Total synthesis of the natural, medium‐length, peptaibol pentadecaibin and study of the chemical features responsible for its membrane activity Article Swipe
 YOU?
·
· 2023
· Open Access
·
· DOI: https://doi.org/10.1002/psc.3479
YOU?
·
· 2023
· Open Access
·
· DOI: https://doi.org/10.1002/psc.3479
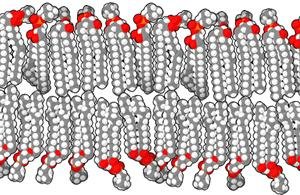
Peptaibols are naturally occurring, antimicrobial peptides endowed with well‐defined helical conformations and resistance to proteolysis. Both features stem from the presence in their sequence of several, C α ‐tetrasubstituted, α‐aminoisobutyric acid (Aib) residues. Peptaibols interact with biological membranes, usually causing their leakage. All of the peptaibol–membrane interaction mechanisms proposed so far begin with peptide aggregation or accumulation. The long‐length alamethicin, the most studied peptaibol, acts by forming pores in the membranes. Conversely, the carpet mechanism has been claimed for short‐length peptaibols, such as trichogin. The mechanism of medium‐length peptaibols is far less studied, and this is partly due to the difficulties of their synthesis. They are believed to perturb membrane permeability in different ways, depending on the membrane properties. The present work focuses on pentadecaibin, a recently discovered, medium‐length peptaibol. In contrast to the majority of its family members, its sequence does not comprise hydroxyprolines or prolines, and its helix is not kinked. A reliable and effective synthesis procedure is described that allowed us to produce also two shorter analogs. By a combination of techniques, we were able to establish a 3D‐structure–activity relationship. In particular, the membrane activity of pentadecaibin heavily depends on the presence of three consecutive Aib residues that are responsible for the clear, albeit modest, amphiphilic character of its helix. The shortest analog, devoid of two of these three Aib residues, preserves a well‐defined helical conformation, but not its amphipathicity, and loses almost completely the ability to cause membrane leakage. We conclude that pentadecaibin amphiphilicity is probably needed for the peptide ability to perturb model membranes.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.1002/psc.3479
- https://onlinelibrary.wiley.com/doi/pdfdirect/10.1002/psc.3479
- OA Status
- hybrid
- Cited By
- 5
- References
- 13
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4317233749
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4317233749Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1002/psc.3479Digital Object Identifier
- Title
-
Total synthesis of the natural, medium‐length, peptaibol pentadecaibin and study of the chemical features responsible for its membrane activityWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2023Year of publication
- Publication date
-
2023-01-18Full publication date if available
- Authors
-
Laura Morbiato, David S. A. Haneen, Fernando Formaggio, Marta De ZottiList of authors in order
- Landing page
-
https://doi.org/10.1002/psc.3479Publisher landing page
- PDF URL
-
https://onlinelibrary.wiley.com/doi/pdfdirect/10.1002/psc.3479Direct link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
hybridOpen access status per OpenAlex
- OA URL
-
https://onlinelibrary.wiley.com/doi/pdfdirect/10.1002/psc.3479Direct OA link when available
- Concepts
-
Alamethicin, Membrane, Chemistry, Peptide, Amino acid, Helix (gastropod), Proteolysis, Biophysics, Stereochemistry, Biochemistry, Lipid bilayer, Biology, Ecology, Snail, EnzymeTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
5Total citation count in OpenAlex
- Citations by year (recent)
-
2025: 1, 2024: 2, 2023: 2Per-year citation counts (last 5 years)
- References (count)
-
13Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4317233749 |
|---|---|
| doi | https://doi.org/10.1002/psc.3479 |
| ids.doi | https://doi.org/10.1002/psc.3479 |
| ids.pmid | https://pubmed.ncbi.nlm.nih.gov/36652104 |
| ids.openalex | https://openalex.org/W4317233749 |
| fwci | 1.35071936 |
| mesh[0].qualifier_ui | Q000032 |
| mesh[0].descriptor_ui | D054848 |
| mesh[0].is_major_topic | True |
| mesh[0].qualifier_name | analysis |
| mesh[0].descriptor_name | Peptaibols |
| mesh[1].qualifier_ui | Q000737 |
| mesh[1].descriptor_ui | D054848 |
| mesh[1].is_major_topic | True |
| mesh[1].qualifier_name | chemistry |
| mesh[1].descriptor_name | Peptaibols |
| mesh[2].qualifier_ui | Q000378 |
| mesh[2].descriptor_ui | D054848 |
| mesh[2].is_major_topic | True |
| mesh[2].qualifier_name | metabolism |
| mesh[2].descriptor_name | Peptaibols |
| mesh[3].qualifier_ui | Q000032 |
| mesh[3].descriptor_ui | D000408 |
| mesh[3].is_major_topic | True |
| mesh[3].qualifier_name | analysis |
| mesh[3].descriptor_name | Alamethicin |
| mesh[4].qualifier_ui | Q000737 |
| mesh[4].descriptor_ui | D000408 |
| mesh[4].is_major_topic | True |
| mesh[4].qualifier_name | chemistry |
| mesh[4].descriptor_name | Alamethicin |
| mesh[5].qualifier_ui | Q000378 |
| mesh[5].descriptor_ui | D000408 |
| mesh[5].is_major_topic | True |
| mesh[5].qualifier_name | metabolism |
| mesh[5].descriptor_name | Alamethicin |
| mesh[6].qualifier_ui | Q000737 |
| mesh[6].descriptor_ui | D002462 |
| mesh[6].is_major_topic | False |
| mesh[6].qualifier_name | chemistry |
| mesh[6].descriptor_name | Cell Membrane |
| mesh[7].qualifier_ui | |
| mesh[7].descriptor_ui | D008968 |
| mesh[7].is_major_topic | False |
| mesh[7].qualifier_name | |
| mesh[7].descriptor_name | Molecular Conformation |
| mesh[8].qualifier_ui | |
| mesh[8].descriptor_ui | D001692 |
| mesh[8].is_major_topic | False |
| mesh[8].qualifier_name | |
| mesh[8].descriptor_name | Biological Transport |
| mesh[9].qualifier_ui | Q000494 |
| mesh[9].descriptor_ui | D000900 |
| mesh[9].is_major_topic | False |
| mesh[9].qualifier_name | pharmacology |
| mesh[9].descriptor_name | Anti-Bacterial Agents |
| mesh[10].qualifier_ui | Q000737 |
| mesh[10].descriptor_ui | D000900 |
| mesh[10].is_major_topic | False |
| mesh[10].qualifier_name | chemistry |
| mesh[10].descriptor_name | Anti-Bacterial Agents |
| mesh[11].qualifier_ui | Q000032 |
| mesh[11].descriptor_ui | D054848 |
| mesh[11].is_major_topic | True |
| mesh[11].qualifier_name | analysis |
| mesh[11].descriptor_name | Peptaibols |
| mesh[12].qualifier_ui | Q000737 |
| mesh[12].descriptor_ui | D054848 |
| mesh[12].is_major_topic | True |
| mesh[12].qualifier_name | chemistry |
| mesh[12].descriptor_name | Peptaibols |
| mesh[13].qualifier_ui | Q000378 |
| mesh[13].descriptor_ui | D054848 |
| mesh[13].is_major_topic | True |
| mesh[13].qualifier_name | metabolism |
| mesh[13].descriptor_name | Peptaibols |
| mesh[14].qualifier_ui | Q000032 |
| mesh[14].descriptor_ui | D000408 |
| mesh[14].is_major_topic | True |
| mesh[14].qualifier_name | analysis |
| mesh[14].descriptor_name | Alamethicin |
| mesh[15].qualifier_ui | Q000737 |
| mesh[15].descriptor_ui | D000408 |
| mesh[15].is_major_topic | True |
| mesh[15].qualifier_name | chemistry |
| mesh[15].descriptor_name | Alamethicin |
| mesh[16].qualifier_ui | Q000378 |
| mesh[16].descriptor_ui | D000408 |
| mesh[16].is_major_topic | True |
| mesh[16].qualifier_name | metabolism |
| mesh[16].descriptor_name | Alamethicin |
| mesh[17].qualifier_ui | Q000737 |
| mesh[17].descriptor_ui | D002462 |
| mesh[17].is_major_topic | False |
| mesh[17].qualifier_name | chemistry |
| mesh[17].descriptor_name | Cell Membrane |
| mesh[18].qualifier_ui | |
| mesh[18].descriptor_ui | D008968 |
| mesh[18].is_major_topic | False |
| mesh[18].qualifier_name | |
| mesh[18].descriptor_name | Molecular Conformation |
| mesh[19].qualifier_ui | |
| mesh[19].descriptor_ui | D001692 |
| mesh[19].is_major_topic | False |
| mesh[19].qualifier_name | |
| mesh[19].descriptor_name | Biological Transport |
| mesh[20].qualifier_ui | Q000494 |
| mesh[20].descriptor_ui | D000900 |
| mesh[20].is_major_topic | False |
| mesh[20].qualifier_name | pharmacology |
| mesh[20].descriptor_name | Anti-Bacterial Agents |
| mesh[21].qualifier_ui | Q000737 |
| mesh[21].descriptor_ui | D000900 |
| mesh[21].is_major_topic | False |
| mesh[21].qualifier_name | chemistry |
| mesh[21].descriptor_name | Anti-Bacterial Agents |
| type | article |
| title | Total synthesis of the natural, medium‐length, peptaibol pentadecaibin and study of the chemical features responsible for its membrane activity |
| awards[0].id | https://openalex.org/G7996902757 |
| awards[0].funder_id | https://openalex.org/F4320321966 |
| awards[0].display_name | |
| awards[0].funder_award_id | Uni‐Impresa 2020‐UNPD0Z9‐0069266 |
| awards[0].funder_display_name | Università degli Studi di Padova |
| awards[1].id | https://openalex.org/G3229059922 |
| awards[1].funder_id | https://openalex.org/F4320321966 |
| awards[1].display_name | |
| awards[1].funder_award_id | Project P‐DiSC#04BIRD2019‐UNIPD |
| awards[1].funder_display_name | Università degli Studi di Padova |
| biblio.issue | 8 |
| biblio.volume | 29 |
| biblio.last_page | e3479 |
| biblio.first_page | e3479 |
| topics[0].id | https://openalex.org/T11103 |
| topics[0].field.id | https://openalex.org/fields/24 |
| topics[0].field.display_name | Immunology and Microbiology |
| topics[0].score | 0.9998999834060669 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/2404 |
| topics[0].subfield.display_name | Microbiology |
| topics[0].display_name | Antimicrobial Peptides and Activities |
| topics[1].id | https://openalex.org/T10814 |
| topics[1].field.id | https://openalex.org/fields/28 |
| topics[1].field.display_name | Neuroscience |
| topics[1].score | 0.9943000078201294 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/2804 |
| topics[1].subfield.display_name | Cellular and Molecular Neuroscience |
| topics[1].display_name | Neuropeptides and Animal Physiology |
| topics[2].id | https://openalex.org/T13326 |
| topics[2].field.id | https://openalex.org/fields/13 |
| topics[2].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[2].score | 0.9930999875068665 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1312 |
| topics[2].subfield.display_name | Molecular Biology |
| topics[2].display_name | Biochemical and Structural Characterization |
| funders[0].id | https://openalex.org/F4320321966 |
| funders[0].ror | https://ror.org/00240q980 |
| funders[0].display_name | Università degli Studi di Padova |
| is_xpac | False |
| apc_list.value | 3660 |
| apc_list.currency | USD |
| apc_list.value_usd | 3660 |
| apc_paid.value | 3660 |
| apc_paid.currency | USD |
| apc_paid.value_usd | 3660 |
| concepts[0].id | https://openalex.org/C2780985075 |
| concepts[0].level | 4 |
| concepts[0].score | 0.926140546798706 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q4705902 |
| concepts[0].display_name | Alamethicin |
| concepts[1].id | https://openalex.org/C41625074 |
| concepts[1].level | 2 |
| concepts[1].score | 0.733604907989502 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q176088 |
| concepts[1].display_name | Membrane |
| concepts[2].id | https://openalex.org/C185592680 |
| concepts[2].level | 0 |
| concepts[2].score | 0.6616705656051636 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q2329 |
| concepts[2].display_name | Chemistry |
| concepts[3].id | https://openalex.org/C2779281246 |
| concepts[3].level | 2 |
| concepts[3].score | 0.557047963142395 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q172847 |
| concepts[3].display_name | Peptide |
| concepts[4].id | https://openalex.org/C515207424 |
| concepts[4].level | 2 |
| concepts[4].score | 0.4726913273334503 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q8066 |
| concepts[4].display_name | Amino acid |
| concepts[5].id | https://openalex.org/C2778530040 |
| concepts[5].level | 3 |
| concepts[5].score | 0.4608141779899597 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q1937315 |
| concepts[5].display_name | Helix (gastropod) |
| concepts[6].id | https://openalex.org/C2781307694 |
| concepts[6].level | 3 |
| concepts[6].score | 0.45643389225006104 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q33123 |
| concepts[6].display_name | Proteolysis |
| concepts[7].id | https://openalex.org/C12554922 |
| concepts[7].level | 1 |
| concepts[7].score | 0.4117743670940399 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q7100 |
| concepts[7].display_name | Biophysics |
| concepts[8].id | https://openalex.org/C71240020 |
| concepts[8].level | 1 |
| concepts[8].score | 0.40292888879776 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q186011 |
| concepts[8].display_name | Stereochemistry |
| concepts[9].id | https://openalex.org/C55493867 |
| concepts[9].level | 1 |
| concepts[9].score | 0.3488990068435669 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q7094 |
| concepts[9].display_name | Biochemistry |
| concepts[10].id | https://openalex.org/C39944091 |
| concepts[10].level | 3 |
| concepts[10].score | 0.24479448795318604 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q423279 |
| concepts[10].display_name | Lipid bilayer |
| concepts[11].id | https://openalex.org/C86803240 |
| concepts[11].level | 0 |
| concepts[11].score | 0.24151840806007385 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[11].display_name | Biology |
| concepts[12].id | https://openalex.org/C18903297 |
| concepts[12].level | 1 |
| concepts[12].score | 0.0 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q7150 |
| concepts[12].display_name | Ecology |
| concepts[13].id | https://openalex.org/C2779965526 |
| concepts[13].level | 2 |
| concepts[13].score | 0.0 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q308841 |
| concepts[13].display_name | Snail |
| concepts[14].id | https://openalex.org/C181199279 |
| concepts[14].level | 2 |
| concepts[14].score | 0.0 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q8047 |
| concepts[14].display_name | Enzyme |
| keywords[0].id | https://openalex.org/keywords/alamethicin |
| keywords[0].score | 0.926140546798706 |
| keywords[0].display_name | Alamethicin |
| keywords[1].id | https://openalex.org/keywords/membrane |
| keywords[1].score | 0.733604907989502 |
| keywords[1].display_name | Membrane |
| keywords[2].id | https://openalex.org/keywords/chemistry |
| keywords[2].score | 0.6616705656051636 |
| keywords[2].display_name | Chemistry |
| keywords[3].id | https://openalex.org/keywords/peptide |
| keywords[3].score | 0.557047963142395 |
| keywords[3].display_name | Peptide |
| keywords[4].id | https://openalex.org/keywords/amino-acid |
| keywords[4].score | 0.4726913273334503 |
| keywords[4].display_name | Amino acid |
| keywords[5].id | https://openalex.org/keywords/helix |
| keywords[5].score | 0.4608141779899597 |
| keywords[5].display_name | Helix (gastropod) |
| keywords[6].id | https://openalex.org/keywords/proteolysis |
| keywords[6].score | 0.45643389225006104 |
| keywords[6].display_name | Proteolysis |
| keywords[7].id | https://openalex.org/keywords/biophysics |
| keywords[7].score | 0.4117743670940399 |
| keywords[7].display_name | Biophysics |
| keywords[8].id | https://openalex.org/keywords/stereochemistry |
| keywords[8].score | 0.40292888879776 |
| keywords[8].display_name | Stereochemistry |
| keywords[9].id | https://openalex.org/keywords/biochemistry |
| keywords[9].score | 0.3488990068435669 |
| keywords[9].display_name | Biochemistry |
| keywords[10].id | https://openalex.org/keywords/lipid-bilayer |
| keywords[10].score | 0.24479448795318604 |
| keywords[10].display_name | Lipid bilayer |
| keywords[11].id | https://openalex.org/keywords/biology |
| keywords[11].score | 0.24151840806007385 |
| keywords[11].display_name | Biology |
| language | en |
| locations[0].id | doi:10.1002/psc.3479 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S184443419 |
| locations[0].source.issn | 1075-2617, 1099-1387 |
| locations[0].source.type | journal |
| locations[0].source.is_oa | False |
| locations[0].source.issn_l | 1075-2617 |
| locations[0].source.is_core | True |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | Journal of Peptide Science |
| locations[0].source.host_organization | https://openalex.org/P4310320595 |
| locations[0].source.host_organization_name | Wiley |
| locations[0].source.host_organization_lineage | https://openalex.org/P4310320595 |
| locations[0].source.host_organization_lineage_names | Wiley |
| locations[0].license | cc-by |
| locations[0].pdf_url | https://onlinelibrary.wiley.com/doi/pdfdirect/10.1002/psc.3479 |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | Journal of Peptide Science |
| locations[0].landing_page_url | https://doi.org/10.1002/psc.3479 |
| locations[1].id | pmid:36652104 |
| locations[1].is_oa | False |
| locations[1].source.id | https://openalex.org/S4306525036 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | PubMed |
| locations[1].source.host_organization | https://openalex.org/I1299303238 |
| locations[1].source.host_organization_name | National Institutes of Health |
| locations[1].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[1].license | |
| locations[1].pdf_url | |
| locations[1].version | publishedVersion |
| locations[1].raw_type | |
| locations[1].license_id | |
| locations[1].is_accepted | True |
| locations[1].is_published | True |
| locations[1].raw_source_name | Journal of peptide science : an official publication of the European Peptide Society |
| locations[1].landing_page_url | https://pubmed.ncbi.nlm.nih.gov/36652104 |
| locations[2].id | pmh:oai:www.research.unipd.it:11577/3466089 |
| locations[2].is_oa | True |
| locations[2].source.id | https://openalex.org/S4306402448 |
| locations[2].source.issn | |
| locations[2].source.type | repository |
| locations[2].source.is_oa | False |
| locations[2].source.issn_l | |
| locations[2].source.is_core | False |
| locations[2].source.is_in_doaj | False |
| locations[2].source.display_name | Padua Research Archive (University of Padua) |
| locations[2].source.host_organization | https://openalex.org/I138689650 |
| locations[2].source.host_organization_name | University of Padua |
| locations[2].source.host_organization_lineage | https://openalex.org/I138689650 |
| locations[2].license | cc-by |
| locations[2].pdf_url | |
| locations[2].version | submittedVersion |
| locations[2].raw_type | info:eu-repo/semantics/article |
| locations[2].license_id | https://openalex.org/licenses/cc-by |
| locations[2].is_accepted | False |
| locations[2].is_published | False |
| locations[2].raw_source_name | |
| locations[2].landing_page_url | https://hdl.handle.net/11577/3466089 |
| indexed_in | crossref, pubmed |
| authorships[0].author.id | https://openalex.org/A5047657095 |
| authorships[0].author.orcid | https://orcid.org/0009-0006-7670-0638 |
| authorships[0].author.display_name | Laura Morbiato |
| authorships[0].countries | IT |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I138689650 |
| authorships[0].affiliations[0].raw_affiliation_string | Department of Chemical Sciences, University of Padova, Padova, Italy |
| authorships[0].institutions[0].id | https://openalex.org/I138689650 |
| authorships[0].institutions[0].ror | https://ror.org/00240q980 |
| authorships[0].institutions[0].type | education |
| authorships[0].institutions[0].lineage | https://openalex.org/I138689650 |
| authorships[0].institutions[0].country_code | IT |
| authorships[0].institutions[0].display_name | University of Padua |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Laura Morbiato |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Department of Chemical Sciences, University of Padova, Padova, Italy |
| authorships[1].author.id | https://openalex.org/A5066080588 |
| authorships[1].author.orcid | |
| authorships[1].author.display_name | David S. A. Haneen |
| authorships[1].countries | EG |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I107720978 |
| authorships[1].affiliations[0].raw_affiliation_string | Chemistry Department, Faculty of Science, Ain Shams University, Cairo, Abbassia, 11566 Egypt |
| authorships[1].institutions[0].id | https://openalex.org/I107720978 |
| authorships[1].institutions[0].ror | https://ror.org/00cb9w016 |
| authorships[1].institutions[0].type | education |
| authorships[1].institutions[0].lineage | https://openalex.org/I107720978 |
| authorships[1].institutions[0].country_code | EG |
| authorships[1].institutions[0].display_name | Ain Shams University |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | David S. A. Haneen |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | Chemistry Department, Faculty of Science, Ain Shams University, Cairo, Abbassia, 11566 Egypt |
| authorships[2].author.id | https://openalex.org/A5040500402 |
| authorships[2].author.orcid | https://orcid.org/0000-0003-2008-4961 |
| authorships[2].author.display_name | Fernando Formaggio |
| authorships[2].countries | IT |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I138689650 |
| authorships[2].affiliations[0].raw_affiliation_string | Department of Chemical Sciences, University of Padova, Padova, Italy |
| authorships[2].affiliations[1].institution_ids | https://openalex.org/I138689650 |
| authorships[2].affiliations[1].raw_affiliation_string | Padova Unit, CNR Institute of Biomolecular Chemistry, University of Padova, Padova, Italy |
| authorships[2].institutions[0].id | https://openalex.org/I138689650 |
| authorships[2].institutions[0].ror | https://ror.org/00240q980 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I138689650 |
| authorships[2].institutions[0].country_code | IT |
| authorships[2].institutions[0].display_name | University of Padua |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Fernando Formaggio |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | Department of Chemical Sciences, University of Padova, Padova, Italy, Padova Unit, CNR Institute of Biomolecular Chemistry, University of Padova, Padova, Italy |
| authorships[3].author.id | https://openalex.org/A5003688394 |
| authorships[3].author.orcid | https://orcid.org/0000-0002-3302-6499 |
| authorships[3].author.display_name | Marta De Zotti |
| authorships[3].countries | IT |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I138689650 |
| authorships[3].affiliations[0].raw_affiliation_string | Department of Chemical Sciences, University of Padova, Padova, Italy |
| authorships[3].affiliations[1].institution_ids | https://openalex.org/I138689650 |
| authorships[3].affiliations[1].raw_affiliation_string | Padova Unit, CNR Institute of Biomolecular Chemistry, University of Padova, Padova, Italy |
| authorships[3].institutions[0].id | https://openalex.org/I138689650 |
| authorships[3].institutions[0].ror | https://ror.org/00240q980 |
| authorships[3].institutions[0].type | education |
| authorships[3].institutions[0].lineage | https://openalex.org/I138689650 |
| authorships[3].institutions[0].country_code | IT |
| authorships[3].institutions[0].display_name | University of Padua |
| authorships[3].author_position | last |
| authorships[3].raw_author_name | Marta De Zotti |
| authorships[3].is_corresponding | True |
| authorships[3].raw_affiliation_strings | Department of Chemical Sciences, University of Padova, Padova, Italy, Padova Unit, CNR Institute of Biomolecular Chemistry, University of Padova, Padova, Italy |
| has_content.pdf | True |
| has_content.grobid_xml | True |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://onlinelibrary.wiley.com/doi/pdfdirect/10.1002/psc.3479 |
| open_access.oa_status | hybrid |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Total synthesis of the natural, medium‐length, peptaibol pentadecaibin and study of the chemical features responsible for its membrane activity |
| has_fulltext | True |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T11103 |
| primary_topic.field.id | https://openalex.org/fields/24 |
| primary_topic.field.display_name | Immunology and Microbiology |
| primary_topic.score | 0.9998999834060669 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/2404 |
| primary_topic.subfield.display_name | Microbiology |
| primary_topic.display_name | Antimicrobial Peptides and Activities |
| related_works | https://openalex.org/W1970092799, https://openalex.org/W2134155512, https://openalex.org/W2055225856, https://openalex.org/W2343150819, https://openalex.org/W4385273854, https://openalex.org/W4382198899, https://openalex.org/W1965301914, https://openalex.org/W2496196519, https://openalex.org/W2315950124, https://openalex.org/W1973744249 |
| cited_by_count | 5 |
| counts_by_year[0].year | 2025 |
| counts_by_year[0].cited_by_count | 1 |
| counts_by_year[1].year | 2024 |
| counts_by_year[1].cited_by_count | 2 |
| counts_by_year[2].year | 2023 |
| counts_by_year[2].cited_by_count | 2 |
| locations_count | 3 |
| best_oa_location.id | doi:10.1002/psc.3479 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S184443419 |
| best_oa_location.source.issn | 1075-2617, 1099-1387 |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | False |
| best_oa_location.source.issn_l | 1075-2617 |
| best_oa_location.source.is_core | True |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | Journal of Peptide Science |
| best_oa_location.source.host_organization | https://openalex.org/P4310320595 |
| best_oa_location.source.host_organization_name | Wiley |
| best_oa_location.source.host_organization_lineage | https://openalex.org/P4310320595 |
| best_oa_location.source.host_organization_lineage_names | Wiley |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | https://onlinelibrary.wiley.com/doi/pdfdirect/10.1002/psc.3479 |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | journal-article |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | Journal of Peptide Science |
| best_oa_location.landing_page_url | https://doi.org/10.1002/psc.3479 |
| primary_location.id | doi:10.1002/psc.3479 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S184443419 |
| primary_location.source.issn | 1075-2617, 1099-1387 |
| primary_location.source.type | journal |
| primary_location.source.is_oa | False |
| primary_location.source.issn_l | 1075-2617 |
| primary_location.source.is_core | True |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | Journal of Peptide Science |
| primary_location.source.host_organization | https://openalex.org/P4310320595 |
| primary_location.source.host_organization_name | Wiley |
| primary_location.source.host_organization_lineage | https://openalex.org/P4310320595 |
| primary_location.source.host_organization_lineage_names | Wiley |
| primary_location.license | cc-by |
| primary_location.pdf_url | https://onlinelibrary.wiley.com/doi/pdfdirect/10.1002/psc.3479 |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | Journal of Peptide Science |
| primary_location.landing_page_url | https://doi.org/10.1002/psc.3479 |
| publication_date | 2023-01-18 |
| publication_year | 2023 |
| referenced_works | https://openalex.org/W2150107572, https://openalex.org/W2164222756, https://openalex.org/W2046704370, https://openalex.org/W2128726798, https://openalex.org/W2079287985, https://openalex.org/W2031695819, https://openalex.org/W2178922091, https://openalex.org/W4281785935, https://openalex.org/W3132357962, https://openalex.org/W1979528014, https://openalex.org/W2038252927, https://openalex.org/W2160780417, https://openalex.org/W2054659127 |
| referenced_works_count | 13 |
| abstract_inverted_index.A | 153 |
| abstract_inverted_index.C | 26 |
| abstract_inverted_index.a | 125, 171, 180, 225 |
| abstract_inverted_index.By | 170 |
| abstract_inverted_index.In | 130, 183 |
| abstract_inverted_index.We | 243 |
| abstract_inverted_index.as | 82 |
| abstract_inverted_index.by | 65 |
| abstract_inverted_index.in | 21, 68, 111 |
| abstract_inverted_index.is | 89, 95, 150, 159, 248 |
| abstract_inverted_index.of | 24, 43, 86, 101, 135, 173, 188, 195, 210, 217, 219 |
| abstract_inverted_index.on | 115, 123, 192 |
| abstract_inverted_index.or | 55, 145 |
| abstract_inverted_index.so | 49 |
| abstract_inverted_index.to | 13, 98, 107, 132, 164, 178, 239, 255 |
| abstract_inverted_index.us | 163 |
| abstract_inverted_index.we | 175 |
| abstract_inverted_index.α | 27 |
| abstract_inverted_index.Aib | 198, 222 |
| abstract_inverted_index.All | 42 |
| abstract_inverted_index.The | 57, 84, 119, 213 |
| abstract_inverted_index.and | 11, 93, 147, 155, 233 |
| abstract_inverted_index.are | 1, 105, 201 |
| abstract_inverted_index.but | 229 |
| abstract_inverted_index.due | 97 |
| abstract_inverted_index.far | 50, 90 |
| abstract_inverted_index.for | 78, 203, 251 |
| abstract_inverted_index.has | 75 |
| abstract_inverted_index.its | 136, 139, 148, 211, 231 |
| abstract_inverted_index.not | 142, 151, 230 |
| abstract_inverted_index.the | 19, 44, 60, 69, 72, 99, 116, 133, 185, 193, 204, 237, 252 |
| abstract_inverted_index.two | 167, 218 |
| abstract_inverted_index.Both | 15 |
| abstract_inverted_index.They | 104 |
| abstract_inverted_index.able | 177 |
| abstract_inverted_index.acid | 30 |
| abstract_inverted_index.acts | 64 |
| abstract_inverted_index.also | 166 |
| abstract_inverted_index.been | 76 |
| abstract_inverted_index.does | 141 |
| abstract_inverted_index.from | 18 |
| abstract_inverted_index.less | 91 |
| abstract_inverted_index.most | 61 |
| abstract_inverted_index.stem | 17 |
| abstract_inverted_index.such | 81 |
| abstract_inverted_index.that | 161, 200, 245 |
| abstract_inverted_index.this | 94 |
| abstract_inverted_index.were | 176 |
| abstract_inverted_index.with | 7, 35, 52 |
| abstract_inverted_index.work | 121 |
| abstract_inverted_index.(Aib) | 31 |
| abstract_inverted_index.begin | 51 |
| abstract_inverted_index.cause | 240 |
| abstract_inverted_index.helix | 149 |
| abstract_inverted_index.loses | 234 |
| abstract_inverted_index.model | 257 |
| abstract_inverted_index.pores | 67 |
| abstract_inverted_index.their | 22, 40, 102 |
| abstract_inverted_index.these | 220 |
| abstract_inverted_index.three | 196, 221 |
| abstract_inverted_index.ways, | 113 |
| abstract_inverted_index.albeit | 206 |
| abstract_inverted_index.almost | 235 |
| abstract_inverted_index.carpet | 73 |
| abstract_inverted_index.clear, | 205 |
| abstract_inverted_index.devoid | 216 |
| abstract_inverted_index.family | 137 |
| abstract_inverted_index.helix. | 212 |
| abstract_inverted_index.needed | 250 |
| abstract_inverted_index.partly | 96 |
| abstract_inverted_index.ability | 238, 254 |
| abstract_inverted_index.allowed | 162 |
| abstract_inverted_index.analog, | 215 |
| abstract_inverted_index.causing | 39 |
| abstract_inverted_index.claimed | 77 |
| abstract_inverted_index.depends | 191 |
| abstract_inverted_index.endowed | 6 |
| abstract_inverted_index.focuses | 122 |
| abstract_inverted_index.forming | 66 |
| abstract_inverted_index.heavily | 190 |
| abstract_inverted_index.helical | 9, 227 |
| abstract_inverted_index.kinked. | 152 |
| abstract_inverted_index.modest, | 207 |
| abstract_inverted_index.peptide | 53, 253 |
| abstract_inverted_index.perturb | 108, 256 |
| abstract_inverted_index.present | 120 |
| abstract_inverted_index.produce | 165 |
| abstract_inverted_index.shorter | 168 |
| abstract_inverted_index.studied | 62 |
| abstract_inverted_index.usually | 38 |
| abstract_inverted_index.activity | 187 |
| abstract_inverted_index.analogs. | 169 |
| abstract_inverted_index.believed | 106 |
| abstract_inverted_index.comprise | 143 |
| abstract_inverted_index.conclude | 244 |
| abstract_inverted_index.contrast | 131 |
| abstract_inverted_index.features | 16 |
| abstract_inverted_index.interact | 34 |
| abstract_inverted_index.leakage. | 41, 242 |
| abstract_inverted_index.majority | 134 |
| abstract_inverted_index.members, | 138 |
| abstract_inverted_index.membrane | 109, 117, 186, 241 |
| abstract_inverted_index.peptides | 5 |
| abstract_inverted_index.presence | 20, 194 |
| abstract_inverted_index.probably | 249 |
| abstract_inverted_index.proposed | 48 |
| abstract_inverted_index.recently | 126 |
| abstract_inverted_index.reliable | 154 |
| abstract_inverted_index.residues | 199 |
| abstract_inverted_index.sequence | 23, 140 |
| abstract_inverted_index.several, | 25 |
| abstract_inverted_index.shortest | 214 |
| abstract_inverted_index.studied, | 92 |
| abstract_inverted_index.character | 209 |
| abstract_inverted_index.depending | 114 |
| abstract_inverted_index.described | 160 |
| abstract_inverted_index.different | 112 |
| abstract_inverted_index.effective | 156 |
| abstract_inverted_index.establish | 179 |
| abstract_inverted_index.mechanism | 74, 85 |
| abstract_inverted_index.naturally | 2 |
| abstract_inverted_index.preserves | 224 |
| abstract_inverted_index.procedure | 158 |
| abstract_inverted_index.prolines, | 146 |
| abstract_inverted_index.residues, | 223 |
| abstract_inverted_index.residues. | 32 |
| abstract_inverted_index.synthesis | 157 |
| abstract_inverted_index.Peptaibols | 0, 33 |
| abstract_inverted_index.biological | 36 |
| abstract_inverted_index.completely | 236 |
| abstract_inverted_index.mechanisms | 47 |
| abstract_inverted_index.membranes, | 37 |
| abstract_inverted_index.membranes. | 70, 258 |
| abstract_inverted_index.occurring, | 3 |
| abstract_inverted_index.peptaibol, | 63 |
| abstract_inverted_index.peptaibol. | 129 |
| abstract_inverted_index.peptaibols | 88 |
| abstract_inverted_index.resistance | 12 |
| abstract_inverted_index.synthesis. | 103 |
| abstract_inverted_index.trichogin. | 83 |
| abstract_inverted_index.Conversely, | 71 |
| abstract_inverted_index.aggregation | 54 |
| abstract_inverted_index.amphiphilic | 208 |
| abstract_inverted_index.combination | 172 |
| abstract_inverted_index.consecutive | 197 |
| abstract_inverted_index.discovered, | 127 |
| abstract_inverted_index.interaction | 46 |
| abstract_inverted_index.particular, | 184 |
| abstract_inverted_index.peptaibols, | 80 |
| abstract_inverted_index.properties. | 118 |
| abstract_inverted_index.responsible | 202 |
| abstract_inverted_index.techniques, | 174 |
| abstract_inverted_index.alamethicin, | 59 |
| abstract_inverted_index.difficulties | 100 |
| abstract_inverted_index.permeability | 110 |
| abstract_inverted_index.proteolysis. | 14 |
| abstract_inverted_index.accumulation. | 56 |
| abstract_inverted_index.antimicrobial | 4 |
| abstract_inverted_index.conformation, | 228 |
| abstract_inverted_index.conformations | 10 |
| abstract_inverted_index.long‐length | 58 |
| abstract_inverted_index.pentadecaibin | 189, 246 |
| abstract_inverted_index.relationship. | 182 |
| abstract_inverted_index.amphiphilicity | 247 |
| abstract_inverted_index.pentadecaibin, | 124 |
| abstract_inverted_index.short‐length | 79 |
| abstract_inverted_index.well‐defined | 8, 226 |
| abstract_inverted_index.amphipathicity, | 232 |
| abstract_inverted_index.hydroxyprolines | 144 |
| abstract_inverted_index.medium‐length | 87, 128 |
| abstract_inverted_index.peptaibol–membrane | 45 |
| abstract_inverted_index.α‐aminoisobutyric | 29 |
| abstract_inverted_index.‐tetrasubstituted, | 28 |
| abstract_inverted_index.3D‐structure–activity | 181 |
| cited_by_percentile_year.max | 96 |
| cited_by_percentile_year.min | 91 |
| corresponding_author_ids | https://openalex.org/A5003688394 |
| countries_distinct_count | 2 |
| institutions_distinct_count | 4 |
| corresponding_institution_ids | https://openalex.org/I138689650 |
| citation_normalized_percentile.value | 0.73675531 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |