study | 145 |
| abstract_inverted_index.there | 129 |
| abstract_inverted_index.these | 29, 125 |
| abstract_inverted_index.which | 79, 150 |
| abstract_inverted_index.(i.e., | 53 |
| abstract_inverted_index.cycle. | 60 |
| abstract_inverted_index.cycle: | 13 |
| abstract_inverted_index.cyclic | 38 |
| abstract_inverted_index.di-AMP | 39 |
| abstract_inverted_index.during | 10 |
| abstract_inverted_index.factor | 44 |
| abstract_inverted_index.genes, | 70, 100 |
| abstract_inverted_index.levels | 76, 115 |
| abstract_inverted_index.linked | 81 |
| abstract_inverted_index.needed | 136 |
| abstract_inverted_index.second | 164 |
| abstract_inverted_index.between | 6, 28 |
| abstract_inverted_index.changes | 83 |
| abstract_inverted_index.earlier | 94, 107 |
| abstract_inverted_index.further | 104 |
| abstract_inverted_index.present | 35 |
| abstract_inverted_index.reveals | 158 |
| abstract_inverted_index.status. | 87 |
| abstract_inverted_index.strains | 73 |
| abstract_inverted_index.trigger | 138 |
| abstract_inverted_index.However, | 21 |
| abstract_inverted_index.c-di-AMP | 68, 90, 114, 135, 168 |
| abstract_inverted_index.conclude | 128 |
| abstract_inverted_index.critical | 160 |
| abstract_inverted_index.delayed. | 122 |
| abstract_inverted_index.evidence | 36 |
| abstract_inverted_index.increase | 95 |
| abstract_inverted_index.knocking | 64 |
| abstract_inverted_index.molecule | 167 |
| abstract_inverted_index.obligate | 1 |
| abstract_inverted_index.process. | 171 |
| abstract_inverted_index.resulted | 91 |
| abstract_inverted_index.synthase | 69 |
| abstract_inverted_index.unknown. | 32 |
| abstract_inverted_index.Chlamydia | 4, 142, 156 |
| abstract_inverted_index.Increases | 88 |
| abstract_inverted_index.bacterium | 3 |
| abstract_inverted_index.c-di-AMP, | 78 |
| abstract_inverted_index.contrast, | 112 |
| abstract_inverted_index.different | 75 |
| abstract_inverted_index.governing | 25 |
| abstract_inverted_index.initiated | 154 |
| abstract_inverted_index.mechanism | 148 |
| abstract_inverted_index.messenger | 165 |
| abstract_inverted_index.molecular | 23 |
| abstract_inverted_index.producing | 74 |
| abstract_inverted_index.secondary | 54, 85, 139, 151 |
| abstract_inverted_index.signaling | 166 |
| abstract_inverted_index.threshold | 132 |
| abstract_inverted_index.(c-di-AMP) | 40 |
| abstract_inverted_index.alternates | 5 |
| abstract_inverted_index.chlamydial | 58 |
| abstract_inverted_index.decreased, | 117 |
| abstract_inverted_index.elementary | 14 |
| abstract_inverted_index.expression | 66 |
| abstract_inverted_index.functional | 8 |
| abstract_inverted_index.identifies | 146 |
| abstract_inverted_index.manifested | 105 |
| abstract_inverted_index.mechanisms | 24 |
| abstract_inverted_index.production | 108 |
| abstract_inverted_index.reticulate | 18 |
| abstract_inverted_index.transition | 48 |
| abstract_inverted_index.triggering | 46 |
| abstract_inverted_index.progression | 120 |
| abstract_inverted_index.transitions | 27 |
| abstract_inverted_index.EB-associated | 99 |
| abstract_inverted_index.developmental | 12, 59, 118 |
| abstract_inverted_index.intracellular | 2 |
| abstract_inverted_index.transcription | 97 |
| abstract_inverted_index.overexpressing | 62 |
| abstract_inverted_index.differentiation | 86, 140, 152 |
| abstract_inverted_index.differentiation) | 55 |
| cited_by_percentile_year | |
| countries_distinct_count | 1 |
| institutions_distinct_count | 2 |
| citation_normalized_percentile.value | 0.02731267 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |