Using Deep Learning Techniques to Enhance Blood Cell Detection in Patients with Leukemia Article Swipe
 YOU?
·
· 2024
· Open Access
·
· DOI: https://doi.org/10.3390/info15120787
YOU?
·
· 2024
· Open Access
·
· DOI: https://doi.org/10.3390/info15120787
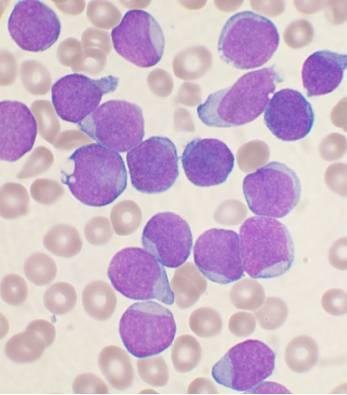
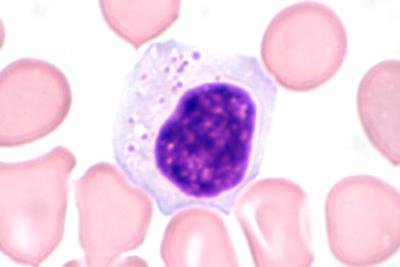
Medical diagnosis plays a critical role in the early detection and treatment of diseases by examining symptoms and supporting findings through advanced laboratory testing. Early and accurate diagnosis is essential for detecting medical problems and then prescribing the most effective treatment strategies, especially in life-threatening diseases such as leukemia. Leukemia, a blood malignancy, is one of the most prevalent cancer types affecting both adults and children. It is caused by the rapid and uncontrolled growth of abnormal white blood cells in the bone marrow. This accumulation interferes with the production of normal blood cells, leading to a weakened immune deficiency, anemia, and bleeding disorders. Conventional leukemia diagnostic methods are time-consuming, manually intensive, and inefficient. This research study proposes an automatic diagnostics prediction of leukemia by analyzing blood images according to the shape of the blast cells using digital image processing and machine learning. The purpose of blood cell detection is to precisely identify and classify diverse blood cells, detecting anomalies associated with blood cancers like leukemia. This supports early diagnosis and monitoring, which leads to more effective treatments and improved results for cancer patients. To accomplish this task, we use digital image processing techniques and then apply the convolutional neural network (CNN) deep learning algorithm to blood sample images. This research employs a multi-stage methodology, including data preparation, data preprocessing, feature extraction, and then classification. While our model is built on a typical CNN architecture, we make significant advances by using preprocessing techniques and hyperparameter tuning. We have modified its layers combination to include convolutional, pooling, and fully connected layers that are optimized for image characteristics. These layers are fine-tuned for better feature extraction and classification accuracy. This study showed that blood cell detection for diagnosing acute leukemia based on images had 99% accuracy and outperformed other advanced models, including DenseNet121, ResNet-50, Incep-tionv3, MobileNet, and EfficientNet. The comprehensive analysis of the results reveals the highest accuracy of leukemia detection as compared to existing studies in the relevant literature.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.3390/info15120787
- OA Status
- gold
- Cited By
- 7
- References
- 45
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4405184441
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4405184441Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.3390/info15120787Digital Object Identifier
- Title
-
Using Deep Learning Techniques to Enhance Blood Cell Detection in Patients with LeukemiaWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2024Year of publication
- Publication date
-
2024-12-08Full publication date if available
- Authors
-
Mahwish Ilyas, Muhammad Bilal, Nadia Shamshad Malik, Hikmat Ullah Khan, Muhammad Ramzan, Anam NazList of authors in order
- Landing page
-
https://doi.org/10.3390/info15120787Publisher landing page
- Open access
-
YesWhether a free full text is available
- OA status
-
goldOpen access status per OpenAlex
- OA URL
-
https://doi.org/10.3390/info15120787Direct OA link when available
- Concepts
-
Leukemia, Artificial intelligence, Convolutional neural network, Preprocessor, Computer science, Feature extraction, Deep learning, Cancer, Blood cancer, White blood cell, Medicine, Pattern recognition (psychology), Machine learning, Immunology, Internal medicineTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
7Total citation count in OpenAlex
- Citations by year (recent)
-
2025: 7Per-year citation counts (last 5 years)
- References (count)
-
45Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4405184441 |
|---|---|
| doi | https://doi.org/10.3390/info15120787 |
| ids.doi | https://doi.org/10.3390/info15120787 |
| ids.openalex | https://openalex.org/W4405184441 |
| fwci | 3.71110292 |
| type | article |
| title | Using Deep Learning Techniques to Enhance Blood Cell Detection in Patients with Leukemia |
| biblio.issue | 12 |
| biblio.volume | 15 |
| biblio.last_page | 787 |
| biblio.first_page | 787 |
| topics[0].id | https://openalex.org/T12874 |
| topics[0].field.id | https://openalex.org/fields/17 |
| topics[0].field.display_name | Computer Science |
| topics[0].score | 1.0 |
| topics[0].domain.id | https://openalex.org/domains/3 |
| topics[0].domain.display_name | Physical Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1707 |
| topics[0].subfield.display_name | Computer Vision and Pattern Recognition |
| topics[0].display_name | Digital Imaging for Blood Diseases |
| topics[1].id | https://openalex.org/T11775 |
| topics[1].field.id | https://openalex.org/fields/27 |
| topics[1].field.display_name | Medicine |
| topics[1].score | 0.9556999802589417 |
| topics[1].domain.id | https://openalex.org/domains/4 |
| topics[1].domain.display_name | Health Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/2741 |
| topics[1].subfield.display_name | Radiology, Nuclear Medicine and Imaging |
| topics[1].display_name | COVID-19 diagnosis using AI |
| topics[2].id | https://openalex.org/T13324 |
| topics[2].field.id | https://openalex.org/fields/35 |
| topics[2].field.display_name | Dentistry |
| topics[2].score | 0.9401999711990356 |
| topics[2].domain.id | https://openalex.org/domains/4 |
| topics[2].domain.display_name | Health Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/3506 |
| topics[2].subfield.display_name | Periodontics |
| topics[2].display_name | Scientific and Engineering Research Topics |
| is_xpac | False |
| apc_list.value | 1400 |
| apc_list.currency | CHF |
| apc_list.value_usd | 1515 |
| apc_paid.value | 1400 |
| apc_paid.currency | CHF |
| apc_paid.value_usd | 1515 |
| concepts[0].id | https://openalex.org/C2778461978 |
| concepts[0].level | 2 |
| concepts[0].score | 0.6319610476493835 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q29496 |
| concepts[0].display_name | Leukemia |
| concepts[1].id | https://openalex.org/C154945302 |
| concepts[1].level | 1 |
| concepts[1].score | 0.6148098707199097 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q11660 |
| concepts[1].display_name | Artificial intelligence |
| concepts[2].id | https://openalex.org/C81363708 |
| concepts[2].level | 2 |
| concepts[2].score | 0.6001704931259155 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q17084460 |
| concepts[2].display_name | Convolutional neural network |
| concepts[3].id | https://openalex.org/C34736171 |
| concepts[3].level | 2 |
| concepts[3].score | 0.5392789244651794 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q918333 |
| concepts[3].display_name | Preprocessor |
| concepts[4].id | https://openalex.org/C41008148 |
| concepts[4].level | 0 |
| concepts[4].score | 0.5257930159568787 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q21198 |
| concepts[4].display_name | Computer science |
| concepts[5].id | https://openalex.org/C52622490 |
| concepts[5].level | 2 |
| concepts[5].score | 0.5110749006271362 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q1026626 |
| concepts[5].display_name | Feature extraction |
| concepts[6].id | https://openalex.org/C108583219 |
| concepts[6].level | 2 |
| concepts[6].score | 0.4659712314605713 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q197536 |
| concepts[6].display_name | Deep learning |
| concepts[7].id | https://openalex.org/C121608353 |
| concepts[7].level | 2 |
| concepts[7].score | 0.4512920379638672 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q12078 |
| concepts[7].display_name | Cancer |
| concepts[8].id | https://openalex.org/C2992972558 |
| concepts[8].level | 3 |
| concepts[8].score | 0.42586055397987366 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q2509220 |
| concepts[8].display_name | Blood cancer |
| concepts[9].id | https://openalex.org/C2778488018 |
| concepts[9].level | 2 |
| concepts[9].score | 0.42082852125167847 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q42395 |
| concepts[9].display_name | White blood cell |
| concepts[10].id | https://openalex.org/C71924100 |
| concepts[10].level | 0 |
| concepts[10].score | 0.38696998357772827 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q11190 |
| concepts[10].display_name | Medicine |
| concepts[11].id | https://openalex.org/C153180895 |
| concepts[11].level | 2 |
| concepts[11].score | 0.36729246377944946 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q7148389 |
| concepts[11].display_name | Pattern recognition (psychology) |
| concepts[12].id | https://openalex.org/C119857082 |
| concepts[12].level | 1 |
| concepts[12].score | 0.3647017776966095 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q2539 |
| concepts[12].display_name | Machine learning |
| concepts[13].id | https://openalex.org/C203014093 |
| concepts[13].level | 1 |
| concepts[13].score | 0.19295263290405273 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q101929 |
| concepts[13].display_name | Immunology |
| concepts[14].id | https://openalex.org/C126322002 |
| concepts[14].level | 1 |
| concepts[14].score | 0.16741982102394104 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q11180 |
| concepts[14].display_name | Internal medicine |
| keywords[0].id | https://openalex.org/keywords/leukemia |
| keywords[0].score | 0.6319610476493835 |
| keywords[0].display_name | Leukemia |
| keywords[1].id | https://openalex.org/keywords/artificial-intelligence |
| keywords[1].score | 0.6148098707199097 |
| keywords[1].display_name | Artificial intelligence |
| keywords[2].id | https://openalex.org/keywords/convolutional-neural-network |
| keywords[2].score | 0.6001704931259155 |
| keywords[2].display_name | Convolutional neural network |
| keywords[3].id | https://openalex.org/keywords/preprocessor |
| keywords[3].score | 0.5392789244651794 |
| keywords[3].display_name | Preprocessor |
| keywords[4].id | https://openalex.org/keywords/computer-science |
| keywords[4].score | 0.5257930159568787 |
| keywords[4].display_name | Computer science |
| keywords[5].id | https://openalex.org/keywords/feature-extraction |
| keywords[5].score | 0.5110749006271362 |
| keywords[5].display_name | Feature extraction |
| keywords[6].id | https://openalex.org/keywords/deep-learning |
| keywords[6].score | 0.4659712314605713 |
| keywords[6].display_name | Deep learning |
| keywords[7].id | https://openalex.org/keywords/cancer |
| keywords[7].score | 0.4512920379638672 |
| keywords[7].display_name | Cancer |
| keywords[8].id | https://openalex.org/keywords/blood-cancer |
| keywords[8].score | 0.42586055397987366 |
| keywords[8].display_name | Blood cancer |
| keywords[9].id | https://openalex.org/keywords/white-blood-cell |
| keywords[9].score | 0.42082852125167847 |
| keywords[9].display_name | White blood cell |
| keywords[10].id | https://openalex.org/keywords/medicine |
| keywords[10].score | 0.38696998357772827 |
| keywords[10].display_name | Medicine |
| keywords[11].id | https://openalex.org/keywords/pattern-recognition |
| keywords[11].score | 0.36729246377944946 |
| keywords[11].display_name | Pattern recognition (psychology) |
| keywords[12].id | https://openalex.org/keywords/machine-learning |
| keywords[12].score | 0.3647017776966095 |
| keywords[12].display_name | Machine learning |
| keywords[13].id | https://openalex.org/keywords/immunology |
| keywords[13].score | 0.19295263290405273 |
| keywords[13].display_name | Immunology |
| keywords[14].id | https://openalex.org/keywords/internal-medicine |
| keywords[14].score | 0.16741982102394104 |
| keywords[14].display_name | Internal medicine |
| language | en |
| locations[0].id | doi:10.3390/info15120787 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4210219776 |
| locations[0].source.issn | 2078-2489 |
| locations[0].source.type | journal |
| locations[0].source.is_oa | True |
| locations[0].source.issn_l | 2078-2489 |
| locations[0].source.is_core | True |
| locations[0].source.is_in_doaj | True |
| locations[0].source.display_name | Information |
| locations[0].source.host_organization | https://openalex.org/P4310310987 |
| locations[0].source.host_organization_name | Multidisciplinary Digital Publishing Institute |
| locations[0].source.host_organization_lineage | https://openalex.org/P4310310987 |
| locations[0].source.host_organization_lineage_names | Multidisciplinary Digital Publishing Institute |
| locations[0].license | cc-by |
| locations[0].pdf_url | |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | Information |
| locations[0].landing_page_url | https://doi.org/10.3390/info15120787 |
| locations[1].id | pmh:oai:doaj.org/article:b4b457e733ef4753b2572bddbb5520df |
| locations[1].is_oa | False |
| locations[1].source.id | https://openalex.org/S4306401280 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | DOAJ (DOAJ: Directory of Open Access Journals) |
| locations[1].source.host_organization | |
| locations[1].source.host_organization_name | |
| locations[1].license | |
| locations[1].pdf_url | |
| locations[1].version | submittedVersion |
| locations[1].raw_type | article |
| locations[1].license_id | |
| locations[1].is_accepted | False |
| locations[1].is_published | False |
| locations[1].raw_source_name | Information, Vol 15, Iss 12, p 787 (2024) |
| locations[1].landing_page_url | https://doaj.org/article/b4b457e733ef4753b2572bddbb5520df |
| indexed_in | crossref, doaj |
| authorships[0].author.id | https://openalex.org/A5087956264 |
| authorships[0].author.orcid | https://orcid.org/0000-0001-9644-1537 |
| authorships[0].author.display_name | Mahwish Ilyas |
| authorships[0].countries | PK |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I160968435 |
| authorships[0].affiliations[0].raw_affiliation_string | Department of Computer Science, Faculty of Computing and IT, University of Sargodha, Sargodha 40100, Pakistan |
| authorships[0].affiliations[1].institution_ids | https://openalex.org/I142204992, https://openalex.org/I160968435 |
| authorships[0].affiliations[1].raw_affiliation_string | Department of Computer Science, Faculty of IT, The University of Lahore, Sargodha Campus, Sargodha 40100, Pakistan |
| authorships[0].institutions[0].id | https://openalex.org/I142204992 |
| authorships[0].institutions[0].ror | https://ror.org/051jrjw38 |
| authorships[0].institutions[0].type | education |
| authorships[0].institutions[0].lineage | https://openalex.org/I142204992 |
| authorships[0].institutions[0].country_code | PK |
| authorships[0].institutions[0].display_name | University of Lahore |
| authorships[0].institutions[1].id | https://openalex.org/I160968435 |
| authorships[0].institutions[1].ror | https://ror.org/0086rpr26 |
| authorships[0].institutions[1].type | education |
| authorships[0].institutions[1].lineage | https://openalex.org/I160968435 |
| authorships[0].institutions[1].country_code | PK |
| authorships[0].institutions[1].display_name | University of Sargodha |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Mahwish Ilyas |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Department of Computer Science, Faculty of Computing and IT, University of Sargodha, Sargodha 40100, Pakistan, Department of Computer Science, Faculty of IT, The University of Lahore, Sargodha Campus, Sargodha 40100, Pakistan |
| authorships[1].author.id | https://openalex.org/A5012666416 |
| authorships[1].author.orcid | https://orcid.org/0000-0002-9827-5023 |
| authorships[1].author.display_name | Muhammad Bilal |
| authorships[1].countries | US |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I33213144 |
| authorships[1].affiliations[0].raw_affiliation_string | Department of Pharmaceutical Outcomes and Policy, Malachowsky Hall for Data Science and Information Technology, University of Florida, Gainesville, FL 32611, USA |
| authorships[1].institutions[0].id | https://openalex.org/I33213144 |
| authorships[1].institutions[0].ror | https://ror.org/02y3ad647 |
| authorships[1].institutions[0].type | education |
| authorships[1].institutions[0].lineage | https://openalex.org/I33213144 |
| authorships[1].institutions[0].country_code | US |
| authorships[1].institutions[0].display_name | University of Florida |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Muhammad Bilal |
| authorships[1].is_corresponding | True |
| authorships[1].raw_affiliation_strings | Department of Pharmaceutical Outcomes and Policy, Malachowsky Hall for Data Science and Information Technology, University of Florida, Gainesville, FL 32611, USA |
| authorships[2].author.id | https://openalex.org/A5086046413 |
| authorships[2].author.orcid | https://orcid.org/0000-0002-0474-3448 |
| authorships[2].author.display_name | Nadia Shamshad Malik |
| authorships[2].countries | PK |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I160968435 |
| authorships[2].affiliations[0].raw_affiliation_string | Department of Software Engineering, Faculty of Computing and IT, University of Sargodha, Sargodha 40100, Pakistan |
| authorships[2].institutions[0].id | https://openalex.org/I160968435 |
| authorships[2].institutions[0].ror | https://ror.org/0086rpr26 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I160968435 |
| authorships[2].institutions[0].country_code | PK |
| authorships[2].institutions[0].display_name | University of Sargodha |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Nadia Malik |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | Department of Software Engineering, Faculty of Computing and IT, University of Sargodha, Sargodha 40100, Pakistan |
| authorships[3].author.id | https://openalex.org/A5073826651 |
| authorships[3].author.orcid | https://orcid.org/0000-0002-8178-6652 |
| authorships[3].author.display_name | Hikmat Ullah Khan |
| authorships[3].countries | PK |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I160968435 |
| authorships[3].affiliations[0].raw_affiliation_string | Department of Information Technology, Faculty of Computing and IT, University of Sargodha, Sargodha 40100, Pakistan |
| authorships[3].institutions[0].id | https://openalex.org/I160968435 |
| authorships[3].institutions[0].ror | https://ror.org/0086rpr26 |
| authorships[3].institutions[0].type | education |
| authorships[3].institutions[0].lineage | https://openalex.org/I160968435 |
| authorships[3].institutions[0].country_code | PK |
| authorships[3].institutions[0].display_name | University of Sargodha |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Hikmat Ullah Khan |
| authorships[3].is_corresponding | False |
| authorships[3].raw_affiliation_strings | Department of Information Technology, Faculty of Computing and IT, University of Sargodha, Sargodha 40100, Pakistan |
| authorships[4].author.id | https://openalex.org/A5059496118 |
| authorships[4].author.orcid | https://orcid.org/0000-0003-1770-8905 |
| authorships[4].author.display_name | Muhammad Ramzan |
| authorships[4].countries | PK |
| authorships[4].affiliations[0].institution_ids | https://openalex.org/I160968435 |
| authorships[4].affiliations[0].raw_affiliation_string | Department of Software Engineering, Faculty of Computing and IT, University of Sargodha, Sargodha 40100, Pakistan |
| authorships[4].institutions[0].id | https://openalex.org/I160968435 |
| authorships[4].institutions[0].ror | https://ror.org/0086rpr26 |
| authorships[4].institutions[0].type | education |
| authorships[4].institutions[0].lineage | https://openalex.org/I160968435 |
| authorships[4].institutions[0].country_code | PK |
| authorships[4].institutions[0].display_name | University of Sargodha |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Muhammad Ramzan |
| authorships[4].is_corresponding | False |
| authorships[4].raw_affiliation_strings | Department of Software Engineering, Faculty of Computing and IT, University of Sargodha, Sargodha 40100, Pakistan |
| authorships[5].author.id | https://openalex.org/A5003223453 |
| authorships[5].author.orcid | https://orcid.org/0000-0002-6514-5417 |
| authorships[5].author.display_name | Anam Naz |
| authorships[5].countries | PK |
| authorships[5].affiliations[0].institution_ids | https://openalex.org/I160968435 |
| authorships[5].affiliations[0].raw_affiliation_string | Department of Computer Science, Faculty of Computing and IT, University of Sargodha, Sargodha 40100, Pakistan |
| authorships[5].institutions[0].id | https://openalex.org/I160968435 |
| authorships[5].institutions[0].ror | https://ror.org/0086rpr26 |
| authorships[5].institutions[0].type | education |
| authorships[5].institutions[0].lineage | https://openalex.org/I160968435 |
| authorships[5].institutions[0].country_code | PK |
| authorships[5].institutions[0].display_name | University of Sargodha |
| authorships[5].author_position | last |
| authorships[5].raw_author_name | Anam Naz |
| authorships[5].is_corresponding | False |
| authorships[5].raw_affiliation_strings | Department of Computer Science, Faculty of Computing and IT, University of Sargodha, Sargodha 40100, Pakistan |
| has_content.pdf | False |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://doi.org/10.3390/info15120787 |
| open_access.oa_status | gold |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Using Deep Learning Techniques to Enhance Blood Cell Detection in Patients with Leukemia |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T12874 |
| primary_topic.field.id | https://openalex.org/fields/17 |
| primary_topic.field.display_name | Computer Science |
| primary_topic.score | 1.0 |
| primary_topic.domain.id | https://openalex.org/domains/3 |
| primary_topic.domain.display_name | Physical Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1707 |
| primary_topic.subfield.display_name | Computer Vision and Pattern Recognition |
| primary_topic.display_name | Digital Imaging for Blood Diseases |
| related_works | https://openalex.org/W4255693317, https://openalex.org/W2397288865, https://openalex.org/W4321487865, https://openalex.org/W4313906399, https://openalex.org/W2354314470, https://openalex.org/W4239306820, https://openalex.org/W2095030957, https://openalex.org/W2066827917, https://openalex.org/W69076820, https://openalex.org/W3212731298 |
| cited_by_count | 7 |
| counts_by_year[0].year | 2025 |
| counts_by_year[0].cited_by_count | 7 |
| locations_count | 2 |
| best_oa_location.id | doi:10.3390/info15120787 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4210219776 |
| best_oa_location.source.issn | 2078-2489 |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | True |
| best_oa_location.source.issn_l | 2078-2489 |
| best_oa_location.source.is_core | True |
| best_oa_location.source.is_in_doaj | True |
| best_oa_location.source.display_name | Information |
| best_oa_location.source.host_organization | https://openalex.org/P4310310987 |
| best_oa_location.source.host_organization_name | Multidisciplinary Digital Publishing Institute |
| best_oa_location.source.host_organization_lineage | https://openalex.org/P4310310987 |
| best_oa_location.source.host_organization_lineage_names | Multidisciplinary Digital Publishing Institute |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | journal-article |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | Information |
| best_oa_location.landing_page_url | https://doi.org/10.3390/info15120787 |
| primary_location.id | doi:10.3390/info15120787 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4210219776 |
| primary_location.source.issn | 2078-2489 |
| primary_location.source.type | journal |
| primary_location.source.is_oa | True |
| primary_location.source.issn_l | 2078-2489 |
| primary_location.source.is_core | True |
| primary_location.source.is_in_doaj | True |
| primary_location.source.display_name | Information |
| primary_location.source.host_organization | https://openalex.org/P4310310987 |
| primary_location.source.host_organization_name | Multidisciplinary Digital Publishing Institute |
| primary_location.source.host_organization_lineage | https://openalex.org/P4310310987 |
| primary_location.source.host_organization_lineage_names | Multidisciplinary Digital Publishing Institute |
| primary_location.license | cc-by |
| primary_location.pdf_url | |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | Information |
| primary_location.landing_page_url | https://doi.org/10.3390/info15120787 |
| publication_date | 2024-12-08 |
| publication_year | 2024 |
| referenced_works | https://openalex.org/W4400387574, https://openalex.org/W4400398067, https://openalex.org/W4399872760, https://openalex.org/W4385234734, https://openalex.org/W4392238047, https://openalex.org/W4400433452, https://openalex.org/W4399883454, https://openalex.org/W4394596621, https://openalex.org/W4380369697, https://openalex.org/W4394620245, https://openalex.org/W4391693196, https://openalex.org/W4391693580, https://openalex.org/W4386073812, https://openalex.org/W4401018336, https://openalex.org/W3162222275, https://openalex.org/W4399486097, https://openalex.org/W4384559078, https://openalex.org/W3039893203, https://openalex.org/W3082931514, https://openalex.org/W3202295523, https://openalex.org/W2991053226, https://openalex.org/W3046874697, https://openalex.org/W3185157968, https://openalex.org/W3132583611, https://openalex.org/W2180133023, https://openalex.org/W4296009403, https://openalex.org/W3045683009, https://openalex.org/W4318571800, https://openalex.org/W6758385990, https://openalex.org/W2946363866, https://openalex.org/W3211592493, https://openalex.org/W3170978670, https://openalex.org/W3201653094, https://openalex.org/W3011939472, https://openalex.org/W6804242616, https://openalex.org/W4205696711, https://openalex.org/W4220797240, https://openalex.org/W4387450324, https://openalex.org/W4400226202, https://openalex.org/W3093305443, https://openalex.org/W4402634077, https://openalex.org/W3016457468, https://openalex.org/W2549139847, https://openalex.org/W2912661780, https://openalex.org/W3214415653 |
| referenced_works_count | 45 |
| abstract_inverted_index.a | 3, 50, 96, 212, 231 |
| abstract_inverted_index.It | 66 |
| abstract_inverted_index.To | 184 |
| abstract_inverted_index.We | 246 |
| abstract_inverted_index.an | 118 |
| abstract_inverted_index.as | 47, 319 |
| abstract_inverted_index.by | 14, 69, 124, 239 |
| abstract_inverted_index.in | 6, 43, 80, 324 |
| abstract_inverted_index.is | 28, 53, 67, 149, 228 |
| abstract_inverted_index.of | 12, 55, 75, 90, 122, 132, 145, 309, 316 |
| abstract_inverted_index.on | 230, 289 |
| abstract_inverted_index.to | 95, 129, 150, 174, 205, 252, 321 |
| abstract_inverted_index.we | 188, 235 |
| abstract_inverted_index.99% | 292 |
| abstract_inverted_index.CNN | 233 |
| abstract_inverted_index.The | 143, 306 |
| abstract_inverted_index.and | 10, 17, 25, 34, 64, 72, 101, 112, 140, 153, 170, 178, 194, 222, 243, 256, 274, 294, 304 |
| abstract_inverted_index.are | 108, 261, 268 |
| abstract_inverted_index.for | 30, 181, 263, 270, 284 |
| abstract_inverted_index.had | 291 |
| abstract_inverted_index.its | 249 |
| abstract_inverted_index.one | 54 |
| abstract_inverted_index.our | 226 |
| abstract_inverted_index.the | 7, 37, 56, 70, 81, 88, 130, 133, 197, 310, 313, 325 |
| abstract_inverted_index.use | 189 |
| abstract_inverted_index.This | 84, 114, 166, 209, 277 |
| abstract_inverted_index.bone | 82 |
| abstract_inverted_index.both | 62 |
| abstract_inverted_index.cell | 147, 282 |
| abstract_inverted_index.data | 216, 218 |
| abstract_inverted_index.deep | 202 |
| abstract_inverted_index.have | 247 |
| abstract_inverted_index.like | 164 |
| abstract_inverted_index.make | 236 |
| abstract_inverted_index.more | 175 |
| abstract_inverted_index.most | 38, 57 |
| abstract_inverted_index.role | 5 |
| abstract_inverted_index.such | 46 |
| abstract_inverted_index.that | 260, 280 |
| abstract_inverted_index.then | 35, 195, 223 |
| abstract_inverted_index.this | 186 |
| abstract_inverted_index.with | 87, 161 |
| abstract_inverted_index.(CNN) | 201 |
| abstract_inverted_index.Early | 24 |
| abstract_inverted_index.These | 266 |
| abstract_inverted_index.While | 225 |
| abstract_inverted_index.acute | 286 |
| abstract_inverted_index.apply | 196 |
| abstract_inverted_index.based | 288 |
| abstract_inverted_index.blast | 134 |
| abstract_inverted_index.blood | 51, 78, 92, 126, 146, 156, 162, 206, 281 |
| abstract_inverted_index.built | 229 |
| abstract_inverted_index.cells | 79, 135 |
| abstract_inverted_index.early | 8, 168 |
| abstract_inverted_index.fully | 257 |
| abstract_inverted_index.image | 138, 191, 264 |
| abstract_inverted_index.leads | 173 |
| abstract_inverted_index.model | 227 |
| abstract_inverted_index.other | 296 |
| abstract_inverted_index.plays | 2 |
| abstract_inverted_index.rapid | 71 |
| abstract_inverted_index.shape | 131 |
| abstract_inverted_index.study | 116, 278 |
| abstract_inverted_index.task, | 187 |
| abstract_inverted_index.types | 60 |
| abstract_inverted_index.using | 136, 240 |
| abstract_inverted_index.which | 172 |
| abstract_inverted_index.white | 77 |
| abstract_inverted_index.adults | 63 |
| abstract_inverted_index.better | 271 |
| abstract_inverted_index.cancer | 59, 182 |
| abstract_inverted_index.caused | 68 |
| abstract_inverted_index.cells, | 93, 157 |
| abstract_inverted_index.growth | 74 |
| abstract_inverted_index.images | 127, 290 |
| abstract_inverted_index.immune | 98 |
| abstract_inverted_index.layers | 250, 259, 267 |
| abstract_inverted_index.neural | 199 |
| abstract_inverted_index.normal | 91 |
| abstract_inverted_index.sample | 207 |
| abstract_inverted_index.showed | 279 |
| abstract_inverted_index.Medical | 0 |
| abstract_inverted_index.anemia, | 100 |
| abstract_inverted_index.cancers | 163 |
| abstract_inverted_index.digital | 137, 190 |
| abstract_inverted_index.diverse | 155 |
| abstract_inverted_index.employs | 211 |
| abstract_inverted_index.feature | 220, 272 |
| abstract_inverted_index.highest | 314 |
| abstract_inverted_index.images. | 208 |
| abstract_inverted_index.include | 253 |
| abstract_inverted_index.leading | 94 |
| abstract_inverted_index.machine | 141 |
| abstract_inverted_index.marrow. | 83 |
| abstract_inverted_index.medical | 32 |
| abstract_inverted_index.methods | 107 |
| abstract_inverted_index.models, | 298 |
| abstract_inverted_index.network | 200 |
| abstract_inverted_index.purpose | 144 |
| abstract_inverted_index.results | 180, 311 |
| abstract_inverted_index.reveals | 312 |
| abstract_inverted_index.studies | 323 |
| abstract_inverted_index.through | 20 |
| abstract_inverted_index.tuning. | 245 |
| abstract_inverted_index.typical | 232 |
| abstract_inverted_index.abnormal | 76 |
| abstract_inverted_index.accuracy | 293, 315 |
| abstract_inverted_index.accurate | 26 |
| abstract_inverted_index.advanced | 21, 297 |
| abstract_inverted_index.advances | 238 |
| abstract_inverted_index.analysis | 308 |
| abstract_inverted_index.bleeding | 102 |
| abstract_inverted_index.classify | 154 |
| abstract_inverted_index.compared | 320 |
| abstract_inverted_index.critical | 4 |
| abstract_inverted_index.diseases | 13, 45 |
| abstract_inverted_index.existing | 322 |
| abstract_inverted_index.findings | 19 |
| abstract_inverted_index.identify | 152 |
| abstract_inverted_index.improved | 179 |
| abstract_inverted_index.learning | 203 |
| abstract_inverted_index.leukemia | 105, 123, 287, 317 |
| abstract_inverted_index.manually | 110 |
| abstract_inverted_index.modified | 248 |
| abstract_inverted_index.pooling, | 255 |
| abstract_inverted_index.problems | 33 |
| abstract_inverted_index.proposes | 117 |
| abstract_inverted_index.relevant | 326 |
| abstract_inverted_index.research | 115, 210 |
| abstract_inverted_index.supports | 167 |
| abstract_inverted_index.symptoms | 16 |
| abstract_inverted_index.testing. | 23 |
| abstract_inverted_index.weakened | 97 |
| abstract_inverted_index.Leukemia, | 49 |
| abstract_inverted_index.according | 128 |
| abstract_inverted_index.accuracy. | 276 |
| abstract_inverted_index.affecting | 61 |
| abstract_inverted_index.algorithm | 204 |
| abstract_inverted_index.analyzing | 125 |
| abstract_inverted_index.anomalies | 159 |
| abstract_inverted_index.automatic | 119 |
| abstract_inverted_index.children. | 65 |
| abstract_inverted_index.connected | 258 |
| abstract_inverted_index.detecting | 31, 158 |
| abstract_inverted_index.detection | 9, 148, 283, 318 |
| abstract_inverted_index.diagnosis | 1, 27, 169 |
| abstract_inverted_index.effective | 39, 176 |
| abstract_inverted_index.essential | 29 |
| abstract_inverted_index.examining | 15 |
| abstract_inverted_index.including | 215, 299 |
| abstract_inverted_index.learning. | 142 |
| abstract_inverted_index.leukemia. | 48, 165 |
| abstract_inverted_index.optimized | 262 |
| abstract_inverted_index.patients. | 183 |
| abstract_inverted_index.precisely | 151 |
| abstract_inverted_index.prevalent | 58 |
| abstract_inverted_index.treatment | 11, 40 |
| abstract_inverted_index.MobileNet, | 303 |
| abstract_inverted_index.ResNet-50, | 301 |
| abstract_inverted_index.accomplish | 185 |
| abstract_inverted_index.associated | 160 |
| abstract_inverted_index.diagnosing | 285 |
| abstract_inverted_index.diagnostic | 106 |
| abstract_inverted_index.disorders. | 103 |
| abstract_inverted_index.especially | 42 |
| abstract_inverted_index.extraction | 273 |
| abstract_inverted_index.fine-tuned | 269 |
| abstract_inverted_index.intensive, | 111 |
| abstract_inverted_index.interferes | 86 |
| abstract_inverted_index.laboratory | 22 |
| abstract_inverted_index.prediction | 121 |
| abstract_inverted_index.processing | 139, 192 |
| abstract_inverted_index.production | 89 |
| abstract_inverted_index.supporting | 18 |
| abstract_inverted_index.techniques | 193, 242 |
| abstract_inverted_index.treatments | 177 |
| abstract_inverted_index.combination | 251 |
| abstract_inverted_index.deficiency, | 99 |
| abstract_inverted_index.diagnostics | 120 |
| abstract_inverted_index.extraction, | 221 |
| abstract_inverted_index.literature. | 327 |
| abstract_inverted_index.malignancy, | 52 |
| abstract_inverted_index.monitoring, | 171 |
| abstract_inverted_index.multi-stage | 213 |
| abstract_inverted_index.prescribing | 36 |
| abstract_inverted_index.significant | 237 |
| abstract_inverted_index.strategies, | 41 |
| abstract_inverted_index.Conventional | 104 |
| abstract_inverted_index.DenseNet121, | 300 |
| abstract_inverted_index.accumulation | 85 |
| abstract_inverted_index.inefficient. | 113 |
| abstract_inverted_index.methodology, | 214 |
| abstract_inverted_index.outperformed | 295 |
| abstract_inverted_index.preparation, | 217 |
| abstract_inverted_index.uncontrolled | 73 |
| abstract_inverted_index.EfficientNet. | 305 |
| abstract_inverted_index.Incep-tionv3, | 302 |
| abstract_inverted_index.architecture, | 234 |
| abstract_inverted_index.comprehensive | 307 |
| abstract_inverted_index.convolutional | 198 |
| abstract_inverted_index.preprocessing | 241 |
| abstract_inverted_index.classification | 275 |
| abstract_inverted_index.convolutional, | 254 |
| abstract_inverted_index.hyperparameter | 244 |
| abstract_inverted_index.preprocessing, | 219 |
| abstract_inverted_index.classification. | 224 |
| abstract_inverted_index.time-consuming, | 109 |
| abstract_inverted_index.characteristics. | 265 |
| abstract_inverted_index.life-threatening | 44 |
| cited_by_percentile_year.max | 99 |
| cited_by_percentile_year.min | 98 |
| corresponding_author_ids | https://openalex.org/A5012666416 |
| countries_distinct_count | 2 |
| institutions_distinct_count | 6 |
| corresponding_institution_ids | https://openalex.org/I33213144 |
| sustainable_development_goals[0].id | https://metadata.un.org/sdg/3 |
| sustainable_development_goals[0].score | 0.5400000214576721 |
| sustainable_development_goals[0].display_name | Good health and well-being |
| citation_normalized_percentile.value | 0.90679946 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | True |