Validation of a Computationally Efficient Model of the Mu-Opioid Receptor Article Swipe
 YOU?
·
· 2023
· Open Access
·
· DOI: https://doi.org/10.33697/ajur.2023.085
YOU?
·
· 2023
· Open Access
·
· DOI: https://doi.org/10.33697/ajur.2023.085
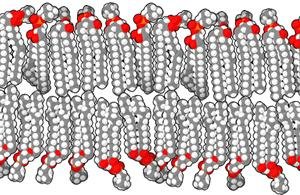
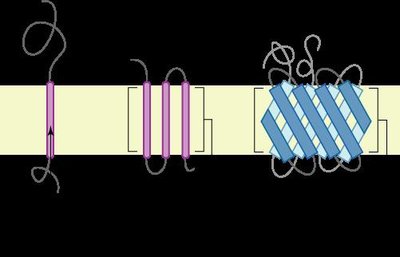
The mu-opioid receptor (MOR) is a transmembrane protein and the primary target for pain-modulating drugs. Opioid drugs come with detrimental side-effects such as physical dependence and addiction. However, recent studies show that understanding structural properties and dynamics of MOR may aid in the design of opioid drugs with reduced side-effects. Molecular dynamics simulations allow researchers to study changes in protein conformation at an atomistic level. However, modeling systems including MOR embedded in a lipid bilayer can be computationally expensive. This study evaluates a modeling approach that uses harmonic restraints on the transmembrane regions of MOR to model the rigidity of the lipid bilayer without explicitly simulating lipid molecules, reducing the number of atoms in the simulation. The proposed model allows MOR to be simulated 49% faster than a simulation explicitly including the lipid bilayer. To assess the accuracy of the proposed model, simulations were performed of MOR in a lipid bilayer, the free MOR in water and MOR in water with harmonic restraints applied to all transmembrane residues using NAMD 3.0 alpha and the CHARMM36 force field. Dynamic properties of MOR were shown to be different in each system, with the free MOR having a higher root mean square deviation (RMSD) than MOR with an explicitly modeled lipid bilayer. The systems with harmonic restraint constants of 0.001 kcal/mol/Å2 applied to the transmembrane residues had RMSD values comparable to those in an explicitly modeled lipid bilayer. This study demonstrates that using restraints on the transmembrane residues of MOR is a feasible way of modeling the ligand-free receptor with reduced computational costs. This model could allow the dynamics of MOR in a lipid bilayer environment to be studied more efficiently. KEYWORDS: Molecular Dynamics; Atomistic Simulations; Computational Modeling; Mu-Opioid Receptor; G-Protein Coupled Receptor; Lipid Bilayer, Opioid, Transmembrane Protein
Related Topics
- Type
- article
- Language
- en
- Landing Page
- http://doi.org/10.33697/ajur.2023.085
- https://doi.org/10.33697/ajur.2023.085
- OA Status
- diamond
- References
- 43
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4387422117
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4387422117Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.33697/ajur.2023.085Digital Object Identifier
- Title
-
Validation of a Computationally Efficient Model of the Mu-Opioid ReceptorWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2023Year of publication
- Publication date
-
2023-10-07Full publication date if available
- Authors
-
Allison Barkdull, Lexin Chen, Akash Mathavan, Karina Martínez‐Mayorga, Coray M. ColinaList of authors in order
- Landing page
-
https://doi.org/10.33697/ajur.2023.085Publisher landing page
- PDF URL
-
https://doi.org/10.33697/ajur.2023.085Direct link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
diamondOpen access status per OpenAlex
- OA URL
-
https://doi.org/10.33697/ajur.2023.085Direct OA link when available
- Concepts
-
Lipid bilayer, Transmembrane protein, Molecular dynamics, Bilayer, Transmembrane domain, Chemistry, Receptor, Computational chemistry, Membrane, BiochemistryTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
0Total citation count in OpenAlex
- References (count)
-
43Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4387422117 |
|---|---|
| doi | https://doi.org/10.33697/ajur.2023.085 |
| ids.doi | https://doi.org/10.33697/ajur.2023.085 |
| ids.openalex | https://openalex.org/W4387422117 |
| fwci | 0.0 |
| type | article |
| title | Validation of a Computationally Efficient Model of the Mu-Opioid Receptor |
| biblio.issue | 2 |
| biblio.volume | 20 |
| biblio.last_page | 44 |
| biblio.first_page | 29 |
| topics[0].id | https://openalex.org/T10814 |
| topics[0].field.id | https://openalex.org/fields/28 |
| topics[0].field.display_name | Neuroscience |
| topics[0].score | 0.9997000098228455 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/2804 |
| topics[0].subfield.display_name | Cellular and Molecular Neuroscience |
| topics[0].display_name | Neuropeptides and Animal Physiology |
| topics[1].id | https://openalex.org/T13338 |
| topics[1].field.id | https://openalex.org/fields/11 |
| topics[1].field.display_name | Agricultural and Biological Sciences |
| topics[1].score | 0.9969000220298767 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1103 |
| topics[1].subfield.display_name | Animal Science and Zoology |
| topics[1].display_name | Pharmacological Effects and Assays |
| topics[2].id | https://openalex.org/T13047 |
| topics[2].field.id | https://openalex.org/fields/13 |
| topics[2].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[2].score | 0.9958999752998352 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1312 |
| topics[2].subfield.display_name | Molecular Biology |
| topics[2].display_name | Pharmacological Receptor Mechanisms and Effects |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C39944091 |
| concepts[0].level | 3 |
| concepts[0].score | 0.7477751970291138 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q423279 |
| concepts[0].display_name | Lipid bilayer |
| concepts[1].id | https://openalex.org/C24530287 |
| concepts[1].level | 3 |
| concepts[1].score | 0.5910434126853943 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q424204 |
| concepts[1].display_name | Transmembrane protein |
| concepts[2].id | https://openalex.org/C59593255 |
| concepts[2].level | 2 |
| concepts[2].score | 0.5771881937980652 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q901663 |
| concepts[2].display_name | Molecular dynamics |
| concepts[3].id | https://openalex.org/C192157962 |
| concepts[3].level | 3 |
| concepts[3].score | 0.5681434869766235 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q4087243 |
| concepts[3].display_name | Bilayer |
| concepts[4].id | https://openalex.org/C118892022 |
| concepts[4].level | 3 |
| concepts[4].score | 0.5385239124298096 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q7834587 |
| concepts[4].display_name | Transmembrane domain |
| concepts[5].id | https://openalex.org/C185592680 |
| concepts[5].level | 0 |
| concepts[5].score | 0.5221588611602783 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q2329 |
| concepts[5].display_name | Chemistry |
| concepts[6].id | https://openalex.org/C170493617 |
| concepts[6].level | 2 |
| concepts[6].score | 0.22397539019584656 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q208467 |
| concepts[6].display_name | Receptor |
| concepts[7].id | https://openalex.org/C147597530 |
| concepts[7].level | 1 |
| concepts[7].score | 0.2052033245563507 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q369472 |
| concepts[7].display_name | Computational chemistry |
| concepts[8].id | https://openalex.org/C41625074 |
| concepts[8].level | 2 |
| concepts[8].score | 0.19533655047416687 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q176088 |
| concepts[8].display_name | Membrane |
| concepts[9].id | https://openalex.org/C55493867 |
| concepts[9].level | 1 |
| concepts[9].score | 0.09253701567649841 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q7094 |
| concepts[9].display_name | Biochemistry |
| keywords[0].id | https://openalex.org/keywords/lipid-bilayer |
| keywords[0].score | 0.7477751970291138 |
| keywords[0].display_name | Lipid bilayer |
| keywords[1].id | https://openalex.org/keywords/transmembrane-protein |
| keywords[1].score | 0.5910434126853943 |
| keywords[1].display_name | Transmembrane protein |
| keywords[2].id | https://openalex.org/keywords/molecular-dynamics |
| keywords[2].score | 0.5771881937980652 |
| keywords[2].display_name | Molecular dynamics |
| keywords[3].id | https://openalex.org/keywords/bilayer |
| keywords[3].score | 0.5681434869766235 |
| keywords[3].display_name | Bilayer |
| keywords[4].id | https://openalex.org/keywords/transmembrane-domain |
| keywords[4].score | 0.5385239124298096 |
| keywords[4].display_name | Transmembrane domain |
| keywords[5].id | https://openalex.org/keywords/chemistry |
| keywords[5].score | 0.5221588611602783 |
| keywords[5].display_name | Chemistry |
| keywords[6].id | https://openalex.org/keywords/receptor |
| keywords[6].score | 0.22397539019584656 |
| keywords[6].display_name | Receptor |
| keywords[7].id | https://openalex.org/keywords/computational-chemistry |
| keywords[7].score | 0.2052033245563507 |
| keywords[7].display_name | Computational chemistry |
| keywords[8].id | https://openalex.org/keywords/membrane |
| keywords[8].score | 0.19533655047416687 |
| keywords[8].display_name | Membrane |
| keywords[9].id | https://openalex.org/keywords/biochemistry |
| keywords[9].score | 0.09253701567649841 |
| keywords[9].display_name | Biochemistry |
| language | en |
| locations[0].id | doi:10.33697/ajur.2023.085 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4210172541 |
| locations[0].source.issn | 1536-4585, 2375-8732 |
| locations[0].source.type | journal |
| locations[0].source.is_oa | True |
| locations[0].source.issn_l | 1536-4585 |
| locations[0].source.is_core | True |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | American Journal of Undergraduate Research |
| locations[0].source.host_organization | |
| locations[0].source.host_organization_name | |
| locations[0].license | |
| locations[0].pdf_url | https://doi.org/10.33697/ajur.2023.085 |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | American Journal of Undergraduate Research |
| locations[0].landing_page_url | http://doi.org/10.33697/ajur.2023.085 |
| indexed_in | crossref |
| authorships[0].author.id | https://openalex.org/A5023580484 |
| authorships[0].author.orcid | https://orcid.org/0000-0001-6813-7191 |
| authorships[0].author.display_name | Allison Barkdull |
| authorships[0].countries | US |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I33213144 |
| authorships[0].affiliations[0].raw_affiliation_string | Department of Biomedical Engineering, University of Florida, Gainesville, FL |
| authorships[0].institutions[0].id | https://openalex.org/I33213144 |
| authorships[0].institutions[0].ror | https://ror.org/02y3ad647 |
| authorships[0].institutions[0].type | education |
| authorships[0].institutions[0].lineage | https://openalex.org/I33213144 |
| authorships[0].institutions[0].country_code | US |
| authorships[0].institutions[0].display_name | University of Florida |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Allison Barkdull |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Department of Biomedical Engineering, University of Florida, Gainesville, FL |
| authorships[1].author.id | https://openalex.org/A5111052820 |
| authorships[1].author.orcid | |
| authorships[1].author.display_name | Lexin Chen |
| authorships[1].countries | US |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I33213144 |
| authorships[1].affiliations[0].raw_affiliation_string | Department of Chemistry, University of Florida, Gainesville, FL |
| authorships[1].institutions[0].id | https://openalex.org/I33213144 |
| authorships[1].institutions[0].ror | https://ror.org/02y3ad647 |
| authorships[1].institutions[0].type | education |
| authorships[1].institutions[0].lineage | https://openalex.org/I33213144 |
| authorships[1].institutions[0].country_code | US |
| authorships[1].institutions[0].display_name | University of Florida |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Lexin Chen |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | Department of Chemistry, University of Florida, Gainesville, FL |
| authorships[2].author.id | https://openalex.org/A5073934550 |
| authorships[2].author.orcid | https://orcid.org/0000-0003-3496-2542 |
| authorships[2].author.display_name | Akash Mathavan |
| authorships[2].countries | US |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I33213144 |
| authorships[2].affiliations[0].raw_affiliation_string | College of Medicine, University of Florida, Gainesville, FL |
| authorships[2].institutions[0].id | https://openalex.org/I33213144 |
| authorships[2].institutions[0].ror | https://ror.org/02y3ad647 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I33213144 |
| authorships[2].institutions[0].country_code | US |
| authorships[2].institutions[0].display_name | University of Florida |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Akash Mathavan |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | College of Medicine, University of Florida, Gainesville, FL |
| authorships[3].author.id | https://openalex.org/A5057869875 |
| authorships[3].author.orcid | https://orcid.org/0000-0002-6974-7941 |
| authorships[3].author.display_name | Karina Martínez‐Mayorga |
| authorships[3].countries | MX |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I8961855 |
| authorships[3].affiliations[0].raw_affiliation_string | UAEY-IIMAS, Universidad Nacional Autónoma de México, Mérida, Yucatán, México |
| authorships[3].affiliations[1].institution_ids | https://openalex.org/I8961855 |
| authorships[3].affiliations[1].raw_affiliation_string | Instituto de Química, Unidad Mérida, Universidad Nacional Autónoma de México, Ucú, Yucatán, México |
| authorships[3].institutions[0].id | https://openalex.org/I8961855 |
| authorships[3].institutions[0].ror | https://ror.org/01tmp8f25 |
| authorships[3].institutions[0].type | education |
| authorships[3].institutions[0].lineage | https://openalex.org/I8961855 |
| authorships[3].institutions[0].country_code | MX |
| authorships[3].institutions[0].display_name | Universidad Nacional Autónoma de México |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Karina Martinez-Mayorga |
| authorships[3].is_corresponding | False |
| authorships[3].raw_affiliation_strings | Instituto de Química, Unidad Mérida, Universidad Nacional Autónoma de México, Ucú, Yucatán, México, UAEY-IIMAS, Universidad Nacional Autónoma de México, Mérida, Yucatán, México |
| authorships[4].author.id | https://openalex.org/A5059420993 |
| authorships[4].author.orcid | https://orcid.org/0000-0003-2367-1352 |
| authorships[4].author.display_name | Coray M. Colina |
| authorships[4].countries | MX, US |
| authorships[4].affiliations[0].institution_ids | https://openalex.org/I8961855 |
| authorships[4].affiliations[0].raw_affiliation_string | UAEY-IIMAS, Universidad Nacional Autónoma de México, Mérida, Yucatán, México |
| authorships[4].affiliations[1].institution_ids | https://openalex.org/I33213144 |
| authorships[4].affiliations[1].raw_affiliation_string | Departments of Materials Science and Nuclear Engineering, University of Florida, Gainesville, FL |
| authorships[4].affiliations[2].institution_ids | https://openalex.org/I33213144 |
| authorships[4].affiliations[2].raw_affiliation_string | Department of Chemistry, University of Florida, Gainesville, FL |
| authorships[4].institutions[0].id | https://openalex.org/I8961855 |
| authorships[4].institutions[0].ror | https://ror.org/01tmp8f25 |
| authorships[4].institutions[0].type | education |
| authorships[4].institutions[0].lineage | https://openalex.org/I8961855 |
| authorships[4].institutions[0].country_code | MX |
| authorships[4].institutions[0].display_name | Universidad Nacional Autónoma de México |
| authorships[4].institutions[1].id | https://openalex.org/I33213144 |
| authorships[4].institutions[1].ror | https://ror.org/02y3ad647 |
| authorships[4].institutions[1].type | education |
| authorships[4].institutions[1].lineage | https://openalex.org/I33213144 |
| authorships[4].institutions[1].country_code | US |
| authorships[4].institutions[1].display_name | University of Florida |
| authorships[4].author_position | last |
| authorships[4].raw_author_name | Coray Colina |
| authorships[4].is_corresponding | False |
| authorships[4].raw_affiliation_strings | Department of Chemistry, University of Florida, Gainesville, FL, Departments of Materials Science and Nuclear Engineering, University of Florida, Gainesville, FL, UAEY-IIMAS, Universidad Nacional Autónoma de México, Mérida, Yucatán, México |
| has_content.pdf | True |
| has_content.grobid_xml | True |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://doi.org/10.33697/ajur.2023.085 |
| open_access.oa_status | diamond |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Validation of a Computationally Efficient Model of the Mu-Opioid Receptor |
| has_fulltext | True |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T10814 |
| primary_topic.field.id | https://openalex.org/fields/28 |
| primary_topic.field.display_name | Neuroscience |
| primary_topic.score | 0.9997000098228455 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/2804 |
| primary_topic.subfield.display_name | Cellular and Molecular Neuroscience |
| primary_topic.display_name | Neuropeptides and Animal Physiology |
| related_works | https://openalex.org/W3099772455, https://openalex.org/W3172198228, https://openalex.org/W1986078734, https://openalex.org/W2129301794, https://openalex.org/W1540599686, https://openalex.org/W2389992822, https://openalex.org/W2016396528, https://openalex.org/W2033001120, https://openalex.org/W2261012589, https://openalex.org/W2061481432 |
| cited_by_count | 0 |
| locations_count | 1 |
| best_oa_location.id | doi:10.33697/ajur.2023.085 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4210172541 |
| best_oa_location.source.issn | 1536-4585, 2375-8732 |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | True |
| best_oa_location.source.issn_l | 1536-4585 |
| best_oa_location.source.is_core | True |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | American Journal of Undergraduate Research |
| best_oa_location.source.host_organization | |
| best_oa_location.source.host_organization_name | |
| best_oa_location.license | |
| best_oa_location.pdf_url | https://doi.org/10.33697/ajur.2023.085 |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | journal-article |
| best_oa_location.license_id | |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | American Journal of Undergraduate Research |
| best_oa_location.landing_page_url | http://doi.org/10.33697/ajur.2023.085 |
| primary_location.id | doi:10.33697/ajur.2023.085 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4210172541 |
| primary_location.source.issn | 1536-4585, 2375-8732 |
| primary_location.source.type | journal |
| primary_location.source.is_oa | True |
| primary_location.source.issn_l | 1536-4585 |
| primary_location.source.is_core | True |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | American Journal of Undergraduate Research |
| primary_location.source.host_organization | |
| primary_location.source.host_organization_name | |
| primary_location.license | |
| primary_location.pdf_url | https://doi.org/10.33697/ajur.2023.085 |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | American Journal of Undergraduate Research |
| primary_location.landing_page_url | http://doi.org/10.33697/ajur.2023.085 |
| publication_date | 2023-10-07 |
| publication_year | 2023 |
| referenced_works | https://openalex.org/W1974674594, https://openalex.org/W3160915496, https://openalex.org/W2051746492, https://openalex.org/W2000716491, https://openalex.org/W2107153432, https://openalex.org/W2052191785, https://openalex.org/W2132164381, https://openalex.org/W2005174659, https://openalex.org/W2067973462, https://openalex.org/W2058373165, https://openalex.org/W3153533584, https://openalex.org/W1918796765, https://openalex.org/W2083725367, https://openalex.org/W2321948417, https://openalex.org/W3211057556, https://openalex.org/W2166660130, https://openalex.org/W2036690970, https://openalex.org/W2312741033, https://openalex.org/W6907608579, https://openalex.org/W2977764209, https://openalex.org/W2066254477, https://openalex.org/W1704565512, https://openalex.org/W2905232362, https://openalex.org/W1976499671, https://openalex.org/W2150981663, https://openalex.org/W2555870966, https://openalex.org/W2066414494, https://openalex.org/W2103945336, https://openalex.org/W2332712348, https://openalex.org/W3123481860, https://openalex.org/W2101870582, https://openalex.org/W2943037937, https://openalex.org/W2337440865, https://openalex.org/W2062576240, https://openalex.org/W3168436232, https://openalex.org/W2134967712, https://openalex.org/W2060433756, https://openalex.org/W1997772366, https://openalex.org/W2070753604, https://openalex.org/W2029667189, https://openalex.org/W2111917512, https://openalex.org/W2074231493, https://openalex.org/W2602696732 |
| referenced_works_count | 43 |
| abstract_inverted_index.a | 5, 72, 82, 127, 148, 194, 248, 269 |
| abstract_inverted_index.To | 134 |
| abstract_inverted_index.an | 62, 204, 230 |
| abstract_inverted_index.as | 22 |
| abstract_inverted_index.at | 61 |
| abstract_inverted_index.be | 76, 122, 184, 274 |
| abstract_inverted_index.in | 41, 58, 71, 113, 147, 154, 158, 186, 229, 268 |
| abstract_inverted_index.is | 4, 247 |
| abstract_inverted_index.of | 37, 44, 93, 99, 111, 138, 145, 179, 215, 245, 251, 266 |
| abstract_inverted_index.on | 89, 241 |
| abstract_inverted_index.to | 55, 95, 121, 164, 183, 219, 227, 273 |
| abstract_inverted_index.3.0 | 170 |
| abstract_inverted_index.49% | 124 |
| abstract_inverted_index.MOR | 38, 69, 94, 120, 146, 153, 157, 180, 192, 202, 246, 267 |
| abstract_inverted_index.The | 0, 116, 209 |
| abstract_inverted_index.aid | 40 |
| abstract_inverted_index.all | 165 |
| abstract_inverted_index.and | 8, 25, 35, 156, 172 |
| abstract_inverted_index.can | 75 |
| abstract_inverted_index.for | 12 |
| abstract_inverted_index.had | 223 |
| abstract_inverted_index.may | 39 |
| abstract_inverted_index.the | 9, 42, 90, 97, 100, 109, 114, 131, 136, 139, 151, 173, 190, 220, 242, 253, 264 |
| abstract_inverted_index.way | 250 |
| abstract_inverted_index.NAMD | 169 |
| abstract_inverted_index.RMSD | 224 |
| abstract_inverted_index.This | 79, 235, 260 |
| abstract_inverted_index.come | 17 |
| abstract_inverted_index.each | 187 |
| abstract_inverted_index.free | 152, 191 |
| abstract_inverted_index.mean | 197 |
| abstract_inverted_index.more | 276 |
| abstract_inverted_index.root | 196 |
| abstract_inverted_index.show | 30 |
| abstract_inverted_index.such | 21 |
| abstract_inverted_index.than | 126, 201 |
| abstract_inverted_index.that | 31, 85, 238 |
| abstract_inverted_index.uses | 86 |
| abstract_inverted_index.were | 143, 181 |
| abstract_inverted_index.with | 18, 47, 160, 189, 203, 211, 256 |
| abstract_inverted_index.(MOR) | 3 |
| abstract_inverted_index.0.001 | 216 |
| abstract_inverted_index.Lipid | 290 |
| abstract_inverted_index.allow | 53, 263 |
| abstract_inverted_index.alpha | 171 |
| abstract_inverted_index.atoms | 112 |
| abstract_inverted_index.could | 262 |
| abstract_inverted_index.drugs | 16, 46 |
| abstract_inverted_index.force | 175 |
| abstract_inverted_index.lipid | 73, 101, 106, 132, 149, 207, 233, 270 |
| abstract_inverted_index.model | 96, 118, 261 |
| abstract_inverted_index.shown | 182 |
| abstract_inverted_index.study | 56, 80, 236 |
| abstract_inverted_index.those | 228 |
| abstract_inverted_index.using | 168, 239 |
| abstract_inverted_index.water | 155, 159 |
| abstract_inverted_index.(RMSD) | 200 |
| abstract_inverted_index.Opioid | 15 |
| abstract_inverted_index.allows | 119 |
| abstract_inverted_index.assess | 135 |
| abstract_inverted_index.costs. | 259 |
| abstract_inverted_index.design | 43 |
| abstract_inverted_index.drugs. | 14 |
| abstract_inverted_index.faster | 125 |
| abstract_inverted_index.field. | 176 |
| abstract_inverted_index.having | 193 |
| abstract_inverted_index.higher | 195 |
| abstract_inverted_index.level. | 64 |
| abstract_inverted_index.model, | 141 |
| abstract_inverted_index.number | 110 |
| abstract_inverted_index.opioid | 45 |
| abstract_inverted_index.recent | 28 |
| abstract_inverted_index.square | 198 |
| abstract_inverted_index.target | 11 |
| abstract_inverted_index.values | 225 |
| abstract_inverted_index.Coupled | 288 |
| abstract_inverted_index.Dynamic | 177 |
| abstract_inverted_index.Opioid, | 292 |
| abstract_inverted_index.Protein | 294 |
| abstract_inverted_index.applied | 163, 218 |
| abstract_inverted_index.bilayer | 74, 102, 271 |
| abstract_inverted_index.changes | 57 |
| abstract_inverted_index.modeled | 206, 232 |
| abstract_inverted_index.primary | 10 |
| abstract_inverted_index.protein | 7, 59 |
| abstract_inverted_index.reduced | 48, 257 |
| abstract_inverted_index.regions | 92 |
| abstract_inverted_index.studied | 275 |
| abstract_inverted_index.studies | 29 |
| abstract_inverted_index.system, | 188 |
| abstract_inverted_index.systems | 67, 210 |
| abstract_inverted_index.without | 103 |
| abstract_inverted_index.Bilayer, | 291 |
| abstract_inverted_index.CHARMM36 | 174 |
| abstract_inverted_index.However, | 27, 65 |
| abstract_inverted_index.accuracy | 137 |
| abstract_inverted_index.approach | 84 |
| abstract_inverted_index.bilayer, | 150 |
| abstract_inverted_index.bilayer. | 133, 208, 234 |
| abstract_inverted_index.dynamics | 36, 51, 265 |
| abstract_inverted_index.embedded | 70 |
| abstract_inverted_index.feasible | 249 |
| abstract_inverted_index.harmonic | 87, 161, 212 |
| abstract_inverted_index.modeling | 66, 83, 252 |
| abstract_inverted_index.physical | 23 |
| abstract_inverted_index.proposed | 117, 140 |
| abstract_inverted_index.receptor | 2, 255 |
| abstract_inverted_index.reducing | 108 |
| abstract_inverted_index.residues | 167, 222, 244 |
| abstract_inverted_index.rigidity | 98 |
| abstract_inverted_index.Atomistic | 281 |
| abstract_inverted_index.Dynamics; | 280 |
| abstract_inverted_index.G-Protein | 287 |
| abstract_inverted_index.KEYWORDS: | 278 |
| abstract_inverted_index.Modeling; | 284 |
| abstract_inverted_index.Molecular | 50, 279 |
| abstract_inverted_index.Mu-Opioid | 285 |
| abstract_inverted_index.Receptor; | 286, 289 |
| abstract_inverted_index.atomistic | 63 |
| abstract_inverted_index.constants | 214 |
| abstract_inverted_index.deviation | 199 |
| abstract_inverted_index.different | 185 |
| abstract_inverted_index.evaluates | 81 |
| abstract_inverted_index.including | 68, 130 |
| abstract_inverted_index.mu-opioid | 1 |
| abstract_inverted_index.performed | 144 |
| abstract_inverted_index.restraint | 213 |
| abstract_inverted_index.simulated | 123 |
| abstract_inverted_index.addiction. | 26 |
| abstract_inverted_index.comparable | 226 |
| abstract_inverted_index.dependence | 24 |
| abstract_inverted_index.expensive. | 78 |
| abstract_inverted_index.explicitly | 104, 129, 205, 231 |
| abstract_inverted_index.molecules, | 107 |
| abstract_inverted_index.properties | 34, 178 |
| abstract_inverted_index.restraints | 88, 162, 240 |
| abstract_inverted_index.simulating | 105 |
| abstract_inverted_index.simulation | 128 |
| abstract_inverted_index.structural | 33 |
| abstract_inverted_index.detrimental | 19 |
| abstract_inverted_index.environment | 272 |
| abstract_inverted_index.ligand-free | 254 |
| abstract_inverted_index.researchers | 54 |
| abstract_inverted_index.simulation. | 115 |
| abstract_inverted_index.simulations | 52, 142 |
| abstract_inverted_index.Simulations; | 282 |
| abstract_inverted_index.conformation | 60 |
| abstract_inverted_index.demonstrates | 237 |
| abstract_inverted_index.efficiently. | 277 |
| abstract_inverted_index.kcal/mol/Å2 | 217 |
| abstract_inverted_index.side-effects | 20 |
| abstract_inverted_index.Computational | 283 |
| abstract_inverted_index.Transmembrane | 293 |
| abstract_inverted_index.computational | 258 |
| abstract_inverted_index.side-effects. | 49 |
| abstract_inverted_index.transmembrane | 6, 91, 166, 221, 243 |
| abstract_inverted_index.understanding | 32 |
| abstract_inverted_index.computationally | 77 |
| abstract_inverted_index.pain-modulating | 13 |
| cited_by_percentile_year | |
| countries_distinct_count | 2 |
| institutions_distinct_count | 5 |
| sustainable_development_goals[0].id | https://metadata.un.org/sdg/3 |
| sustainable_development_goals[0].score | 0.6399999856948853 |
| sustainable_development_goals[0].display_name | Good health and well-being |
| citation_normalized_percentile.value | 0.20868574 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |