Using the Quantum Interface in Phenix to improve macromolecular model and ligand structures Article Swipe
 YOU?
·
· 2025
· Open Access
·
· DOI: https://doi.org/10.1063/4.0000364
YOU?
·
· 2025
· Open Access
·
· DOI: https://doi.org/10.1063/4.0000364
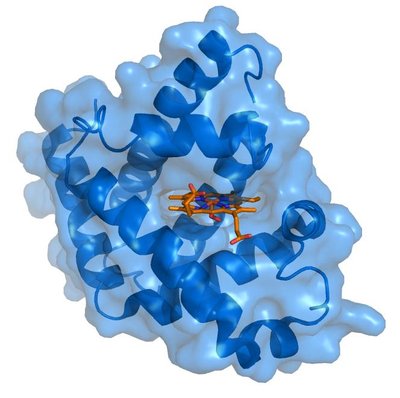
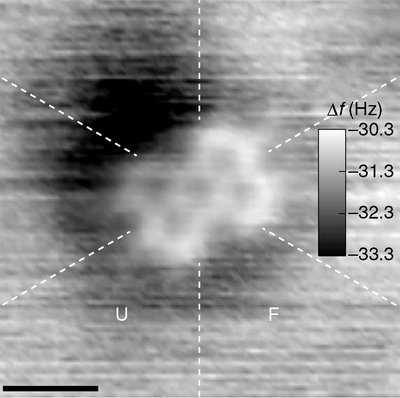
Quantum Mechanical methods provide geometries and energies of molecules using just the atomic and electronic positions. Being independent of the experimental data, they provide complementary information. Furthermore, allowing the experimental and QM methods to share information leads to better results. The Quantum Interface (QI) in Phenix (Liebschner et al., 2019) provides close integration with MOPAC (Moussa & Stewart, 2024) allowing the calculation of in situ restraints for drug candidates. Known as QM Restraints (QMR) (Liebschner et al., 2023), this method will provide protein binding pocket specific restraints during a refinement or in a stand-alone program. Another QI procedure is a novel approach for predicting histidine protonation states using QM methods. Historically, determining histidine protonation has been challenging due to limited resolution in X-ray crystallography and the inherent difficulty in detecting hydrogen atoms. Previous methods relied on empirical or geometric models, which provided some insights but had limitations. The proposed method, Quantum Mechanical Flipping (QMF) (Moriarty et al., In review), employs quantum mechanical calculations to predict protonation states based on the molecular environment. This approach considers all possible configurations of histidine protonation and assesses their feasibility by minimising geometry and energy calculations. QMF accounts for factors such as hydrogen bonding and molecular interactions providing a more accurate prediction of the most likely protonation state particularly in the binding pocket. The study demonstrates the effectiveness of QMF through comparisons with existing methods and validation using high- resolution protein structures. Results show that QMF can accurately predict histidine protonation states, even in cases with limited experimental data. Additionally, QMF's versatility allows it to be applied to various macromolecular environments, including ligand interactions and non-standard amino acids. Other applications of QI include metal coordination and ligand strain energies, both of which are being pursued.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.1063/4.0000364
- OA Status
- gold
- References
- 2
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4409325174
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4409325174Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1063/4.0000364Digital Object Identifier
- Title
-
Using the Quantum Interface in Phenix to improve macromolecular model and ligand structuresWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2025Year of publication
- Publication date
-
2025-03-01Full publication date if available
- Authors
-
Nigel W. Moriarty, Jonathan E. Moussa, Paul D. AdamsList of authors in order
- Landing page
-
https://doi.org/10.1063/4.0000364Publisher landing page
- Open access
-
YesWhether a free full text is available
- OA status
-
goldOpen access status per OpenAlex
- OA URL
-
https://doi.org/10.1063/4.0000364Direct OA link when available
- Concepts
-
Interface (matter), Ligand (biochemistry), Quantum, Macromolecule, Quantum chemical, Computer science, Physics, Chemical physics, Chemistry, Molecule, Quantum mechanics, Gibbs isotherm, Receptor, BiochemistryTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
0Total citation count in OpenAlex
- References (count)
-
2Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4409325174 |
|---|---|
| doi | https://doi.org/10.1063/4.0000364 |
| ids.doi | https://doi.org/10.1063/4.0000364 |
| ids.openalex | https://openalex.org/W4409325174 |
| fwci | 0.0 |
| type | article |
| title | Using the Quantum Interface in Phenix to improve macromolecular model and ligand structures |
| biblio.issue | 2_Supplement |
| biblio.volume | 12 |
| biblio.last_page | A55 |
| biblio.first_page | A55 |
| topics[0].id | https://openalex.org/T12285 |
| topics[0].field.id | https://openalex.org/fields/22 |
| topics[0].field.display_name | Engineering |
| topics[0].score | 0.9994999766349792 |
| topics[0].domain.id | https://openalex.org/domains/3 |
| topics[0].domain.display_name | Physical Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/2204 |
| topics[0].subfield.display_name | Biomedical Engineering |
| topics[0].display_name | Surface Chemistry and Catalysis |
| topics[1].id | https://openalex.org/T10903 |
| topics[1].field.id | https://openalex.org/fields/25 |
| topics[1].field.display_name | Materials Science |
| topics[1].score | 0.9976999759674072 |
| topics[1].domain.id | https://openalex.org/domains/3 |
| topics[1].domain.display_name | Physical Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/2505 |
| topics[1].subfield.display_name | Materials Chemistry |
| topics[1].display_name | Porphyrin and Phthalocyanine Chemistry |
| topics[2].id | https://openalex.org/T10973 |
| topics[2].field.id | https://openalex.org/fields/16 |
| topics[2].field.display_name | Chemistry |
| topics[2].score | 0.9958000183105469 |
| topics[2].domain.id | https://openalex.org/domains/3 |
| topics[2].domain.display_name | Physical Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1604 |
| topics[2].subfield.display_name | Inorganic Chemistry |
| topics[2].display_name | Radioactive element chemistry and processing |
| is_xpac | False |
| apc_list.value | 2200 |
| apc_list.currency | USD |
| apc_list.value_usd | 2200 |
| apc_paid.value | 2200 |
| apc_paid.currency | USD |
| apc_paid.value_usd | 2200 |
| concepts[0].id | https://openalex.org/C113843644 |
| concepts[0].level | 4 |
| concepts[0].score | 0.6993957757949829 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q901882 |
| concepts[0].display_name | Interface (matter) |
| concepts[1].id | https://openalex.org/C116569031 |
| concepts[1].level | 3 |
| concepts[1].score | 0.6379793286323547 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q899107 |
| concepts[1].display_name | Ligand (biochemistry) |
| concepts[2].id | https://openalex.org/C84114770 |
| concepts[2].level | 2 |
| concepts[2].score | 0.536300539970398 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q46344 |
| concepts[2].display_name | Quantum |
| concepts[3].id | https://openalex.org/C204389451 |
| concepts[3].level | 2 |
| concepts[3].score | 0.5249100923538208 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q178593 |
| concepts[3].display_name | Macromolecule |
| concepts[4].id | https://openalex.org/C2991951333 |
| concepts[4].level | 3 |
| concepts[4].score | 0.4675869941711426 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q188403 |
| concepts[4].display_name | Quantum chemical |
| concepts[5].id | https://openalex.org/C41008148 |
| concepts[5].level | 0 |
| concepts[5].score | 0.4339885115623474 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q21198 |
| concepts[5].display_name | Computer science |
| concepts[6].id | https://openalex.org/C121332964 |
| concepts[6].level | 0 |
| concepts[6].score | 0.3734140396118164 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q413 |
| concepts[6].display_name | Physics |
| concepts[7].id | https://openalex.org/C159467904 |
| concepts[7].level | 1 |
| concepts[7].score | 0.32697874307632446 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q2001702 |
| concepts[7].display_name | Chemical physics |
| concepts[8].id | https://openalex.org/C185592680 |
| concepts[8].level | 0 |
| concepts[8].score | 0.27683353424072266 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q2329 |
| concepts[8].display_name | Chemistry |
| concepts[9].id | https://openalex.org/C32909587 |
| concepts[9].level | 2 |
| concepts[9].score | 0.18204575777053833 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q11369 |
| concepts[9].display_name | Molecule |
| concepts[10].id | https://openalex.org/C62520636 |
| concepts[10].level | 1 |
| concepts[10].score | 0.1723124384880066 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q944 |
| concepts[10].display_name | Quantum mechanics |
| concepts[11].id | https://openalex.org/C55352822 |
| concepts[11].level | 3 |
| concepts[11].score | 0.0 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q5558978 |
| concepts[11].display_name | Gibbs isotherm |
| concepts[12].id | https://openalex.org/C170493617 |
| concepts[12].level | 2 |
| concepts[12].score | 0.0 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q208467 |
| concepts[12].display_name | Receptor |
| concepts[13].id | https://openalex.org/C55493867 |
| concepts[13].level | 1 |
| concepts[13].score | 0.0 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q7094 |
| concepts[13].display_name | Biochemistry |
| keywords[0].id | https://openalex.org/keywords/interface |
| keywords[0].score | 0.6993957757949829 |
| keywords[0].display_name | Interface (matter) |
| keywords[1].id | https://openalex.org/keywords/ligand |
| keywords[1].score | 0.6379793286323547 |
| keywords[1].display_name | Ligand (biochemistry) |
| keywords[2].id | https://openalex.org/keywords/quantum |
| keywords[2].score | 0.536300539970398 |
| keywords[2].display_name | Quantum |
| keywords[3].id | https://openalex.org/keywords/macromolecule |
| keywords[3].score | 0.5249100923538208 |
| keywords[3].display_name | Macromolecule |
| keywords[4].id | https://openalex.org/keywords/quantum-chemical |
| keywords[4].score | 0.4675869941711426 |
| keywords[4].display_name | Quantum chemical |
| keywords[5].id | https://openalex.org/keywords/computer-science |
| keywords[5].score | 0.4339885115623474 |
| keywords[5].display_name | Computer science |
| keywords[6].id | https://openalex.org/keywords/physics |
| keywords[6].score | 0.3734140396118164 |
| keywords[6].display_name | Physics |
| keywords[7].id | https://openalex.org/keywords/chemical-physics |
| keywords[7].score | 0.32697874307632446 |
| keywords[7].display_name | Chemical physics |
| keywords[8].id | https://openalex.org/keywords/chemistry |
| keywords[8].score | 0.27683353424072266 |
| keywords[8].display_name | Chemistry |
| keywords[9].id | https://openalex.org/keywords/molecule |
| keywords[9].score | 0.18204575777053833 |
| keywords[9].display_name | Molecule |
| keywords[10].id | https://openalex.org/keywords/quantum-mechanics |
| keywords[10].score | 0.1723124384880066 |
| keywords[10].display_name | Quantum mechanics |
| language | en |
| locations[0].id | doi:10.1063/4.0000364 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S2737888654 |
| locations[0].source.issn | 2329-7778 |
| locations[0].source.type | journal |
| locations[0].source.is_oa | True |
| locations[0].source.issn_l | 2329-7778 |
| locations[0].source.is_core | True |
| locations[0].source.is_in_doaj | True |
| locations[0].source.display_name | Structural Dynamics |
| locations[0].source.host_organization | https://openalex.org/P4310320061 |
| locations[0].source.host_organization_name | AIP Publishing |
| locations[0].source.host_organization_lineage | https://openalex.org/P4310320061, https://openalex.org/P4310320257 |
| locations[0].source.host_organization_lineage_names | AIP Publishing, American Institute of Physics |
| locations[0].license | cc-by |
| locations[0].pdf_url | |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | Structural Dynamics |
| locations[0].landing_page_url | https://doi.org/10.1063/4.0000364 |
| indexed_in | crossref, doaj |
| authorships[0].author.id | https://openalex.org/A5084490031 |
| authorships[0].author.orcid | https://orcid.org/0000-0001-8857-9464 |
| authorships[0].author.display_name | Nigel W. Moriarty |
| authorships[0].countries | US |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I148283060 |
| authorships[0].affiliations[0].raw_affiliation_string | Lawrence Berkeley Lab 1 , Berkeley, CA, |
| authorships[0].institutions[0].id | https://openalex.org/I148283060 |
| authorships[0].institutions[0].ror | https://ror.org/02jbv0t02 |
| authorships[0].institutions[0].type | facility |
| authorships[0].institutions[0].lineage | https://openalex.org/I1330989302, https://openalex.org/I148283060, https://openalex.org/I39565521 |
| authorships[0].institutions[0].country_code | US |
| authorships[0].institutions[0].display_name | Lawrence Berkeley National Laboratory |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Nigel Moriarty |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Lawrence Berkeley Lab 1 , Berkeley, CA, |
| authorships[1].author.id | https://openalex.org/A5031127321 |
| authorships[1].author.orcid | https://orcid.org/0000-0003-3701-1830 |
| authorships[1].author.display_name | Jonathan E. Moussa |
| authorships[1].countries | US |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I859038795 |
| authorships[1].affiliations[0].raw_affiliation_string | Virginia Tech 2 , Blacksberg, VT, |
| authorships[1].institutions[0].id | https://openalex.org/I859038795 |
| authorships[1].institutions[0].ror | https://ror.org/02smfhw86 |
| authorships[1].institutions[0].type | education |
| authorships[1].institutions[0].lineage | https://openalex.org/I859038795 |
| authorships[1].institutions[0].country_code | US |
| authorships[1].institutions[0].display_name | Virginia Tech |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Jonathan Moussa |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | Virginia Tech 2 , Blacksberg, VT, |
| authorships[2].author.id | https://openalex.org/A5027915011 |
| authorships[2].author.orcid | https://orcid.org/0000-0001-9333-8219 |
| authorships[2].author.display_name | Paul D. Adams |
| authorships[2].countries | US |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I95457486 |
| authorships[2].affiliations[0].raw_affiliation_string | University of California Berkeley 3 , Berkeley, CA, |
| authorships[2].affiliations[1].institution_ids | https://openalex.org/I148283060 |
| authorships[2].affiliations[1].raw_affiliation_string | Lawrence Berkeley Lab 1 , Berkeley, CA, |
| authorships[2].institutions[0].id | https://openalex.org/I148283060 |
| authorships[2].institutions[0].ror | https://ror.org/02jbv0t02 |
| authorships[2].institutions[0].type | facility |
| authorships[2].institutions[0].lineage | https://openalex.org/I1330989302, https://openalex.org/I148283060, https://openalex.org/I39565521 |
| authorships[2].institutions[0].country_code | US |
| authorships[2].institutions[0].display_name | Lawrence Berkeley National Laboratory |
| authorships[2].institutions[1].id | https://openalex.org/I95457486 |
| authorships[2].institutions[1].ror | https://ror.org/01an7q238 |
| authorships[2].institutions[1].type | education |
| authorships[2].institutions[1].lineage | https://openalex.org/I95457486 |
| authorships[2].institutions[1].country_code | US |
| authorships[2].institutions[1].display_name | University of California, Berkeley |
| authorships[2].author_position | last |
| authorships[2].raw_author_name | Paul Adams |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | Lawrence Berkeley Lab 1 , Berkeley, CA,, University of California Berkeley 3 , Berkeley, CA, |
| has_content.pdf | False |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://doi.org/10.1063/4.0000364 |
| open_access.oa_status | gold |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Using the Quantum Interface in Phenix to improve macromolecular model and ligand structures |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T12285 |
| primary_topic.field.id | https://openalex.org/fields/22 |
| primary_topic.field.display_name | Engineering |
| primary_topic.score | 0.9994999766349792 |
| primary_topic.domain.id | https://openalex.org/domains/3 |
| primary_topic.domain.display_name | Physical Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/2204 |
| primary_topic.subfield.display_name | Biomedical Engineering |
| primary_topic.display_name | Surface Chemistry and Catalysis |
| related_works | https://openalex.org/W2044014390, https://openalex.org/W1846316342, https://openalex.org/W1964725291, https://openalex.org/W1984670152, https://openalex.org/W4210447090, https://openalex.org/W2043468722, https://openalex.org/W2083665351, https://openalex.org/W2123419409, https://openalex.org/W2003172604, https://openalex.org/W2768531357 |
| cited_by_count | 0 |
| locations_count | 1 |
| best_oa_location.id | doi:10.1063/4.0000364 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S2737888654 |
| best_oa_location.source.issn | 2329-7778 |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | True |
| best_oa_location.source.issn_l | 2329-7778 |
| best_oa_location.source.is_core | True |
| best_oa_location.source.is_in_doaj | True |
| best_oa_location.source.display_name | Structural Dynamics |
| best_oa_location.source.host_organization | https://openalex.org/P4310320061 |
| best_oa_location.source.host_organization_name | AIP Publishing |
| best_oa_location.source.host_organization_lineage | https://openalex.org/P4310320061, https://openalex.org/P4310320257 |
| best_oa_location.source.host_organization_lineage_names | AIP Publishing, American Institute of Physics |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | journal-article |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | Structural Dynamics |
| best_oa_location.landing_page_url | https://doi.org/10.1063/4.0000364 |
| primary_location.id | doi:10.1063/4.0000364 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S2737888654 |
| primary_location.source.issn | 2329-7778 |
| primary_location.source.type | journal |
| primary_location.source.is_oa | True |
| primary_location.source.issn_l | 2329-7778 |
| primary_location.source.is_core | True |
| primary_location.source.is_in_doaj | True |
| primary_location.source.display_name | Structural Dynamics |
| primary_location.source.host_organization | https://openalex.org/P4310320061 |
| primary_location.source.host_organization_name | AIP Publishing |
| primary_location.source.host_organization_lineage | https://openalex.org/P4310320061, https://openalex.org/P4310320257 |
| primary_location.source.host_organization_lineage_names | AIP Publishing, American Institute of Physics |
| primary_location.license | cc-by |
| primary_location.pdf_url | |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | Structural Dynamics |
| primary_location.landing_page_url | https://doi.org/10.1063/4.0000364 |
| publication_date | 2025-03-01 |
| publication_year | 2025 |
| referenced_works | https://openalex.org/W2973457155, https://openalex.org/W4317583457 |
| referenced_works_count | 2 |
| abstract_inverted_index.a | 88, 92, 99, 203 |
| abstract_inverted_index.In | 157 |
| abstract_inverted_index.QI | 96, 276 |
| abstract_inverted_index.QM | 31, 71, 108 |
| abstract_inverted_index.as | 70, 196 |
| abstract_inverted_index.be | 260 |
| abstract_inverted_index.by | 185 |
| abstract_inverted_index.et | 47, 75, 155 |
| abstract_inverted_index.in | 44, 63, 91, 121, 128, 214, 248 |
| abstract_inverted_index.is | 98 |
| abstract_inverted_index.it | 258 |
| abstract_inverted_index.of | 7, 18, 62, 178, 207, 223, 275, 285 |
| abstract_inverted_index.on | 135, 168 |
| abstract_inverted_index.or | 90, 137 |
| abstract_inverted_index.to | 33, 37, 118, 163, 259, 262 |
| abstract_inverted_index.QMF | 191, 224, 240 |
| abstract_inverted_index.The | 40, 147, 218 |
| abstract_inverted_index.all | 175 |
| abstract_inverted_index.and | 5, 13, 30, 124, 181, 188, 199, 230, 269, 280 |
| abstract_inverted_index.are | 287 |
| abstract_inverted_index.but | 144 |
| abstract_inverted_index.can | 241 |
| abstract_inverted_index.due | 117 |
| abstract_inverted_index.for | 66, 102, 193 |
| abstract_inverted_index.had | 145 |
| abstract_inverted_index.has | 114 |
| abstract_inverted_index.the | 11, 19, 28, 60, 125, 169, 208, 215, 221 |
| abstract_inverted_index.(QI) | 43 |
| abstract_inverted_index.This | 172 |
| abstract_inverted_index.al., | 48, 76, 156 |
| abstract_inverted_index.been | 115 |
| abstract_inverted_index.both | 284 |
| abstract_inverted_index.drug | 67 |
| abstract_inverted_index.even | 247 |
| abstract_inverted_index.just | 10 |
| abstract_inverted_index.more | 204 |
| abstract_inverted_index.most | 209 |
| abstract_inverted_index.show | 238 |
| abstract_inverted_index.situ | 64 |
| abstract_inverted_index.some | 142 |
| abstract_inverted_index.such | 195 |
| abstract_inverted_index.that | 239 |
| abstract_inverted_index.they | 22 |
| abstract_inverted_index.this | 78 |
| abstract_inverted_index.will | 80 |
| abstract_inverted_index.with | 53, 227, 250 |
| abstract_inverted_index.& | 56 |
| abstract_inverted_index.(QMF) | 153 |
| abstract_inverted_index.(QMR) | 73 |
| abstract_inverted_index.2019) | 49 |
| abstract_inverted_index.2024) | 58 |
| abstract_inverted_index.Being | 16 |
| abstract_inverted_index.Known | 69 |
| abstract_inverted_index.MOPAC | 54 |
| abstract_inverted_index.Other | 273 |
| abstract_inverted_index.QMF's | 255 |
| abstract_inverted_index.X-ray | 122 |
| abstract_inverted_index.amino | 271 |
| abstract_inverted_index.based | 167 |
| abstract_inverted_index.being | 288 |
| abstract_inverted_index.cases | 249 |
| abstract_inverted_index.close | 51 |
| abstract_inverted_index.data, | 21 |
| abstract_inverted_index.data. | 253 |
| abstract_inverted_index.high- | 233 |
| abstract_inverted_index.leads | 36 |
| abstract_inverted_index.metal | 278 |
| abstract_inverted_index.novel | 100 |
| abstract_inverted_index.share | 34 |
| abstract_inverted_index.state | 212 |
| abstract_inverted_index.study | 219 |
| abstract_inverted_index.their | 183 |
| abstract_inverted_index.using | 9, 107, 232 |
| abstract_inverted_index.which | 140, 286 |
| abstract_inverted_index.2023), | 77 |
| abstract_inverted_index.Phenix | 45 |
| abstract_inverted_index.acids. | 272 |
| abstract_inverted_index.allows | 257 |
| abstract_inverted_index.atomic | 12 |
| abstract_inverted_index.atoms. | 131 |
| abstract_inverted_index.better | 38 |
| abstract_inverted_index.during | 87 |
| abstract_inverted_index.energy | 189 |
| abstract_inverted_index.ligand | 267, 281 |
| abstract_inverted_index.likely | 210 |
| abstract_inverted_index.method | 79 |
| abstract_inverted_index.pocket | 84 |
| abstract_inverted_index.relied | 134 |
| abstract_inverted_index.states | 106, 166 |
| abstract_inverted_index.strain | 282 |
| abstract_inverted_index.(Moussa | 55 |
| abstract_inverted_index.Another | 95 |
| abstract_inverted_index.Quantum | 0, 41, 150 |
| abstract_inverted_index.Results | 237 |
| abstract_inverted_index.applied | 261 |
| abstract_inverted_index.binding | 83, 216 |
| abstract_inverted_index.bonding | 198 |
| abstract_inverted_index.employs | 159 |
| abstract_inverted_index.factors | 194 |
| abstract_inverted_index.include | 277 |
| abstract_inverted_index.limited | 119, 251 |
| abstract_inverted_index.method, | 149 |
| abstract_inverted_index.methods | 2, 32, 133, 229 |
| abstract_inverted_index.models, | 139 |
| abstract_inverted_index.pocket. | 217 |
| abstract_inverted_index.predict | 164, 243 |
| abstract_inverted_index.protein | 82, 235 |
| abstract_inverted_index.provide | 3, 23, 81 |
| abstract_inverted_index.quantum | 160 |
| abstract_inverted_index.states, | 246 |
| abstract_inverted_index.through | 225 |
| abstract_inverted_index.various | 263 |
| abstract_inverted_index.Flipping | 152 |
| abstract_inverted_index.Previous | 132 |
| abstract_inverted_index.Stewart, | 57 |
| abstract_inverted_index.accounts | 192 |
| abstract_inverted_index.accurate | 205 |
| abstract_inverted_index.allowing | 27, 59 |
| abstract_inverted_index.approach | 101, 173 |
| abstract_inverted_index.assesses | 182 |
| abstract_inverted_index.energies | 6 |
| abstract_inverted_index.existing | 228 |
| abstract_inverted_index.geometry | 187 |
| abstract_inverted_index.hydrogen | 130, 197 |
| abstract_inverted_index.inherent | 126 |
| abstract_inverted_index.insights | 143 |
| abstract_inverted_index.methods. | 109 |
| abstract_inverted_index.possible | 176 |
| abstract_inverted_index.program. | 94 |
| abstract_inverted_index.proposed | 148 |
| abstract_inverted_index.provided | 141 |
| abstract_inverted_index.provides | 50 |
| abstract_inverted_index.pursued. | 289 |
| abstract_inverted_index.results. | 39 |
| abstract_inverted_index.review), | 158 |
| abstract_inverted_index.specific | 85 |
| abstract_inverted_index.(Moriarty | 154 |
| abstract_inverted_index.Interface | 42 |
| abstract_inverted_index.considers | 174 |
| abstract_inverted_index.detecting | 129 |
| abstract_inverted_index.empirical | 136 |
| abstract_inverted_index.energies, | 283 |
| abstract_inverted_index.geometric | 138 |
| abstract_inverted_index.histidine | 104, 112, 179, 244 |
| abstract_inverted_index.including | 266 |
| abstract_inverted_index.molecular | 170, 200 |
| abstract_inverted_index.molecules | 8 |
| abstract_inverted_index.procedure | 97 |
| abstract_inverted_index.providing | 202 |
| abstract_inverted_index.Mechanical | 1, 151 |
| abstract_inverted_index.Restraints | 72 |
| abstract_inverted_index.accurately | 242 |
| abstract_inverted_index.difficulty | 127 |
| abstract_inverted_index.electronic | 14 |
| abstract_inverted_index.geometries | 4 |
| abstract_inverted_index.mechanical | 161 |
| abstract_inverted_index.minimising | 186 |
| abstract_inverted_index.positions. | 15 |
| abstract_inverted_index.predicting | 103 |
| abstract_inverted_index.prediction | 206 |
| abstract_inverted_index.refinement | 89 |
| abstract_inverted_index.resolution | 120, 234 |
| abstract_inverted_index.restraints | 65, 86 |
| abstract_inverted_index.validation | 231 |
| abstract_inverted_index.(Liebschner | 46, 74 |
| abstract_inverted_index.calculation | 61 |
| abstract_inverted_index.candidates. | 68 |
| abstract_inverted_index.challenging | 116 |
| abstract_inverted_index.comparisons | 226 |
| abstract_inverted_index.determining | 111 |
| abstract_inverted_index.feasibility | 184 |
| abstract_inverted_index.independent | 17 |
| abstract_inverted_index.information | 35 |
| abstract_inverted_index.integration | 52 |
| abstract_inverted_index.protonation | 105, 113, 165, 180, 211, 245 |
| abstract_inverted_index.stand-alone | 93 |
| abstract_inverted_index.structures. | 236 |
| abstract_inverted_index.versatility | 256 |
| abstract_inverted_index.Furthermore, | 26 |
| abstract_inverted_index.applications | 274 |
| abstract_inverted_index.calculations | 162 |
| abstract_inverted_index.coordination | 279 |
| abstract_inverted_index.demonstrates | 220 |
| abstract_inverted_index.environment. | 171 |
| abstract_inverted_index.experimental | 20, 29, 252 |
| abstract_inverted_index.information. | 25 |
| abstract_inverted_index.interactions | 201, 268 |
| abstract_inverted_index.limitations. | 146 |
| abstract_inverted_index.non-standard | 270 |
| abstract_inverted_index.particularly | 213 |
| abstract_inverted_index.Additionally, | 254 |
| abstract_inverted_index.Historically, | 110 |
| abstract_inverted_index.calculations. | 190 |
| abstract_inverted_index.complementary | 24 |
| abstract_inverted_index.effectiveness | 222 |
| abstract_inverted_index.environments, | 265 |
| abstract_inverted_index.configurations | 177 |
| abstract_inverted_index.macromolecular | 264 |
| abstract_inverted_index.crystallography | 123 |
| cited_by_percentile_year | |
| countries_distinct_count | 1 |
| institutions_distinct_count | 3 |
| citation_normalized_percentile.value | 0.12196031 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |