Use of pus metagenomic next-generation sequencing for efficient identification of pathogens in patients with sepsis Article Swipe
 YOU?
·
· 2024
· Open Access
·
· DOI: https://doi.org/10.1007/s12223-024-01134-7
YOU?
·
· 2024
· Open Access
·
· DOI: https://doi.org/10.1007/s12223-024-01134-7
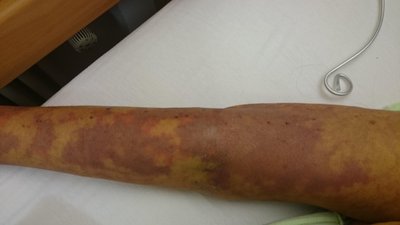
The positive detection rate of blood metagenomic next-generation sequencing (mNGS) was still too low to meet clinical needs, while pus from the site of primary infection may be advantageous for identification of pathogens. To assess the value of mNGS using pus in patients with sepsis, thirty-five samples were collected. Pathogen identification and mixed infection diagnosis obtained by use of mNGS or cultivation methods were compared. Fifty-three aerobic or facultative anaerobes, 59 obligate anaerobes and 7 fungi were identified by the two methods. mNGS increased the accuracy rate of diagnosing aerobic or facultative anaerobic infections from 44.4% to 94.4%; mNGS also increased the sensitivity of diagnosing obligate anaerobic infections from 52.9% to 100.0%; however, mNGS did not show any advantage in terms of fungal infections. Culture and mNGS identified 1 and 24 patients with mixed infection, respectively. For obligate anaerobes, source of microorganisms was analyzed. The odontogenic bacteria all caused empyema (n = 7) or skin and soft tissue infections (n = 5), whereas the gut-derived microbes all caused intra-abdominal infections (n = 7). We also compared the clinical characteristics of non-obligate anaerobic and obligate anaerobic infection groups. The SOFA score [9.0 (7.5, 14.3) vs. 5.0 (3.0, 8.0), P = 0.005], procalcitonin value [4.7 (1.8, 39.9) vs. 2.50 (0.7, 8.0), P = 0.035], the proportion of septic shock (66.7% vs. 35.3%, P = 0.044) and acute liver injury (66.7% vs. 23.5%, P = 0.018) in the non-obligate anaerobic infection group were significantly higher than those in the obligate anaerobic infection group. In patients with sepsis caused by purulent infection, mNGS using pus from the primary lesion may yield more valuable microbiological information.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.1007/s12223-024-01134-7
- https://link.springer.com/content/pdf/10.1007/s12223-024-01134-7.pdf
- OA Status
- hybrid
- Cited By
- 1
- References
- 29
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4391735419
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4391735419Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1007/s12223-024-01134-7Digital Object Identifier
- Title
-
Use of pus metagenomic next-generation sequencing for efficient identification of pathogens in patients with sepsisWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2024Year of publication
- Publication date
-
2024-02-11Full publication date if available
- Authors
-
Zhendong Chen, Tingting Ye, Yuxi He, Aijun Pan, Qing MeiList of authors in order
- Landing page
-
https://doi.org/10.1007/s12223-024-01134-7Publisher landing page
- PDF URL
-
https://link.springer.com/content/pdf/10.1007/s12223-024-01134-7.pdfDirect link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
hybridOpen access status per OpenAlex
- OA URL
-
https://link.springer.com/content/pdf/10.1007/s12223-024-01134-7.pdfDirect OA link when available
- Concepts
-
Metagenomics, Identification (biology), DNA sequencing, Sepsis, Biology, Computational biology, Microbiology, Genetics, Immunology, Gene, BotanyTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
1Total citation count in OpenAlex
- Citations by year (recent)
-
2025: 1Per-year citation counts (last 5 years)
- References (count)
-
29Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4391735419 |
|---|---|
| doi | https://doi.org/10.1007/s12223-024-01134-7 |
| ids.doi | https://doi.org/10.1007/s12223-024-01134-7 |
| ids.pmid | https://pubmed.ncbi.nlm.nih.gov/38341816 |
| ids.openalex | https://openalex.org/W4391735419 |
| fwci | 0.9674516 |
| mesh[0].qualifier_ui | |
| mesh[0].descriptor_ui | D006801 |
| mesh[0].is_major_topic | False |
| mesh[0].qualifier_name | |
| mesh[0].descriptor_name | Humans |
| mesh[1].qualifier_ui | Q000382 |
| mesh[1].descriptor_ui | D018805 |
| mesh[1].is_major_topic | True |
| mesh[1].qualifier_name | microbiology |
| mesh[1].descriptor_name | Sepsis |
| mesh[2].qualifier_ui | Q000175 |
| mesh[2].descriptor_ui | D018805 |
| mesh[2].is_major_topic | True |
| mesh[2].qualifier_name | diagnosis |
| mesh[2].descriptor_name | Sepsis |
| mesh[3].qualifier_ui | |
| mesh[3].descriptor_ui | D059014 |
| mesh[3].is_major_topic | True |
| mesh[3].qualifier_name | |
| mesh[3].descriptor_name | High-Throughput Nucleotide Sequencing |
| mesh[4].qualifier_ui | Q000379 |
| mesh[4].descriptor_ui | D056186 |
| mesh[4].is_major_topic | True |
| mesh[4].qualifier_name | methods |
| mesh[4].descriptor_name | Metagenomics |
| mesh[5].qualifier_ui | |
| mesh[5].descriptor_ui | D008297 |
| mesh[5].is_major_topic | False |
| mesh[5].qualifier_name | |
| mesh[5].descriptor_name | Male |
| mesh[6].qualifier_ui | |
| mesh[6].descriptor_ui | D008875 |
| mesh[6].is_major_topic | False |
| mesh[6].qualifier_name | |
| mesh[6].descriptor_name | Middle Aged |
| mesh[7].qualifier_ui | |
| mesh[7].descriptor_ui | D005260 |
| mesh[7].is_major_topic | False |
| mesh[7].qualifier_name | |
| mesh[7].descriptor_name | Female |
| mesh[8].qualifier_ui | |
| mesh[8].descriptor_ui | D000368 |
| mesh[8].is_major_topic | False |
| mesh[8].qualifier_name | |
| mesh[8].descriptor_name | Aged |
| mesh[9].qualifier_ui | Q000235 |
| mesh[9].descriptor_ui | D001419 |
| mesh[9].is_major_topic | True |
| mesh[9].qualifier_name | genetics |
| mesh[9].descriptor_name | Bacteria |
| mesh[10].qualifier_ui | Q000302 |
| mesh[10].descriptor_ui | D001419 |
| mesh[10].is_major_topic | True |
| mesh[10].qualifier_name | isolation & purification |
| mesh[10].descriptor_name | Bacteria |
| mesh[11].qualifier_ui | Q000145 |
| mesh[11].descriptor_ui | D001419 |
| mesh[11].is_major_topic | True |
| mesh[11].qualifier_name | classification |
| mesh[11].descriptor_name | Bacteria |
| mesh[12].qualifier_ui | Q000235 |
| mesh[12].descriptor_ui | D005658 |
| mesh[12].is_major_topic | False |
| mesh[12].qualifier_name | genetics |
| mesh[12].descriptor_name | Fungi |
| mesh[13].qualifier_ui | Q000302 |
| mesh[13].descriptor_ui | D005658 |
| mesh[13].is_major_topic | False |
| mesh[13].qualifier_name | isolation & purification |
| mesh[13].descriptor_name | Fungi |
| mesh[14].qualifier_ui | Q000145 |
| mesh[14].descriptor_ui | D005658 |
| mesh[14].is_major_topic | False |
| mesh[14].qualifier_name | classification |
| mesh[14].descriptor_name | Fungi |
| mesh[15].qualifier_ui | Q000382 |
| mesh[15].descriptor_ui | D013492 |
| mesh[15].is_major_topic | False |
| mesh[15].qualifier_name | microbiology |
| mesh[15].descriptor_name | Suppuration |
| mesh[16].qualifier_ui | |
| mesh[16].descriptor_ui | D000328 |
| mesh[16].is_major_topic | False |
| mesh[16].qualifier_name | |
| mesh[16].descriptor_name | Adult |
| mesh[17].qualifier_ui | |
| mesh[17].descriptor_ui | D000369 |
| mesh[17].is_major_topic | False |
| mesh[17].qualifier_name | |
| mesh[17].descriptor_name | Aged, 80 and over |
| mesh[18].qualifier_ui | |
| mesh[18].descriptor_ui | D012680 |
| mesh[18].is_major_topic | False |
| mesh[18].qualifier_name | |
| mesh[18].descriptor_name | Sensitivity and Specificity |
| mesh[19].qualifier_ui | |
| mesh[19].descriptor_ui | D006801 |
| mesh[19].is_major_topic | False |
| mesh[19].qualifier_name | |
| mesh[19].descriptor_name | Humans |
| mesh[20].qualifier_ui | Q000382 |
| mesh[20].descriptor_ui | D018805 |
| mesh[20].is_major_topic | True |
| mesh[20].qualifier_name | microbiology |
| mesh[20].descriptor_name | Sepsis |
| mesh[21].qualifier_ui | Q000175 |
| mesh[21].descriptor_ui | D018805 |
| mesh[21].is_major_topic | True |
| mesh[21].qualifier_name | diagnosis |
| mesh[21].descriptor_name | Sepsis |
| mesh[22].qualifier_ui | |
| mesh[22].descriptor_ui | D059014 |
| mesh[22].is_major_topic | True |
| mesh[22].qualifier_name | |
| mesh[22].descriptor_name | High-Throughput Nucleotide Sequencing |
| mesh[23].qualifier_ui | Q000379 |
| mesh[23].descriptor_ui | D056186 |
| mesh[23].is_major_topic | True |
| mesh[23].qualifier_name | methods |
| mesh[23].descriptor_name | Metagenomics |
| mesh[24].qualifier_ui | |
| mesh[24].descriptor_ui | D008297 |
| mesh[24].is_major_topic | False |
| mesh[24].qualifier_name | |
| mesh[24].descriptor_name | Male |
| mesh[25].qualifier_ui | |
| mesh[25].descriptor_ui | D008875 |
| mesh[25].is_major_topic | False |
| mesh[25].qualifier_name | |
| mesh[25].descriptor_name | Middle Aged |
| mesh[26].qualifier_ui | |
| mesh[26].descriptor_ui | D005260 |
| mesh[26].is_major_topic | False |
| mesh[26].qualifier_name | |
| mesh[26].descriptor_name | Female |
| mesh[27].qualifier_ui | |
| mesh[27].descriptor_ui | D000368 |
| mesh[27].is_major_topic | False |
| mesh[27].qualifier_name | |
| mesh[27].descriptor_name | Aged |
| mesh[28].qualifier_ui | Q000235 |
| mesh[28].descriptor_ui | D001419 |
| mesh[28].is_major_topic | True |
| mesh[28].qualifier_name | genetics |
| mesh[28].descriptor_name | Bacteria |
| mesh[29].qualifier_ui | Q000302 |
| mesh[29].descriptor_ui | D001419 |
| mesh[29].is_major_topic | True |
| mesh[29].qualifier_name | isolation & purification |
| mesh[29].descriptor_name | Bacteria |
| mesh[30].qualifier_ui | Q000145 |
| mesh[30].descriptor_ui | D001419 |
| mesh[30].is_major_topic | True |
| mesh[30].qualifier_name | classification |
| mesh[30].descriptor_name | Bacteria |
| mesh[31].qualifier_ui | Q000235 |
| mesh[31].descriptor_ui | D005658 |
| mesh[31].is_major_topic | False |
| mesh[31].qualifier_name | genetics |
| mesh[31].descriptor_name | Fungi |
| mesh[32].qualifier_ui | Q000302 |
| mesh[32].descriptor_ui | D005658 |
| mesh[32].is_major_topic | False |
| mesh[32].qualifier_name | isolation & purification |
| mesh[32].descriptor_name | Fungi |
| mesh[33].qualifier_ui | Q000145 |
| mesh[33].descriptor_ui | D005658 |
| mesh[33].is_major_topic | False |
| mesh[33].qualifier_name | classification |
| mesh[33].descriptor_name | Fungi |
| mesh[34].qualifier_ui | Q000382 |
| mesh[34].descriptor_ui | D013492 |
| mesh[34].is_major_topic | False |
| mesh[34].qualifier_name | microbiology |
| mesh[34].descriptor_name | Suppuration |
| mesh[35].qualifier_ui | |
| mesh[35].descriptor_ui | D000328 |
| mesh[35].is_major_topic | False |
| mesh[35].qualifier_name | |
| mesh[35].descriptor_name | Adult |
| mesh[36].qualifier_ui | |
| mesh[36].descriptor_ui | D000369 |
| mesh[36].is_major_topic | False |
| mesh[36].qualifier_name | |
| mesh[36].descriptor_name | Aged, 80 and over |
| mesh[37].qualifier_ui | |
| mesh[37].descriptor_ui | D012680 |
| mesh[37].is_major_topic | False |
| mesh[37].qualifier_name | |
| mesh[37].descriptor_name | Sensitivity and Specificity |
| mesh[38].qualifier_ui | |
| mesh[38].descriptor_ui | D006801 |
| mesh[38].is_major_topic | False |
| mesh[38].qualifier_name | |
| mesh[38].descriptor_name | Humans |
| mesh[39].qualifier_ui | Q000382 |
| mesh[39].descriptor_ui | D018805 |
| mesh[39].is_major_topic | True |
| mesh[39].qualifier_name | microbiology |
| mesh[39].descriptor_name | Sepsis |
| mesh[40].qualifier_ui | Q000175 |
| mesh[40].descriptor_ui | D018805 |
| mesh[40].is_major_topic | True |
| mesh[40].qualifier_name | diagnosis |
| mesh[40].descriptor_name | Sepsis |
| mesh[41].qualifier_ui | |
| mesh[41].descriptor_ui | D059014 |
| mesh[41].is_major_topic | True |
| mesh[41].qualifier_name | |
| mesh[41].descriptor_name | High-Throughput Nucleotide Sequencing |
| mesh[42].qualifier_ui | Q000379 |
| mesh[42].descriptor_ui | D056186 |
| mesh[42].is_major_topic | True |
| mesh[42].qualifier_name | methods |
| mesh[42].descriptor_name | Metagenomics |
| mesh[43].qualifier_ui | |
| mesh[43].descriptor_ui | D008297 |
| mesh[43].is_major_topic | False |
| mesh[43].qualifier_name | |
| mesh[43].descriptor_name | Male |
| mesh[44].qualifier_ui | |
| mesh[44].descriptor_ui | D008875 |
| mesh[44].is_major_topic | False |
| mesh[44].qualifier_name | |
| mesh[44].descriptor_name | Middle Aged |
| mesh[45].qualifier_ui | |
| mesh[45].descriptor_ui | D005260 |
| mesh[45].is_major_topic | False |
| mesh[45].qualifier_name | |
| mesh[45].descriptor_name | Female |
| mesh[46].qualifier_ui | |
| mesh[46].descriptor_ui | D000368 |
| mesh[46].is_major_topic | False |
| mesh[46].qualifier_name | |
| mesh[46].descriptor_name | Aged |
| mesh[47].qualifier_ui | Q000235 |
| mesh[47].descriptor_ui | D001419 |
| mesh[47].is_major_topic | True |
| mesh[47].qualifier_name | genetics |
| mesh[47].descriptor_name | Bacteria |
| mesh[48].qualifier_ui | Q000302 |
| mesh[48].descriptor_ui | D001419 |
| mesh[48].is_major_topic | True |
| mesh[48].qualifier_name | isolation & purification |
| mesh[48].descriptor_name | Bacteria |
| mesh[49].qualifier_ui | Q000145 |
| mesh[49].descriptor_ui | D001419 |
| mesh[49].is_major_topic | True |
| mesh[49].qualifier_name | classification |
| mesh[49].descriptor_name | Bacteria |
| type | article |
| title | Use of pus metagenomic next-generation sequencing for efficient identification of pathogens in patients with sepsis |
| biblio.issue | 5 |
| biblio.volume | 69 |
| biblio.last_page | 1011 |
| biblio.first_page | 1003 |
| topics[0].id | https://openalex.org/T11681 |
| topics[0].field.id | https://openalex.org/fields/27 |
| topics[0].field.display_name | Medicine |
| topics[0].score | 0.996999979019165 |
| topics[0].domain.id | https://openalex.org/domains/4 |
| topics[0].domain.display_name | Health Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/2706 |
| topics[0].subfield.display_name | Critical Care and Intensive Care Medicine |
| topics[0].display_name | Nosocomial Infections in ICU |
| topics[1].id | https://openalex.org/T11288 |
| topics[1].field.id | https://openalex.org/fields/27 |
| topics[1].field.display_name | Medicine |
| topics[1].score | 0.9961000084877014 |
| topics[1].domain.id | https://openalex.org/domains/4 |
| topics[1].domain.display_name | Health Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/2725 |
| topics[1].subfield.display_name | Infectious Diseases |
| topics[1].display_name | Clostridium difficile and Clostridium perfringens research |
| topics[2].id | https://openalex.org/T12167 |
| topics[2].field.id | https://openalex.org/fields/13 |
| topics[2].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[2].score | 0.9933000206947327 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1308 |
| topics[2].subfield.display_name | Clinical Biochemistry |
| topics[2].display_name | Bacterial Identification and Susceptibility Testing |
| is_xpac | False |
| apc_list.value | 2790 |
| apc_list.currency | EUR |
| apc_list.value_usd | 3590 |
| apc_paid.value | 2790 |
| apc_paid.currency | EUR |
| apc_paid.value_usd | 3590 |
| concepts[0].id | https://openalex.org/C15151743 |
| concepts[0].level | 3 |
| concepts[0].score | 0.8676527738571167 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q903778 |
| concepts[0].display_name | Metagenomics |
| concepts[1].id | https://openalex.org/C116834253 |
| concepts[1].level | 2 |
| concepts[1].score | 0.6160637140274048 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q2039217 |
| concepts[1].display_name | Identification (biology) |
| concepts[2].id | https://openalex.org/C51679486 |
| concepts[2].level | 3 |
| concepts[2].score | 0.5698767304420471 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q380546 |
| concepts[2].display_name | DNA sequencing |
| concepts[3].id | https://openalex.org/C2778384902 |
| concepts[3].level | 2 |
| concepts[3].score | 0.5166528820991516 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q183134 |
| concepts[3].display_name | Sepsis |
| concepts[4].id | https://openalex.org/C86803240 |
| concepts[4].level | 0 |
| concepts[4].score | 0.48409926891326904 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[4].display_name | Biology |
| concepts[5].id | https://openalex.org/C70721500 |
| concepts[5].level | 1 |
| concepts[5].score | 0.4712505638599396 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q177005 |
| concepts[5].display_name | Computational biology |
| concepts[6].id | https://openalex.org/C89423630 |
| concepts[6].level | 1 |
| concepts[6].score | 0.4571644067764282 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q7193 |
| concepts[6].display_name | Microbiology |
| concepts[7].id | https://openalex.org/C54355233 |
| concepts[7].level | 1 |
| concepts[7].score | 0.15584877133369446 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[7].display_name | Genetics |
| concepts[8].id | https://openalex.org/C203014093 |
| concepts[8].level | 1 |
| concepts[8].score | 0.13044270873069763 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q101929 |
| concepts[8].display_name | Immunology |
| concepts[9].id | https://openalex.org/C104317684 |
| concepts[9].level | 2 |
| concepts[9].score | 0.12881404161453247 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[9].display_name | Gene |
| concepts[10].id | https://openalex.org/C59822182 |
| concepts[10].level | 1 |
| concepts[10].score | 0.0 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q441 |
| concepts[10].display_name | Botany |
| keywords[0].id | https://openalex.org/keywords/metagenomics |
| keywords[0].score | 0.8676527738571167 |
| keywords[0].display_name | Metagenomics |
| keywords[1].id | https://openalex.org/keywords/identification |
| keywords[1].score | 0.6160637140274048 |
| keywords[1].display_name | Identification (biology) |
| keywords[2].id | https://openalex.org/keywords/dna-sequencing |
| keywords[2].score | 0.5698767304420471 |
| keywords[2].display_name | DNA sequencing |
| keywords[3].id | https://openalex.org/keywords/sepsis |
| keywords[3].score | 0.5166528820991516 |
| keywords[3].display_name | Sepsis |
| keywords[4].id | https://openalex.org/keywords/biology |
| keywords[4].score | 0.48409926891326904 |
| keywords[4].display_name | Biology |
| keywords[5].id | https://openalex.org/keywords/computational-biology |
| keywords[5].score | 0.4712505638599396 |
| keywords[5].display_name | Computational biology |
| keywords[6].id | https://openalex.org/keywords/microbiology |
| keywords[6].score | 0.4571644067764282 |
| keywords[6].display_name | Microbiology |
| keywords[7].id | https://openalex.org/keywords/genetics |
| keywords[7].score | 0.15584877133369446 |
| keywords[7].display_name | Genetics |
| keywords[8].id | https://openalex.org/keywords/immunology |
| keywords[8].score | 0.13044270873069763 |
| keywords[8].display_name | Immunology |
| keywords[9].id | https://openalex.org/keywords/gene |
| keywords[9].score | 0.12881404161453247 |
| keywords[9].display_name | Gene |
| language | en |
| locations[0].id | doi:10.1007/s12223-024-01134-7 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S68090309 |
| locations[0].source.issn | 0015-5632, 1874-9356 |
| locations[0].source.type | journal |
| locations[0].source.is_oa | False |
| locations[0].source.issn_l | 0015-5632 |
| locations[0].source.is_core | True |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | Folia Microbiologica |
| locations[0].source.host_organization | https://openalex.org/P4310319900 |
| locations[0].source.host_organization_name | Springer Science+Business Media |
| locations[0].source.host_organization_lineage | https://openalex.org/P4310319900, https://openalex.org/P4310319965 |
| locations[0].source.host_organization_lineage_names | Springer Science+Business Media, Springer Nature |
| locations[0].license | cc-by |
| locations[0].pdf_url | https://link.springer.com/content/pdf/10.1007/s12223-024-01134-7.pdf |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | Folia Microbiologica |
| locations[0].landing_page_url | https://doi.org/10.1007/s12223-024-01134-7 |
| locations[1].id | pmid:38341816 |
| locations[1].is_oa | False |
| locations[1].source.id | https://openalex.org/S4306525036 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | PubMed |
| locations[1].source.host_organization | https://openalex.org/I1299303238 |
| locations[1].source.host_organization_name | National Institutes of Health |
| locations[1].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[1].license | |
| locations[1].pdf_url | |
| locations[1].version | publishedVersion |
| locations[1].raw_type | |
| locations[1].license_id | |
| locations[1].is_accepted | True |
| locations[1].is_published | True |
| locations[1].raw_source_name | Folia microbiologica |
| locations[1].landing_page_url | https://pubmed.ncbi.nlm.nih.gov/38341816 |
| locations[2].id | pmh:oai:pubmedcentral.nih.gov:11379781 |
| locations[2].is_oa | True |
| locations[2].source.id | https://openalex.org/S2764455111 |
| locations[2].source.issn | |
| locations[2].source.type | repository |
| locations[2].source.is_oa | False |
| locations[2].source.issn_l | |
| locations[2].source.is_core | False |
| locations[2].source.is_in_doaj | False |
| locations[2].source.display_name | PubMed Central |
| locations[2].source.host_organization | https://openalex.org/I1299303238 |
| locations[2].source.host_organization_name | National Institutes of Health |
| locations[2].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[2].license | other-oa |
| locations[2].pdf_url | |
| locations[2].version | submittedVersion |
| locations[2].raw_type | Text |
| locations[2].license_id | https://openalex.org/licenses/other-oa |
| locations[2].is_accepted | False |
| locations[2].is_published | False |
| locations[2].raw_source_name | Folia Microbiol (Praha) |
| locations[2].landing_page_url | https://www.ncbi.nlm.nih.gov/pmc/articles/11379781 |
| indexed_in | crossref, pubmed |
| authorships[0].author.id | https://openalex.org/A5100755912 |
| authorships[0].author.orcid | https://orcid.org/0000-0002-2035-635X |
| authorships[0].author.display_name | Zhendong Chen |
| authorships[0].countries | CN |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I126520041 |
| authorships[0].affiliations[0].raw_affiliation_string | Department of Intensive Care Unit, The First Affiliated Hospital of USTC, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, Anhui, 230001, China |
| authorships[0].institutions[0].id | https://openalex.org/I126520041 |
| authorships[0].institutions[0].ror | https://ror.org/04c4dkn09 |
| authorships[0].institutions[0].type | education |
| authorships[0].institutions[0].lineage | https://openalex.org/I126520041, https://openalex.org/I19820366 |
| authorships[0].institutions[0].country_code | CN |
| authorships[0].institutions[0].display_name | University of Science and Technology of China |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Zhendong Chen |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Department of Intensive Care Unit, The First Affiliated Hospital of USTC, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, Anhui, 230001, China |
| authorships[1].author.id | https://openalex.org/A5101964832 |
| authorships[1].author.orcid | https://orcid.org/0000-0002-0731-9263 |
| authorships[1].author.display_name | Tingting Ye |
| authorships[1].countries | CN |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I126520041 |
| authorships[1].affiliations[0].raw_affiliation_string | Department of Cardiovascular Medicine, The First Affiliated Hospital of USTC, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, Anhui, 230001, China |
| authorships[1].institutions[0].id | https://openalex.org/I126520041 |
| authorships[1].institutions[0].ror | https://ror.org/04c4dkn09 |
| authorships[1].institutions[0].type | education |
| authorships[1].institutions[0].lineage | https://openalex.org/I126520041, https://openalex.org/I19820366 |
| authorships[1].institutions[0].country_code | CN |
| authorships[1].institutions[0].display_name | University of Science and Technology of China |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Tingting Ye |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | Department of Cardiovascular Medicine, The First Affiliated Hospital of USTC, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, Anhui, 230001, China |
| authorships[2].author.id | https://openalex.org/A5102622299 |
| authorships[2].author.orcid | |
| authorships[2].author.display_name | Yuxi He |
| authorships[2].countries | CN |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I126520041 |
| authorships[2].affiliations[0].raw_affiliation_string | Department of Intensive Care Unit, The First Affiliated Hospital of USTC, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, Anhui, 230001, China |
| authorships[2].institutions[0].id | https://openalex.org/I126520041 |
| authorships[2].institutions[0].ror | https://ror.org/04c4dkn09 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I126520041, https://openalex.org/I19820366 |
| authorships[2].institutions[0].country_code | CN |
| authorships[2].institutions[0].display_name | University of Science and Technology of China |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Yuxi He |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | Department of Intensive Care Unit, The First Affiliated Hospital of USTC, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, Anhui, 230001, China |
| authorships[3].author.id | https://openalex.org/A5031487484 |
| authorships[3].author.orcid | https://orcid.org/0000-0002-3853-3879 |
| authorships[3].author.display_name | Aijun Pan |
| authorships[3].countries | CN |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I197869895 |
| authorships[3].affiliations[0].raw_affiliation_string | Department of Intensive Care Unit, The Affiliated Provincial Hospital of Anhui Medical University, Anhui, 230001, China |
| authorships[3].institutions[0].id | https://openalex.org/I197869895 |
| authorships[3].institutions[0].ror | https://ror.org/03xb04968 |
| authorships[3].institutions[0].type | education |
| authorships[3].institutions[0].lineage | https://openalex.org/I197869895 |
| authorships[3].institutions[0].country_code | CN |
| authorships[3].institutions[0].display_name | Anhui Medical University |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Aijun Pan |
| authorships[3].is_corresponding | True |
| authorships[3].raw_affiliation_strings | Department of Intensive Care Unit, The Affiliated Provincial Hospital of Anhui Medical University, Anhui, 230001, China |
| authorships[4].author.id | https://openalex.org/A5047493010 |
| authorships[4].author.orcid | https://orcid.org/0000-0001-9087-9642 |
| authorships[4].author.display_name | Qing Mei |
| authorships[4].countries | CN |
| authorships[4].affiliations[0].institution_ids | https://openalex.org/I126520041 |
| authorships[4].affiliations[0].raw_affiliation_string | Department of Intensive Care Unit, The First Affiliated Hospital of USTC, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, Anhui, 230001, China |
| authorships[4].institutions[0].id | https://openalex.org/I126520041 |
| authorships[4].institutions[0].ror | https://ror.org/04c4dkn09 |
| authorships[4].institutions[0].type | education |
| authorships[4].institutions[0].lineage | https://openalex.org/I126520041, https://openalex.org/I19820366 |
| authorships[4].institutions[0].country_code | CN |
| authorships[4].institutions[0].display_name | University of Science and Technology of China |
| authorships[4].author_position | last |
| authorships[4].raw_author_name | Qing Mei |
| authorships[4].is_corresponding | False |
| authorships[4].raw_affiliation_strings | Department of Intensive Care Unit, The First Affiliated Hospital of USTC, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, Anhui, 230001, China |
| has_content.pdf | True |
| has_content.grobid_xml | True |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://link.springer.com/content/pdf/10.1007/s12223-024-01134-7.pdf |
| open_access.oa_status | hybrid |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Use of pus metagenomic next-generation sequencing for efficient identification of pathogens in patients with sepsis |
| has_fulltext | True |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T11681 |
| primary_topic.field.id | https://openalex.org/fields/27 |
| primary_topic.field.display_name | Medicine |
| primary_topic.score | 0.996999979019165 |
| primary_topic.domain.id | https://openalex.org/domains/4 |
| primary_topic.domain.display_name | Health Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/2706 |
| primary_topic.subfield.display_name | Critical Care and Intensive Care Medicine |
| primary_topic.display_name | Nosocomial Infections in ICU |
| related_works | https://openalex.org/W2210738511, https://openalex.org/W2487541429, https://openalex.org/W1910109602, https://openalex.org/W2080583581, https://openalex.org/W4297131566, https://openalex.org/W2017364432, https://openalex.org/W2329606903, https://openalex.org/W3156493549, https://openalex.org/W3195915274, https://openalex.org/W3158976456 |
| cited_by_count | 1 |
| counts_by_year[0].year | 2025 |
| counts_by_year[0].cited_by_count | 1 |
| locations_count | 3 |
| best_oa_location.id | doi:10.1007/s12223-024-01134-7 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S68090309 |
| best_oa_location.source.issn | 0015-5632, 1874-9356 |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | False |
| best_oa_location.source.issn_l | 0015-5632 |
| best_oa_location.source.is_core | True |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | Folia Microbiologica |
| best_oa_location.source.host_organization | https://openalex.org/P4310319900 |
| best_oa_location.source.host_organization_name | Springer Science+Business Media |
| best_oa_location.source.host_organization_lineage | https://openalex.org/P4310319900, https://openalex.org/P4310319965 |
| best_oa_location.source.host_organization_lineage_names | Springer Science+Business Media, Springer Nature |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | https://link.springer.com/content/pdf/10.1007/s12223-024-01134-7.pdf |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | journal-article |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | Folia Microbiologica |
| best_oa_location.landing_page_url | https://doi.org/10.1007/s12223-024-01134-7 |
| primary_location.id | doi:10.1007/s12223-024-01134-7 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S68090309 |
| primary_location.source.issn | 0015-5632, 1874-9356 |
| primary_location.source.type | journal |
| primary_location.source.is_oa | False |
| primary_location.source.issn_l | 0015-5632 |
| primary_location.source.is_core | True |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | Folia Microbiologica |
| primary_location.source.host_organization | https://openalex.org/P4310319900 |
| primary_location.source.host_organization_name | Springer Science+Business Media |
| primary_location.source.host_organization_lineage | https://openalex.org/P4310319900, https://openalex.org/P4310319965 |
| primary_location.source.host_organization_lineage_names | Springer Science+Business Media, Springer Nature |
| primary_location.license | cc-by |
| primary_location.pdf_url | https://link.springer.com/content/pdf/10.1007/s12223-024-01134-7.pdf |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | Folia Microbiologica |
| primary_location.landing_page_url | https://doi.org/10.1007/s12223-024-01134-7 |
| publication_date | 2024-02-11 |
| publication_year | 2024 |
| referenced_works | https://openalex.org/W3088268715, https://openalex.org/W3200531480, https://openalex.org/W4295693541, https://openalex.org/W2222314148, https://openalex.org/W3109444600, https://openalex.org/W3101157915, https://openalex.org/W2982825545, https://openalex.org/W4224990193, https://openalex.org/W2997805206, https://openalex.org/W3131536684, https://openalex.org/W3119995551, https://openalex.org/W2049740461, https://openalex.org/W2965279968, https://openalex.org/W2150345049, https://openalex.org/W3012191857, https://openalex.org/W2078517765, https://openalex.org/W2745036943, https://openalex.org/W2761489767, https://openalex.org/W2015550272, https://openalex.org/W2096604736, https://openalex.org/W2900013960, https://openalex.org/W4285391847, https://openalex.org/W3135217424, https://openalex.org/W2895357006, https://openalex.org/W3195463377, https://openalex.org/W4225876158, https://openalex.org/W3198932362, https://openalex.org/W4211237777, https://openalex.org/W4226088977 |
| referenced_works_count | 29 |
| abstract_inverted_index.1 | 129 |
| abstract_inverted_index.7 | 75 |
| abstract_inverted_index.= | 152, 161, 172, 199, 211, 222, 232 |
| abstract_inverted_index.P | 198, 210, 221, 231 |
| abstract_inverted_index.(n | 151, 160, 171 |
| abstract_inverted_index.24 | 131 |
| abstract_inverted_index.59 | 71 |
| abstract_inverted_index.7) | 153 |
| abstract_inverted_index.In | 251 |
| abstract_inverted_index.To | 34 |
| abstract_inverted_index.We | 174 |
| abstract_inverted_index.be | 28 |
| abstract_inverted_index.by | 57, 79, 256 |
| abstract_inverted_index.in | 42, 120, 234, 245 |
| abstract_inverted_index.of | 5, 24, 32, 38, 59, 88, 104, 122, 141, 180, 215 |
| abstract_inverted_index.or | 61, 68, 91, 154 |
| abstract_inverted_index.to | 15, 97, 111 |
| abstract_inverted_index.5), | 162 |
| abstract_inverted_index.5.0 | 195 |
| abstract_inverted_index.7). | 173 |
| abstract_inverted_index.For | 137 |
| abstract_inverted_index.The | 1, 145, 188 |
| abstract_inverted_index.all | 148, 167 |
| abstract_inverted_index.and | 52, 74, 126, 130, 156, 183, 224 |
| abstract_inverted_index.any | 118 |
| abstract_inverted_index.did | 115 |
| abstract_inverted_index.for | 30 |
| abstract_inverted_index.low | 14 |
| abstract_inverted_index.may | 27, 266 |
| abstract_inverted_index.not | 116 |
| abstract_inverted_index.pus | 20, 41, 261 |
| abstract_inverted_index.the | 22, 36, 80, 85, 102, 164, 177, 213, 235, 246, 263 |
| abstract_inverted_index.too | 13 |
| abstract_inverted_index.two | 81 |
| abstract_inverted_index.use | 58 |
| abstract_inverted_index.vs. | 194, 206, 219, 229 |
| abstract_inverted_index.was | 11, 143 |
| abstract_inverted_index.2.50 | 207 |
| abstract_inverted_index.SOFA | 189 |
| abstract_inverted_index.[4.7 | 203 |
| abstract_inverted_index.[9.0 | 191 |
| abstract_inverted_index.also | 100, 175 |
| abstract_inverted_index.from | 21, 95, 109, 262 |
| abstract_inverted_index.mNGS | 39, 60, 83, 99, 114, 127, 259 |
| abstract_inverted_index.meet | 16 |
| abstract_inverted_index.more | 268 |
| abstract_inverted_index.rate | 4, 87 |
| abstract_inverted_index.show | 117 |
| abstract_inverted_index.site | 23 |
| abstract_inverted_index.skin | 155 |
| abstract_inverted_index.soft | 157 |
| abstract_inverted_index.than | 243 |
| abstract_inverted_index.were | 48, 64, 77, 240 |
| abstract_inverted_index.with | 44, 133, 253 |
| abstract_inverted_index.(0.7, | 208 |
| abstract_inverted_index.(1.8, | 204 |
| abstract_inverted_index.(3.0, | 196 |
| abstract_inverted_index.(7.5, | 192 |
| abstract_inverted_index.14.3) | 193 |
| abstract_inverted_index.39.9) | 205 |
| abstract_inverted_index.44.4% | 96 |
| abstract_inverted_index.52.9% | 110 |
| abstract_inverted_index.8.0), | 197, 209 |
| abstract_inverted_index.acute | 225 |
| abstract_inverted_index.blood | 6 |
| abstract_inverted_index.fungi | 76 |
| abstract_inverted_index.group | 239 |
| abstract_inverted_index.liver | 226 |
| abstract_inverted_index.mixed | 53, 134 |
| abstract_inverted_index.score | 190 |
| abstract_inverted_index.shock | 217 |
| abstract_inverted_index.still | 12 |
| abstract_inverted_index.terms | 121 |
| abstract_inverted_index.those | 244 |
| abstract_inverted_index.using | 40, 260 |
| abstract_inverted_index.value | 37, 202 |
| abstract_inverted_index.while | 19 |
| abstract_inverted_index.yield | 267 |
| abstract_inverted_index.(66.7% | 218, 228 |
| abstract_inverted_index.(mNGS) | 10 |
| abstract_inverted_index.0.018) | 233 |
| abstract_inverted_index.0.044) | 223 |
| abstract_inverted_index.23.5%, | 230 |
| abstract_inverted_index.35.3%, | 220 |
| abstract_inverted_index.94.4%; | 98 |
| abstract_inverted_index.assess | 35 |
| abstract_inverted_index.caused | 149, 168, 255 |
| abstract_inverted_index.fungal | 123 |
| abstract_inverted_index.group. | 250 |
| abstract_inverted_index.higher | 242 |
| abstract_inverted_index.injury | 227 |
| abstract_inverted_index.lesion | 265 |
| abstract_inverted_index.needs, | 18 |
| abstract_inverted_index.sepsis | 254 |
| abstract_inverted_index.septic | 216 |
| abstract_inverted_index.source | 140 |
| abstract_inverted_index.tissue | 158 |
| abstract_inverted_index.0.005], | 200 |
| abstract_inverted_index.0.035], | 212 |
| abstract_inverted_index.100.0%; | 112 |
| abstract_inverted_index.Culture | 125 |
| abstract_inverted_index.aerobic | 67, 90 |
| abstract_inverted_index.empyema | 150 |
| abstract_inverted_index.groups. | 187 |
| abstract_inverted_index.methods | 63 |
| abstract_inverted_index.primary | 25, 264 |
| abstract_inverted_index.samples | 47 |
| abstract_inverted_index.sepsis, | 45 |
| abstract_inverted_index.whereas | 163 |
| abstract_inverted_index.Abstract | 0 |
| abstract_inverted_index.Pathogen | 50 |
| abstract_inverted_index.accuracy | 86 |
| abstract_inverted_index.bacteria | 147 |
| abstract_inverted_index.clinical | 17, 178 |
| abstract_inverted_index.compared | 176 |
| abstract_inverted_index.however, | 113 |
| abstract_inverted_index.methods. | 82 |
| abstract_inverted_index.microbes | 166 |
| abstract_inverted_index.obligate | 72, 106, 138, 184, 247 |
| abstract_inverted_index.obtained | 56 |
| abstract_inverted_index.patients | 43, 132, 252 |
| abstract_inverted_index.positive | 2 |
| abstract_inverted_index.purulent | 257 |
| abstract_inverted_index.valuable | 269 |
| abstract_inverted_index.advantage | 119 |
| abstract_inverted_index.anaerobes | 73 |
| abstract_inverted_index.anaerobic | 93, 107, 182, 185, 237, 248 |
| abstract_inverted_index.analyzed. | 144 |
| abstract_inverted_index.compared. | 65 |
| abstract_inverted_index.detection | 3 |
| abstract_inverted_index.diagnosis | 55 |
| abstract_inverted_index.increased | 84, 101 |
| abstract_inverted_index.infection | 26, 54, 186, 238, 249 |
| abstract_inverted_index.anaerobes, | 70, 139 |
| abstract_inverted_index.collected. | 49 |
| abstract_inverted_index.diagnosing | 89, 105 |
| abstract_inverted_index.identified | 78, 128 |
| abstract_inverted_index.infection, | 135, 258 |
| abstract_inverted_index.infections | 94, 108, 159, 170 |
| abstract_inverted_index.pathogens. | 33 |
| abstract_inverted_index.proportion | 214 |
| abstract_inverted_index.sequencing | 9 |
| abstract_inverted_index.Fifty-three | 66 |
| abstract_inverted_index.cultivation | 62 |
| abstract_inverted_index.facultative | 69, 92 |
| abstract_inverted_index.gut-derived | 165 |
| abstract_inverted_index.infections. | 124 |
| abstract_inverted_index.metagenomic | 7 |
| abstract_inverted_index.odontogenic | 146 |
| abstract_inverted_index.sensitivity | 103 |
| abstract_inverted_index.thirty-five | 46 |
| abstract_inverted_index.advantageous | 29 |
| abstract_inverted_index.information. | 271 |
| abstract_inverted_index.non-obligate | 181, 236 |
| abstract_inverted_index.procalcitonin | 201 |
| abstract_inverted_index.respectively. | 136 |
| abstract_inverted_index.significantly | 241 |
| abstract_inverted_index.identification | 31, 51 |
| abstract_inverted_index.microorganisms | 142 |
| abstract_inverted_index.characteristics | 179 |
| abstract_inverted_index.intra-abdominal | 169 |
| abstract_inverted_index.microbiological | 270 |
| abstract_inverted_index.next-generation | 8 |
| cited_by_percentile_year.max | 95 |
| cited_by_percentile_year.min | 91 |
| corresponding_author_ids | https://openalex.org/A5031487484 |
| countries_distinct_count | 1 |
| institutions_distinct_count | 5 |
| corresponding_institution_ids | https://openalex.org/I197869895 |
| sustainable_development_goals[0].id | https://metadata.un.org/sdg/3 |
| sustainable_development_goals[0].score | 0.5099999904632568 |
| sustainable_development_goals[0].display_name | Good health and well-being |
| citation_normalized_percentile.value | 0.60509875 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |